Abstract
The diazotrophic, bacterial endophyte Herbaspirillum frisingense GSF30T has been identified in biomass grasses grown in temperate climate, including the highly nitrogen-efficient grass Miscanthus. Its genome was annotated and compared with related Herbaspirillum species from diverse habitats, including H. seropedicae, and further well-characterized endophytes. The analysis revealed that Herbaspirillum frisingense lacks a type III secretion system that is present in some related Herbaspirillum grass endophytes. Together with the lack of components of the type II secretion system, the genomic inventory indicates distinct interaction scenarios of endophytic Herbaspirillum strains with plants. Differences in respiration, carbon, nitrogen and cell wall metabolism among Herbaspirillum isolates partially correlate with their different habitats. Herbaspirillum frisingense is closely related to strains isolated from the rhizosphere of phragmites and from well water, but these lack nitrogen fixation and metabolism genes. Within grass endophytes, the high diversity in their genomic inventory suggests that even individual plant species provide distinct, highly diverse metabolic niches for successful endophyte-plant associations.
Keywords: microbe, diazotroph, nitrogen fixation, plant associated bacteria, plant growth promoting bacteria
Introduction
Many gramineous species maintain a close association with endophytic bacteria that are often beneficial for plant growth and health (Reinhold-Hurek and Hurek, 1998). Their considerable ecologic importance and agronomic potential is best documented in warm tropical and subtropical climates (Reinhold-Hurek and Hurek, 1998). Plant growth promoting bacteria are often considered as a cost efficient and ecological alternative to improve crop growth on low-nutrient soils (Sturz et al., 2000) and may gain further interest for future large-scale biomass production on marginal land with low-input grasses (Heaton et al., 2008).
Herbaspirillum frisingense belongs to the β-proteobacteria and is a close relative of Herbaspirillum seropedicae SmR1 (HsSmR1) and Herbaspirillum rubrisubalbicans (HrM1), which are both common in tropical and subtropical soils and endophytically colonize various grasses (Monteiro et al., 2012b). Endophytes are referred to here as microorganisms (bacteria) that have low soil competence and spend most of their life cycle within the plant, mostly without causing symptoms of plant damage. Beneficial associations of HsSmR1 and HrM1 with sorghum, sugar cane, rice, and maize have been reported, but HrM1 causes red stripe disease on some sorghum varieties and can cause mottled stripe disease on sugarcane. Other isolates of H. seropedicae from rice (HsOs34, HsOs45) induced disease symptoms (Ye et al., 2012; Zhu et al., 2012). So far, plant growth promoting action, but no disease symptoms, were identified for H. frisingense (Straub et al., unpublished observation), which was originally isolated from the perennial C4-fiber plant Miscanthus in southern Germany (Kirchhof et al., 2001). Other potential N-fixing bacteria, such as Azospirillum doebereinerae (Eckert et al., 2001) and bacterial consortia consisting of N2-fixing clostridia (Miyamoto et al., 2004) has also been isolated from Miscanthus. Herbaspirillum frisingense strains were also recovered from other biomass grasses, Spartina pectinata and Pennisetum purpureum, grown in temperate conditions. Model calculations proposed that Miscanthus x giganteus gained substantial nitrogen from the N-fixation by endophytic symbionts (Davis et al., 2010), but the type of nitrogen fixers remains unclear. H. seropedicae isolates were shown to fix nitrogen in association with wild rice, but not with cultivated rice (Elbeltagy et al., 2001).
The entire HsSmR1 genome (Pedrosa et al., 2011) and various other Herbaspirillum genomes (Table 1) from diverse habitats were recently sequenced, while that of HrM1 was partially sequenced (Monteiro et al., 2012a). Sequenced Herbaspirillum species include plant growth promoting soil bacteria (HGW103) from the rhizosphere of the grass Phragmites australis (Lee et al., 2012), isolates (HlP6-12) from the root nodules of Phaseolus vulgaris (Weiss et al., 2012), strains (HCF444 and HYR522) colonizing poplar (Brown et al., 2012), a strain (HhIAM) isolated from Japanese well water (De Souza et al., 2013) and an isolate (HJC206) from human fecal flora (Lagier et al., 2012).
Table 1.
Bacteria included in the genome/protein comparison.
| Species | Abbreviation | Available sequences | Isolated from | Accession number | References |
|---|---|---|---|---|---|
| Herbaspirillum rubrisubalbicans M1 | HrM1 | SSH library | Various grasses | Monteiro et al., 2012a | |
| Herbaspirillum huttiense subsp. putei IAM 15032 | HhIAM | Contigs | Well water | ANJR00000000 | De Souza et al., 2013 |
| Herbaspirillum lusitanum P6-12 (DSM 17154) | HIP6-12 | Contigs | Root nodules of Phaseolus vulgaris | AJHH00000000 | Weiss et al., 2012 |
| Herbaspirillum sp. GW103 | HGW103 | Contigs 4655 proteins | Rhizosphere of Phragmites australis | AJVC00000000 | Lee et al., 2012 |
| Herbaspirillum sp. JC206 | HJC206 | Contigs | Human fecal flora | CAHF00000000 | Lagier et al., 2012 |
| Herbaspirillum sp. CF444 | HCF444 | Contigs 4974 proteins | Rhizosphere and endosphere of Populus deltoide | AKJW00000000 | Brown et al., 2012 |
| Herbaspirillum sp. YR522 | HYR522 | Contigs 4612 proteins | Rhizosphere and endosphere of Populus deltoide | AKJA00000000 | Brown et al., 2012 |
| Herbaspirillum seropedicae Os45 | HsOs45 | Contigs | Rice roots | AMSA00000000 | Zhu et al., 2012 |
| Herbaspirillum seropedicae Os34 | HsOs34 | Contigs | Rice roots | AMSB00000000 | Ye et al., 2012 |
| Herbaspirillum seropedicae SmR1 | HsSmR1 | Full genome 4735 proteins | Tropical grasses | CP002039 | Pedrosa et al., 2011 |
| Herbaspirillum frisingense GSF30T | HfGSF30 | Contigs 4871 proteins | Various grasses | AEEC00000000 | This work |
| Gluconacetobacter diazotrophicus PAI5 | GdPAI5 | Full genome 3851 proteins | Sugarcane | AM889285–AM889287 | Bertalan et al., 2009 |
| Azoarcus sp. BH72 | AzoaBH72 | Full genome 3989 proteins | Kallar grass | AM406670 | Krause et al., 2006 |
| Klebsiella pneumoniae 342 | Kp342 | Full genome 5768 proteins | Maize | CP000964–CP000966 | Fouts et al., 2008 |
| Azospirillum sp. B510 | AzospB510 | Full genome 6309 proteins | Rice | AP010946–AP0109452 | Kaneko et al., 2010 |
Detailed descriptions of the entire genome sequences from various distant, well-described endophytes with defined endophytic habitats and plant growth promoting capabilities include Azoarcus sp. BH72 (AzoaBH72, a β-proteobacterium) (Krause et al., 2006), Klebsiella pneumoniae 342 (Kp342, γ-proteobacterium) (Fouts et al., 2008), Azospirillum sp. B510 (AzospB510, α-proteobacterium) (Kaneko et al., 2010) and Gluconacetobacter diazotrophicus PAl5 (GdPAI5, α-proteobacterium) (Bertalan et al., 2009). However, fundamental questions regarding their competitiveness, specificity to invade selected hosts, manipulate the plant growth, strategies for nutrition and survival in the plants, and the essential set of genes required for endophytic life, remain unclear.
