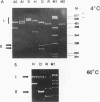
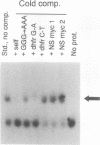
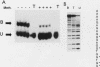
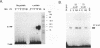Abstract
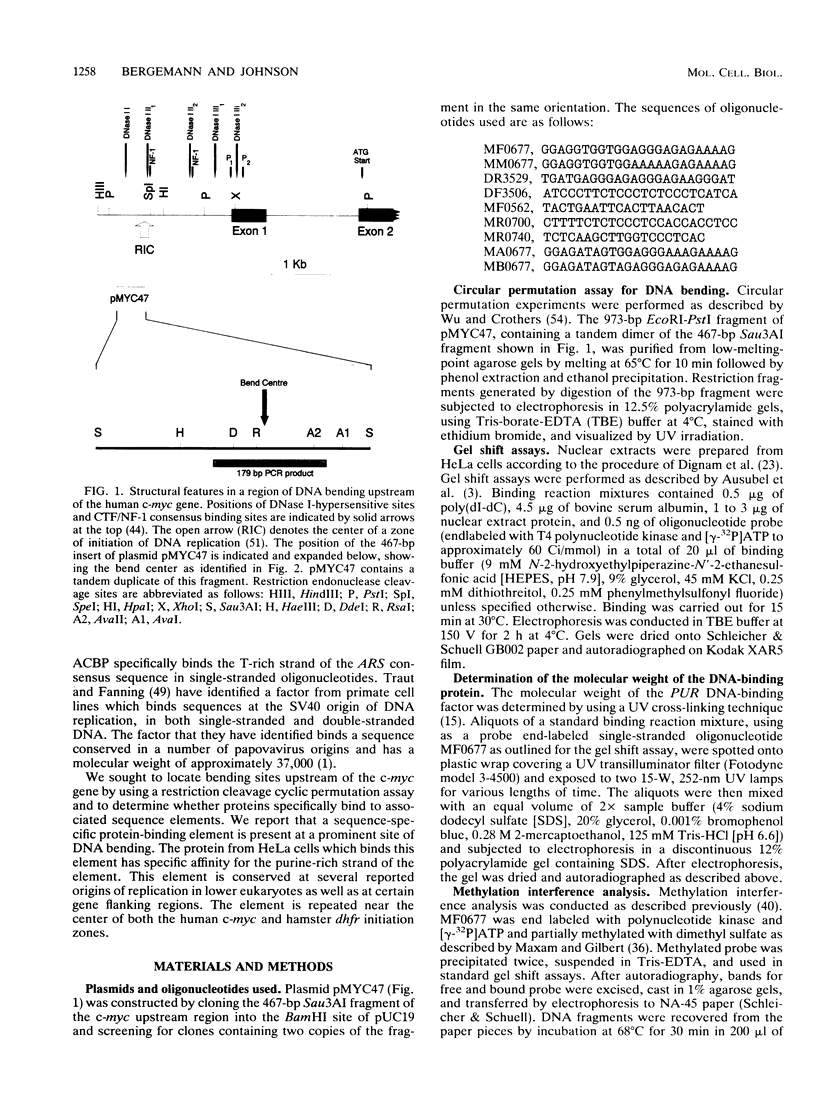
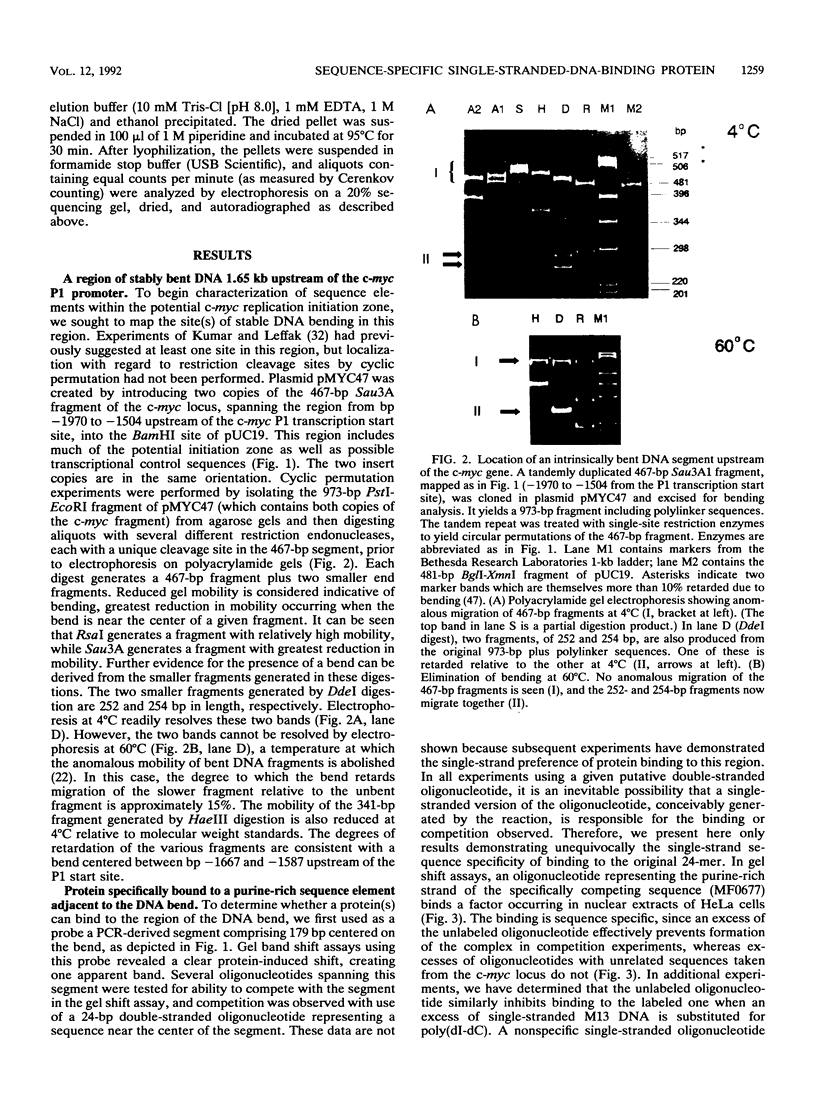
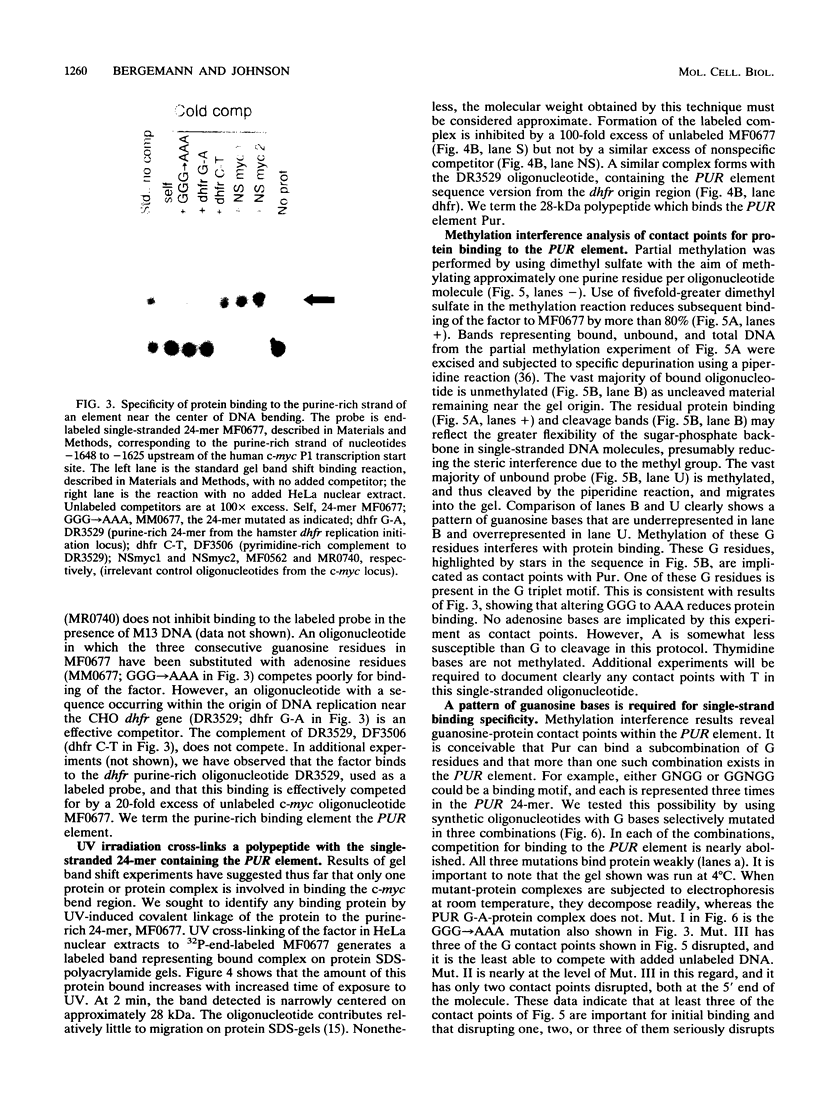
A major site of DNA bending is located 1.6 kb upstream of the P1 transcription start site of the human c-myc gene, near the center of a reported zone of initiation of DNA replication. A repeated, purine-rich element, termed PUR, at the bend site is specifically bound by a protein in HeLa cell nuclear extracts. This protein has specific affinity for the purine-rich single strand of the element. Methylation interference maps a pattern of specific contact points with guanosine bases in a 24-mer oligonucleotide containing the element. UV cross-linking reveals that contact is made by a polypeptide of approximately 28 kDa. The PUR element is present at origins of replication and in gene flanking regions in a variety of eukaryotes from yeasts through humans. The consensus sequence GGNNGAGGGAGARRRR has been derived. This element is present near centers of regions of two mammalian loci (human c-myc and hamster dhfr) recently reported as initiation zones for DNA replication. A 24-mer oligonucleotide representing the hamster dhfr version of the PUR element effectively competes with the human c-myc version for binding to Pur.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alliger P., Traut W., Carstens E., Fanning E. Partial purification and characterization of a cellular protein that binds to the SV40 core origin of DNA replication. Biochim Biophys Acta. 1988 Dec 20;951(2-3):396–402. doi: 10.1016/0167-4781(88)90112-1. [DOI] [PubMed] [Google Scholar]