Although it is desirable to have entire genome sequences available, the comparison of the genomic inventories does not necessarily require completely assembled genomes. Instead, comparisons of incomplete draft genome sequences with related species represents often a sufficient powerful approach for the identification of similarities and differences in their genomic inventory (Almeida et al., 2009; Studholme et al., 2009).
Here, the bacterial genome of Herbaspirillum frisingense GSF30T was sequenced and annotated. The genome (containing a few gaps) was compared to other Herbaspirillum strains and selected, well-described plant endophytes. These served as references to compare the basic genome equipments necessary to colonize the endophytic niche. The lack of the type III secretion system, diversity in other secretion systems and major differences in the basic metabolic capacities characterize Herbaspirillum frisingense as a non-pathogenic, diazotrophic endophytic grass colonizer that is closely related to non-diazotrophic Herbaspirillum strains that were isolated from the rhizosphere and from well water.
Materials and methods
Sequencing
H. frisingense GSF30T was grown over night at 30°C on LB-media containing 50 μg/l kanamycin. Genomic DNA was isolated and sequenced with the Roche/454 GS FLX system and with illumina technology, to increase the coverage and to close gaps. Sequencing and de-novo assembly was performed by GATC Biotech AG (Germany). The entire genome shotgun sequencing project has been deposited at DDBJ/EMBL/GenBank under the accession AEEC02000000 (Accession: PRJNA50373, ID: 50373).
Genome annotation
Open reading frame prediction and annotation were performed by the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) in April 2013.
Phylogeny
16S rRNA sequences of all 14 bacteria were obtained from NCBI and analyzed with MEGA5.2 (Tamura et al., 2011). The sequences were aligned using ClustalW and the phylogeny reconstruction was done using the Maximum Likelihood method with 500 Bootstrap Replications. Marker protein sequences (or proteins predicted from draft genome sequences) were selected with AMPHORA2 (Wu and Scott, 2012). Four sequences were not identified in HsOs45 (rplK, rpoB, rplL, rplA) and were excluded, as well as duplicate sequences. A concatenated tree and phylogenetic analysis was conducted with MEGA5.20.
Genome comparisons
All bacteria included in the genome/protein comparison are shown in Table 1. Among these are six without protein annotation, five have draft genome information, while sufficient publically available data for comparison is lacking for Herbaspirillum rubrisubalbicans M1. The partial, fragmented genomic sequences available for Herbaspirillum sp. isolates B501, B59, and B65 were not included. Herbaspirilla nucleotide sequences were searched with annotated protein sequences, preferably from Herbaspirillum seropedicae SmR1, using NCBI's tblastn algorithm against whole-genome shotgun contigs (wgs) databases. Ambiguous hits (expect value >e-50 or identically predicted amino acids <80%), or multiple hits were reviewed with blastx against the nr protein database.
Results
Genome sequencing and annotation
The genome sequence of H. frisingense GSF30T was obtained using a combined strategy with 454 pyrosequencing (Margulies et al., 2005) and illumina technology. The 454 sequencing produced more than 600000 reads with approximately 48× coverage and 265 Mb, the illumina sequencing more than 25 million reads, ca. 420 times coverage and 2.3 Gb. From these, 93 contigs (>200 bp) were assembled with a total length of ~5.4 Mb, which is in the lower range of endophyte genomes. Compared to the similarly sequenced bacterial draft genome of Pseudomonas syringae pv. tomato T1, a relatively large number of contig gaps was present in the HfGSF30 draft genome. The individual inspection of the contig borders identified that repetitive sequences likely perturbed the total assembly of sequences.
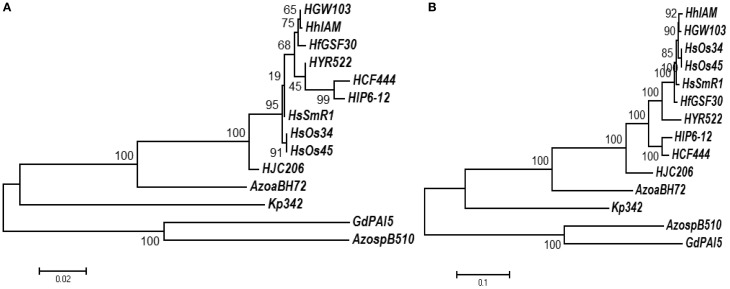
The contig endings were manually compared using NCBI's blastn to the full genome of Herbaspirillum seropedicae SmR1 and a clear colinearity was identified for around 70% of the contigs, leaving 28 gaps. These sequences were used to carry out the analysis. On average, the coverage of contigs with more than 500 bp was ~400, which likely represents >99.9% of the entire sequence. The genomic inventory and its relation to distinct physiological processes is discussed below; the references for the endophyte genomes are given in Table 1. Based on their 16S rRNA sequence, H. frisingense is phylogenetically most closely related to HGW103 and HhIAM (Figure 1A). However, based on the sequence similarity of 27 marker proteins, HfGSF30 groups outside a cluster containing the H. seropedicae strains and HGW103 and HhIAM (Figure 1B).
Figure 1.
Phylogenetic relationship based on 16S rRNA sequences (A) and marker proteins (B). HfGSF30 and other Herbaspririllum strains and diverse endophytes. The scale bar represents 2% (A) or 10% (B) sequence divergence and numbers on the tree represent bootstrap values. HsSmR1, Herbaspirillum seropedicae SmR1; HsOs34, Herbaspirillum seropedicae Os34; HsOs45, Herbaspirillum seropedicae Os45; HfGSF30, Herbaspirillum frisingense GSF30T; HGW103, Herbaspirillum sp. GW103; HhIAM, Herbaspirillum huttiense subsp. putei IAM 15032; HYR522, Herbaspirillum sp. YR522; HCF444, Herbaspirillum sp. CF444; HlP6-12, Herbaspirillum lusitanum P6-12 (DSM 17154); HJC206, Herbaspirillum sp. JC206; AzoaBH72, Azoarcus sp. BH72; AzospB510, Azospirillum sp. B510; Kp342, Klebsiella pneumoniae 342; GdPAI5, Gluconacetobacter diazotrophicus PAl5.
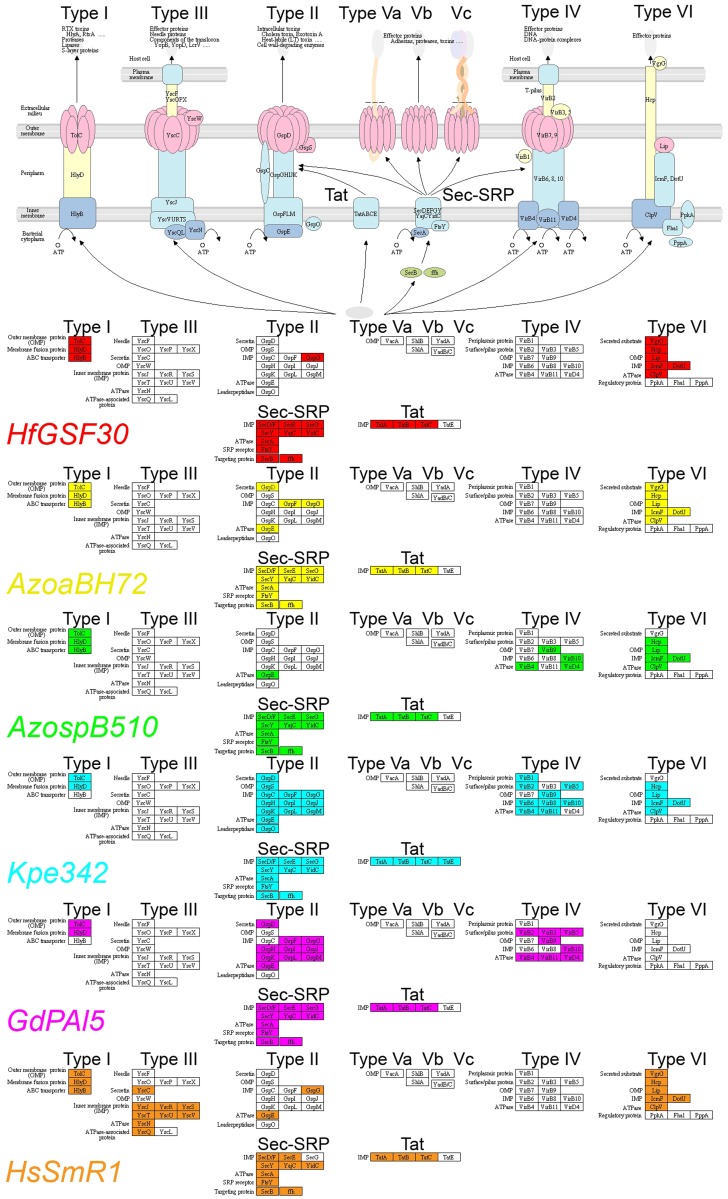
Protein secretion systems
The HfGSF30 genome encodes the type I, type VI, Sec-SRP and the Tat (twin-arginine translocation) systems, but lacks the type III secretion system, as shown in Figure 2. The type III secretion system is typically used by pathogenic bacteria to deliver effector proteins into the plant host cells, but is also used in beneficial interactions for optimization (Viprey et al., 1998). HfGSF30, as well as the reference grass endophytes AzoaBH72, AzospB510, Kp342, and GdPAI5, completely lack the type III secretion system hrp/hrc genes (Figures 2, 3, Table S1). By contrast, other Herbaspirillum grass endophytes and poplar colonizers, namely HsSmR1, H. rubrisubalbicans M1, HsOs34, HsOs45, HCF444, and HYR522, contained that system. It is critical for pathogenicity, but also endophytic invasion of HrM1 (Monteiro et al., 2012a). Pedrosa et al. (2011) found no transposon elements flanking the type III secretion system genes in HsSmR1, suggesting that it was not recently added into the genome. Flanking regions of the type III secretion system genes were only partially conserved among Herbaspirillum strains, suggesting that the type III protein secretion was deleted in some Herbaspirillum strains, including HfGSF30.
Figure 2.
Presence of secretion systems in HfGSF30 and comparison with other endophytes. HfGSF30 genome (red), AzoaBH72 (yellow), AzospB510 (green), Kp342 (blue), GdPAI5 (pink), and HsSmR1 (orange). Missing genes are shown in white.
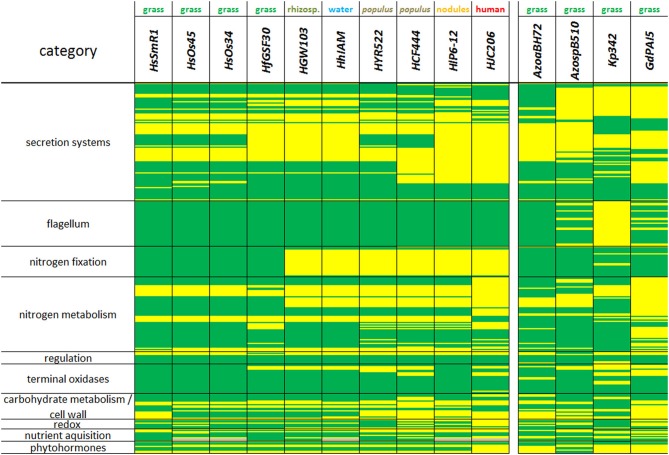
Figure 3.
Similarity and divergence of gene clusters reflecting various cell functions, including secretion systems, cell wall, nitrogen, carbon, and hormone metabolism in Herbaspirillum and endophyte strains. Present genes are schown in green, lacking genes are shown in yellow, missing information is given in gray. Strains from left to right: HsSmR1, Herbaspirillum seropedicae SmR1; HsOs34, Herbaspirillum seropedicae Os34; HsOs45, Herbaspirillum seropedicae Os45; HfGSF30, Herbaspirillum frisingense GSF30T; HGW103, Herbaspirillum sp. GW103; HhIAM, Herbaspirillum huttiense subsp. putei IAM 15032; HYR522, Herbaspirillum sp. YR522; HCF444, Herbaspirillum sp. CF444; HlP6-12, Herbaspirillum lusitanum P6-12 (DSM 17154); HJC206, Herbaspirillum sp. JC206; AzoaBH72, Azoarcus sp. BH72; AzospB510, Azospirillum sp. B510; Kp342, Klebsiella pneumoniae 342; GdPAI5, Gluconacetobacter diazotrophicus PAl5.
All Herbaspirillum strains lack the type IV secretion system, which is involved in virulence and horizontal gene transfer (Juhas et al., 2008), but genes of this system are identified in more distant endophytes, such as AzospB510, Kp342, and GdPAI5 (Figures 2, 3, Table S1). Except for Kp342, all mentioned endophytes and Herbaspirillum strains contain the entire machinery for flagellum export and function (Figure 3).
Furthermore, a reduced set of the type IV pilin secretion/fimbrial assembly genes, members of the type II secretion system, was identified in HfGSF30 (similar as in HGW103 and HhIAM), when compared to H. seropedicae strains. The tree colonizers HCF444 and HYR522 have almost the same set of type IV pilin secretion genes as HsSmR1. These genes were completely absent in AzospB510, Kp342, and GdPAI5, while they were present in AzoaBH72.
HfGSF30 possesses type VI secretion system genes. This system is involved in host-bacteria interaction, both in pathogenic and symbiotic relationships (Filloux a Hachani and Bleves, 2008). These genes are also present in most Herbaspirillum strains and all considered grass endophytes, except GdPAI5. Notably, the type VI system is present in one, but lacking in another Herbaspirillum strain isolated from poplar, and is also absent the strains isolated from nodules (HIP6-12) and from human fecal flora (HJC206). HfGSF30 contains the chaperone-usher system (type I pilus assembly proteins), whereas some Herbaspirillum strains, including H. seropedicae isolated from rice, and only Kp342, but not AzoaBH72, AzospB510, and GdPAI5, contain that system (Figure 3, Table S1).
Nitrogen metabolism
The acetylene reduction assay has suggested nitrogenase activity in HfGSF30 (Kirchhof et al., 2001). Among the Herbaspirillum strains, nitrogen fixation genes were only present in H. seropedicae strains and HfGSF30. The nif-region is very similar to the corresponding region of H. seropedicae SmR1 with 94% nucleotide identity, 96% amino acid identity and identical gene arrangement. Some gene products, nifB, nifX, nifZ1, fdxB, and fix, were even 100% identical between HfGSF30 and HsSmR1. Nif genes are absent in HGW103, HhIAM, HYR522, HCF444, HJC206, and even in HlP6-12, which was isolated from Phaseolus nodules. The AzospB510, AzoaBH72, GdPAI5, and Kp342 grass endophytes contain the entire nif cluster (Figure 3).
HfGSF30 is equipped with an assimilatory nitrate reductase (nasAC) and a NAD(P)H-dependent nitrite reductase (nirBD; EC 1.7.1.4), similar to AzospB510, AzoaBH72, Kp342 and other Herbaspirillum strains, except for HJC206 (and GdPAI5), which completely lack nitrate assimilatory and dissimilartory genes (Figure 3). HfGSF30, HsSmR1, HsOs34, and HsOs45 strains contain the respiratory nitrate reductase (narGHJI), the nitrite/nitrate transporter (narU) and a nitrate/nitrite sensor histidine kinase transcription regulator (narXL) to utilize nitrate in anaerobic respiration. Kp342 has a similar set of genes, but other Herbaspirillum isolates, AzoaBH72, GdPAI5, and AzospB510 apparently cannot utilize nitrate as alternative electron acceptor in anaerobic conditions. The absence of nitrate reductase in HJC206 is consistent with the minor role of nitrate in the human habitat (Lagier et al., 2012), but the endophyte GdPAI5 also lacks the respective genes. The presence of α-, δ- and γ-subunits of a formate dehydrogenase (EC 1.2.1.2) parallels the occurrence of genes for nitrate reduction, and is absent in GdPAI5 and HJC206. However, HJC206 has formate dehydrogenase genes with sequence similarity to Herminiimonas arsenicoxydans that are unique in the Herbaspirillum genus.
HfGSF30 is likely capable to reduce nitrate to NO and further to N2O (EC 1.7.2.1, 1.7.99.7), a feature exclusively present in HfGSF30 among Herbaspirillum strains, but no nitrous oxide reductase (EC 1.7.99.6) to reduce N2O to N2 is identified (Figure 3). This is in line with previous experimental evidence, which showed that NO−3 reduction to N2 did not occur in HfGSF30 (Kirchhof et al., 2001). Nitrogen reduction varies greatly in other diazotrophic endophytes, namely AzoaBH72 appears capable to reduce NO via N2O to N2, but a nitrite reductase is missing. GdPAI5 also lacks nitrate reductase (Cavalcante and Dobereiner, 1988). AzospB510, like HfGSF30, has the possibility to reduce NO2 to N2O, but not to N2.
Amino acids, such as asparagine and aspartic acid, were utilized as nitrogen sources by HfGSF30 (Kirchhof et al., 2001), but the capabilities to synthesize aspartic acid and asparagine differ among Herbaspirillum strains, with only HsSmR1 and HJC206 containing an asparagine synthase gene (Table S1). Although the full urea cycle is present in all strains (except for GdPAI5), differences are identified with respect to the alternative urea degradation pathway, which is partially missing in HfGSF30, although it is present in all other grass endophytes.
Respiration
HfGSF30 contains four terminal oxidases that allow adaptation to different oxygen levels and microhabitats: cytochrome aa3 (coxAB); cytochrome bd-type quinol oxidase (cydAB), cbb3-type cytochrome c oxidase (fixPON), cytochrome o ubiquinol oxidase (cyoABC). Genes for NADH dehydrogenase, succinate dehydrogenase and cytochrome c reductase are ubiquitously identified in all Herbaspirillum strains.
The high affinity cbb3-type cytochrome c oxidase may support ATP-synthesis under oxygen limitation during nitrogen fixation and accordingly, this system is lacking in the non-diazotrophic HCF444. However, the diazotrophic Kp342 and GdPAI5 also lack this oxidase and it is present in other nitrogenase-lacking Herbaspirillum strains. Multiple coxAB copies are only present in H. seropedicae strains and in the strain isolated from nodules (HIP6-12). The cytochrome bd-type quinol oxidase is absent in HCF444 isolated from poplar and Kp342.
Carbohydrate metabolism and cell wall degradation
A broad spectrum of monosaccharides, organic acids and alcohols, but not di-and tri-saccharides, are utilized as carbon sources by HfGSF30 (Kirchhof et al., 2001). This is in line with the identification of metabolizing enzymes for these substrates. HfGSF30 lacks the sucrose-degrading enzyme invertase (EC:3.2.1.26) and α-glucosidase (EC:3.2.1.20), while HsSmR1, HsOs34, HsOs45, HCF444, HhIAM, and AzoaBH72 encode α-glucosidase. AzospB510 and GdPAI5 lack both enzymes, while both are present in Kp342. Except for HCF444 and HJC206, all Herbaspirillum strains had two trehalose synthesis pathways (otsAB and treXYZ). A gene related to the large ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) subunit from plants was also present in several Herbaspirillum strains, including HfGSF30, but no sequence encoding a phosphoribulokinase was found (but present in AzoaBH72 and Kpe342). Therefore, CO2 fixation appears to be impossible for these endophytes and the RubisCO-like proteins are probably involved in sulfur metabolism (Tabita et al., 2007). A few membrane transporters were notably different within the Herbaspirillum strains: the arsenite/antimonite transporter was only present in HfGSF30, HsOs34, HsOs45, HGW103, and HhIAM, differences were also obvious in the number and type of ammonium and iron transporters (Table S1).
There is no evidence that the plant cell wall was affected by H. frisingense colonization (Rothballer et al., 2008), but HfGSF30 (and HsOs34, HsOs45, HGW103, HhIAM, HYR522, and other endophytes) are equipped with an endo-1,4-D-glucanase that may break down cellulose (EC:3.2.1.4, absent in HsSmR1). Two chitin deacetylases (EC:3.5.1.41 and 3.1.1.58) are present in all Herbaspirillum strains, while HsSmR1 and HJC206 possess two additional enzymes. α-glucosidase (EC:3.2.1.20) and α –amylase (EC:3.2.1.1) are absent in HfGSF30 and some Herbaspirillum strains. In HrM1, a large operon involved in cellulose synthesis (or degradation) appears crucial for colonization (Monteiro et al., 2012a); this entire operon was present in HfGSF30, HsOs34, HsOs45, HGW103, HhIAM (and Kp342), but was absent in HsSmR1, HlP6-12, HYR522, HCF444, HJC206.
Survival against the plant defense and environmental stress
The plant defense against bacterial, fungal and viral threats generally involves the production of reactive oxygen species (ROS), nitric oxide and phytoalexins. It has recently been shown that antioxidant enzymes are up-regulated during biological nitrogen fixation to prevent ROS in G. diazotrophicus PAl5 (Alquéres et al., 2010), but compared to the other bacteria under study, this strain, together with the human isolate HJC206, contains the least number of potential detoxification genes. Different strategies to cope with reactive oxygen are apparent within Herbaspirillum strains and other endophytes (Figure 3, Table S1).
Biosynthesis of plant hormones
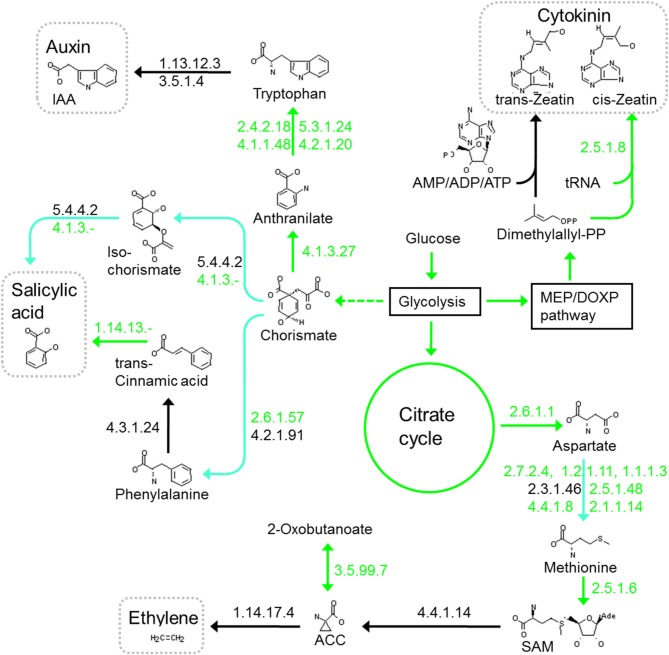
The production of plant hormones, or other beneficial agents, is a common strategy of endophytes to promote plant growth (Hardoim et al., 2008). All Herbaspirillum strains, except for HJC206, contain the genes for 1-aminocyclopropane-1-carboxylate (ACC) deaminase to degrade the ethylene precursor ACC to 2-oxobutanoate and NH3. ACC is taken up by H. frisingense (Rothballer et al., 2008) and its efficient breakdown by ACC deaminase may reduce locally plant ethylene levels at sites of invasion (Hardoim et al., 2008). The endophytes AzoaBH72, Kp342 and GdPAI5 do not contain ACC deaminase and thus appear not capable of modulating plant ethylene signaling.
Auxin (indole acetic acid) synthesis proceeds via several pathways, which are at least partially present in all Herbaspirillum and other grass endophytes. Differences in auxin production are suggested in the Herbaspirillum strains, as only HsSmR1 encodes the amidase that releases NH3 and indole acetic acid from indole-3-acetamide iaaH (and AzoaBH72) (Costacurta and Vanderleyden, 1995). However, the essential tryptophan 2-monooxygenase (iaaM) for decarboxylation of tryptophan to indole-3-acetamide is not unambiguously identified in any Herbaspirillum. All Herbaspirillum strains lack an ipdC homolog, which is present in Kp342, where indole acetic acid may be synthesized by indole-3-pyruvate decarboxylase from tryptophan via indole-3-pyruvic acid. Herbaspirillum strains also lack enzymes for the indole-3-acetonitrile pathway. Tryptophan-independent reactions from indoles to indole acetic acid via transferases are likely and potential genes are abundant, but no gene appears to encode a clearcut prototype indole acetic acid-producing enzyme (Figure 4).
Figure 4.
Biosynthesis and manipulation of plant hormones in HfGSF30. Green arrows or EC numbers indicate possible reactions or present enzymes, turquoise arrows indicate that at least one enzyme is missing in the pathway, while black arrows or EC numbers indicate that this pathway or enzyme is missing.
Lactoserines are utilized by AzospB510 for quorum-sensing, modulate the rhizosphere density competence and the adaptation of the bacteria to the environment. H. frisingense GSF30T failed to produce acyl homoserine lactones (Rothballer et al., 2008). In accordance with these experimental findings, the genes related to acyl homoserine lactone synthase and acylase were absent in all Herbaspirillum strains (Figure 3).
Discussion
The comparison of the draft genome sequence of HfGSF30 with the genetic inventory of related Herbaspirillum strains and more distant diazotrophic grass endophytes revealed a high diversity in their potential capabilities. The well-characterized endophyte H. seropedicae SmR1, which is associated with gramineous species like sorghum, sugarcane, rice and maize (Kirchhof et al., 2001) in warm climates, shares high nucleotide sequence identity with HfGSF30. However, even higher conservation in the genomic equipment was detected with non-diazotrophic Herbaspirillum strains that were not isolated as endophytes, but rather from the rhizosphere of Australian phragmites (HGW103) and well water (HhIAM).
Among the sequenced endophytes, a differential inventory for the nitrogen metabolism is striking. This suggests that a range of different metabolic capabilities allows endophytic colonization of various plant habitats, even within a single plant species. HfGSF30 is closer related to Herbaspirillum seropedicae isolates from rice than to HsSmR1, and among more distant endophytes its metabolic capabilities most closely resemble that of AzoaBH72, but it has little overlap with the metabolic equipment of the sugarcane-associated GdPAI5. Endophytes may colonize different niches within the same plant and interact; despite their contrasting metabolic inventory, different endophytic strains were abundant in sugar cane fields that were inoculated with a bacterial inoculation mixture including Gluconacetobacter diazotrophicus PAI5 and Herbaspirillum (Fischer et al., 2012). Interestingly, even bacteria not present in the inoculum were associated with these sugarcane plants (Fischer et al., 2012).
The metabolic traits discussed above differ widely in the Herbaspirillum genus, in accordance with diverse habitats, manifested, e.g., by the human isolate HJC206 or the nodule isolate HlP6-12. These two bacteria show least overlapping genomic capabilities with Herbaspirillum seropedicae strains (Figure 3). With the exception of the Herbaspirillum strain isolated from human fecal flora, all Herbaspirillum strains are equipped to utilize nitrate as a nutrient and reduce it to ammonium. This is not a common feature of plant endophytes, as GdPAI5 lacks all essential nitrate assimilation genes. The capability of anaerobic respiration using nitrate as an electron acceptor in HfGSF30, HsSmR1, HsOs45, and HsOs34 correlates with the presence of nitrogen fixation genes, suggesting that these strains can adapt to low nitrogen and oxygen availabilities. This is also underscored by the tendency that these strains have higher number of terminal oxidase genes.
H. frisingense GSF30T turned out unique as a potential N2O producer among the Herbaspirillum strains. Significant N2O emissions, exceeding those of a heavily fertilized rye field, but less than those from fertilized maize, have been reported from fertilized M. x giganteus, a host of HfGSF30 (Jørgensen et al., 1997; Gauder et al., 2012). However, not relevant N2O emissions were detected from unfertilized M. x giganteus (Jørgensen et al., 1997; Gauder et al., 2012).
Hormone production and/or degradation may contribute to the variable growth promoting effect of Herbaspirillum strains. The metabolic pathways that produce these metabolites have been identified by analytical tests (Rothballer et al., 2008) and in the sequence. AzoaBH72, a native colonizer of Kallar grass, appears in many aspects similar to HfGSF30. For example, both strains lack the entire type IV secretion system, which is partially present in the other sequenced endophyte genomes, but not in the Herbaspirillum genus. Highly relevant is the lack of the type III system in HfGSF30 (and in HhIAM, HlP6-12, HGW103, HJC206), its presence and importance for colonization in HsSmR1 (and HsOs45, HsOs34, HCF444, HYR522) and HrM1 (Monteiro et al., 2012a); and similar diversity within parts of the type II system. The different sets of secretion systems in HfGSF30 are compatible with the observed broad host ranges and no pathogenicity associated with this strain. Furthermore, several further candidate genes that are potentially involved in plant colonization, e.g., genes encoding attachment proteins of the hemagglutinin-type and genes involved in lipopolysaccharide formation and export differ between individual Herbaspirillum strains (Monteiro et al., 2012a). The absence of flagella that often harbor molecular patterns that are recognized by the plant pathogen defense, may be an advantage for high colonization numbers by Kp342 (Fouts et al., 2008). However, HfGSF30 and the other endophytes contain the entire flagella machinery, and this suggests that the flagellum plays an important role for these organisms, similar as in other root colonizing bacteria. For example, in Azospirillum brasilense Sp7, the flagellum is not only crucial for the chemotactic movement toward the root, but also for the initial attachment and final anchoring to the root surface. Mutants impaired in flagella formation are severely hampered in their colonization efficiency (Croes et al., 1993). However, it is also known that in contact with the plant, Azospirillum brasilense strains undergo substantial pleomorphic changes which also includes the loss of the flagellum (Assmus et al., 1995).
In summary, the HfGSF30 genome shows high similarity to the well known diazotrophic endophyte Herbaspirillum seropedicae, but even higher similarity (except for nitrogen fixation) with genomes from strains isolated from Australian phragmites rhizosphere and Japanese well water. High similarity in secretion systems and cell wall metabolism, among other traits, may suggest that either the respective habitats of these Herbaspirillum strains (HfGSF30, HGW103, HhIAM) are wider or that minor differences can confer different habitat competence. Grass endophytes do not only utilize highly diverse interaction (secretion) and attachment systems, but individual endophytes utilize highly different basic metabolic modules to survive in grasses. Endophytic, rhizosphere-competent and well water Herbaspirillum bacteria have surprising overlap in their genomic equipment.
Author contributions
Daniel Straub carried out all sequence annotations and molecular genetic analysis, Michael Rothballer, Anton Hartmann and Uwe Ludewig participated in the analysis and writing of the manuscript. Uwe Ludewig designed the study, and all authors read and approved the final manuscript.
Acknowledgments and funding
We thank Dr. Marek Dynowski (University of Tübingen, Germany) for software advice, the Deutsche Forschungsgemein- schaft and EU grant “Biofector” for partial financial support and Prof. Ralf Kaldenhoff (Technical University of Darmstadt, Germany) for generous support. We thank S. Demyan for proof reading.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- EC
Enzyme classification
- AzoaBH72
Azoarcus sp. BH72
- Kp342
Klebsiella pneumoniae 342
- AzospB510
Azospirillum sp. B510
- GdPAI5
Gluconacetobacter diazotrophicus PAl5
- HfGSF30
Herbaspirillum frisingense GSF30T
- HsSmR1
Herbaspirillum seropedicae SmR1
- HrM1
Herbaspirillum rubrisubalbicans M1
- HhIAM
Herbaspirillum huttiense subsp. putei IAM 15032
- HlP6-12
Herbaspirillum lusitanum P6-12 (DSM 17154)
- HGW103
Herbaspirillum sp. GW103
- HJC206
Herbaspirillum sp. JC206
- HCF444
Herbaspirillum sp. CF444
- HYR522
Herbaspirillum sp. YR522
- HsOs34
Herbaspirillum seropedicae Os34
- HsOs45
Herbaspirillum seropedicae Os45.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/Evolutionary_and_Genomic_Microbiology/10.3389/fmicb.2013.00168/abstract
References
- Almeida N. F., Yan S., Lindeberg M., Studholme D. J., Schneider D. J., Condon B., et al. (2009). A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a Type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 22, 52–62 10.1094/MPMI-22-1-0052 [DOI] [PubMed] [Google Scholar]
- Alquéres S., Oliveira J., Nogueira E., Guedes H., Oliveira P., Câmara F., et al. (2010). Antioxidant pathways are up-regulated during biological nitrogen fixation to prevent ROS-induced nitrogenase inhibition in Gluconacetobacter diazotrophicus. Arch. Microbiol. 192, 835–841 10.1007/s00203-010-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmus B., Hutzler P., Kirchhof G., Amann R., Lawrence J. R., Hartmann A. (1995). In situ localization of Azospirillum brasilense in the rhizosphere of wheat using fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl. Environ. Microbiol. 61, 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertalan M., Albano R., de Padua V., Rouws L., Rojas C., Hemerly A., et al. (2009). Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450 10.1186/1471-2164-10-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Utturkar S. M., Klingeman D. M., Johnson C. M., Martin S. L., Land M. L., et al. (2012). Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J. Bacteriol. 194, 5991–5993 10.1128/JB.01243-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante V. A., Dobereiner J. (1988). A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 108, 23–31 10.1007/BF02370096 [DOI] [Google Scholar]
- Costacurta A., Vanderleyden J. (1995). Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 21, 1–18 10.3109/10408419509113531 [DOI] [PubMed] [Google Scholar]
- Croes C. L., Moens S., Vanbastelaere E., Vanderleyden J., Michiels K. W. (1993). The polar flagellum mediates azospirillum-brasilense adsorption to wheat roots. J. Gen. Microbiol. 139, 2261–2269 10.1099/00221287-139-9-2261 [DOI] [Google Scholar]
- Davis S. C., Parton W. J., Dohleman F. G., Smith C. M., Del Grosso S., Kent A. D., et al. (2010). Comparative biogeochemical cycles of bioenergy crops reveal nitrogen-fixation and low greenhouse gas emissions in a miscanthus x giganteus agro-ecosystem. Ecosystems 13, 144–156 10.1007/s10021-009-9306-9 [DOI] [Google Scholar]
- De Souza V., Piro V. C., Faoro H., Tadra-Sfeir M. Z., Chicora V. K., Guizelini D., et al. (2013). Draft genome sequence of Herbaspirillum huttiense subsp. putei IAM 15032, a strain isolated from well water. Genome Announc. 1:e00252-12 10.1128/genomeA.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B., Weber O. B., Kirchhof G., Halbritter A., Stoffels M., Hartmann A. (2001). Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int. J. Syst. Evol. Microbiol. 51, 17–26 [DOI] [PubMed] [Google Scholar]
- Elbeltagy A., Nishioka K., Sato T., Suzuki H., Ye B., Hamada T., et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293 10.1128/AEM.67.11.5285-5293.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux a, Hachani A., Bleves S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 10.1099/mic.0.2008/016840-0 [DOI] [PubMed] [Google Scholar]
- Fischer D., Pfitzner B., Schmid M., Simões-Araújo J. L., Reis V. M., Pereira W., et al. (2012). Molecular characterization of the diazotrophic bacterial community in non-inoculated and inoculated field grown sugar cane (Saccharum sp.). Plant Soil 356, 83–99 10.1007/s11104-011-0812-0 [DOI] [Google Scholar]
- Fouts D. E., Tyler H. L., DeBoy R. T., Daugherty S., Ren Q., Badger J. H., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4:e1000141 10.1371/journal.pgen.1000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauder M., Butterbach-Bahl K., Graeff-Hönninger S., Claupein W., Wiegel R. (2012). Soil-derived trace gas fluxes from different energy crops – results from a field experiment in Southwest Germany. GCB Bioenerg. 4, 289–301 10.1111/j.1757-1707.2011.01135.x [DOI] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., Elsas J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Heaton E. A., Flavell R. B., Mascia P. N., Thomas S. R., Dohleman F. G., Long S. P. (2008). Herbaceous energy crop development: recent progress and future prospects. Curr. Opin. Biotechnol. 19, 202–209 10.1016/j.copbio.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Jørgensen R. N., Jørgensen B. J., Nielsen N. E., Maag M., Lind A. M. (1997). N2O emission from energy crop fields of Miscanthus “Giganteus” and winter rye. Atmos. Environ. 31, 2899–2904 10.1016/S1352-2310(97)00128-3 [DOI] [Google Scholar]
- Juhas M., Crook D. W., Hood D. W. (2008). Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10, 2377–2386 10.1111/j.1462-5822.2008.01187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Minamisawa K., Isawa T., Nakatsukasa H., Mitsui H., Kawaharada Y., et al. (2010). Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res. 17, 37–50 10.1093/dnares/dsp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhof G., Eckert B., Stoffels M., Baldani J. I., Reis V. M., Hartmann A. (2001). Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int. J. Syst. Evol. Microbiol. 51, 157–168 [DOI] [PubMed] [Google Scholar]
- Krause A., Ramakumar A., Bartels D., Battistoni F., Bekel T., Boch J., et al. (2006). Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24, 1385–1391 10.1038/nbt1243 [DOI] [PubMed] [Google Scholar]
- Lagier J. C., Gimenez G., Robert C., Raoult D., Fournier P. E. (2012). Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand. Genomic Sci. 7, 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. W., Lee K. J., Chae J. C. (2012). Genome sequence of Herbaspirillum sp. strain GW103, a plant growth-promoting bacterium. J. Bacteriol. 194, 4150 10.1128/JB.00806-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Kawahara M., Minamisawa K. (2004). Novel endophytic nitrogen-fixing clostridia from the grass Miscanthus sinensis as revealed by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70, 6580–6586 10.1128/AEM.70.11.6580-6586.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R. A., Balsanelli E., Tuleski T., Faoro H., Cruz L. M., Wassem R., et al. (2012a). Genomic comparison of the endophyte Herbaspirillum seropedicae SmR1 and the phytopathogen Herbaspirillum rubrisubalbicans M1 by suppressive subtractive hybridization and partial genome sequencing. FEMS Microbiol. Ecol. 80, 441–451 10.1111/j.1574-6941.2012.01309.x [DOI] [PubMed] [Google Scholar]
- Monteiro R. A., Balsanelli E., Wassem R., Marin A. M., Brusamarello-Santos L. C. C., Schmidt M. A., et al. (2012b). Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant Soil 356, 175–196 10.1007/s11104-11012-11125-11107 [DOI] [Google Scholar]
- Pedrosa F. O., Monteiro R. A., Wassem R., Cruz L. M., Ayub R. A., Colauto N. B., et al. (2011). Genome of Herbaspirillum seropedicae Strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 7:e1002064 10.1371/journal.pgen.1002064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Hurek T. (1998). Life in grasses: diazotrophic endophytes. Trends Microbiol. 6, 139–144 10.1016/S0966-842X(98)01229-3 [DOI] [PubMed] [Google Scholar]
- Rothballer M., Schmid M., Klein I., Gattinger A., Grundmann S., Hartmann A., et al. (2008). Endophytic root colonization of gramineous plants by Herbaspirillum frisingense. FEMS Microbiol. Ecol. 66, 85–95 10.1111/j.1574-6941.2008.00582.x [DOI] [PubMed] [Google Scholar]
- Studholme D. J., Ibanez S. G., MacLean D., Dangl J. L., Chang J. H., Rathjen J. P. (2009). A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics 10:395 10.1186/1471-2164-10-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturz A. V., Christie B. R., Nowak J. (2000). Bacterial endophytes: Potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19, 1–30 10.1016/S0735-2689(01)80001-0 [DOI] [Google Scholar]
- Tabita F. R., Hanson T. E., Li H. Y., Satagopan S., Singh J., Chan S. (2007). Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71, 576 10.1128/MMBR.00015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprey V., Del Greco A., Golinowski W., Broughton W., Perret X. (1998). Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28, 1381–1389 10.1046/j.1365-2958.1998.00920.x [DOI] [PubMed] [Google Scholar]
- Weiss V. A., Faoro H., Tadra-Sfeir M. Z., Raittz R. T., de Souza E. M., Monteiro R. A., et al. (2012). Draft genome sequence of Herbaspirillum lusitanum P6-12, an endophyte isolated from root nodules of Phaseolus vulgaris. J. Bacteriol. 194, 4136–4137 10.1128/JB.00657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Scott A. J. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28, 1033–1034 10.1093/bioinformatics/bts079 [DOI] [PubMed] [Google Scholar]
- Ye W., Ye S., Liu J., Chang S., Chen M., Zhu B., et al. (2012). Genome sequence of the pathogenic Herbaspirillum seropedicae strain Os34, isolated from rice roots. J. Bacteriol. 194, 6993–6994 10.1128/JB.01934-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Ye S., Chang S., Chen M., Sun L., An Q. (2012). Genome sequence of the pathogenic Herbaspirillum seropedicae strain Os45, isolated from rice roots. J. Bacteriol. 194, 6995–6996 10.1128/JB.01935-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.