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Brookes S., Smith R., Casey G., Dickson C., Peters G. Sequence organization of the human int-2 gene and its expression in teratocarcinoma cells. Oncogene. 1989 Apr;4(4):429–436. [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986 Jan 28;25(2):441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Dailey L., Heintz N. H. RIP60, a mammalian origin-binding protein, enhances DNA bending near the dihydrofolate reductase origin of replication. Mol Cell Biol. 1990 Dec;10(12):6236–6243. doi: 10.1128/mcb.10.12.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Cheng L., Kelly T. J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989 Nov 3;59(3):541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S., Riley D. E., Stein G., Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Caddle M. S., Heintz N., Heintz N. H. Purification of RIP60 and RIP100, mammalian proteins with origin-specific DNA-binding and ATP-dependent DNA helicase activities. Mol Cell Biol. 1990 Dec;10(12):6225–6235. doi: 10.1128/mcb.10.12.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel D. C., Johnson E. M. Selective initiation of replication at origin sequences of the rDNA molecule of Physarum polycephalum using synchronous plasmodial extracts. Nucleic Acids Res. 1989 Oct 25;17(20):8343–8362. doi: 10.1093/nar/17.20.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J. Nucleotide sequence of the central non-transcribed spacer region of Physarum polycephalum rDNA. Gene. 1985;39(2-3):203–211. doi: 10.1016/0378-1119(85)90314-2. [DOI] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F., Gasser S. M. Identification and purification of a protein that binds the yeast ARS consensus sequence. Cell. 1991 Mar 8;64(5):951–960. doi: 10.1016/0092-8674(91)90319-t. [DOI] [PubMed] [Google Scholar]
- Kumar S., Leffak M. DNA topology of the ordered chromatin domain 5' to the human c-myc gene. Nucleic Acids Res. 1989 Apr 11;17(7):2819–2833. doi: 10.1093/nar/17.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak M., James C. D. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol Cell Biol. 1989 Feb;9(2):586–593. doi: 10.1128/mcb.9.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohia A., Haider N., Biswas B. B. Characterisation of a repetitive DNA family from Entamoeba histolytica containing Saccharomyces cerevisiae ARS consensus sequences. Gene. 1990 Dec 15;96(2):197–203. doi: 10.1016/0378-1119(90)90253-n. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McWhinney C., Leffak M. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 1990 Mar 11;18(5):1233–1242. doi: 10.1093/nar/18.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschonas N., de Boer E., Flavell R. A. The DNA sequence of the 5' flanking region of the human beta-globin gene: evolutionary conservation and polymorphic differences. Nucleic Acids Res. 1982 Mar 25;10(6):2109–2120. doi: 10.1093/nar/10.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. G., Newlon C. S. A yeast replication origin consists of multiple copies of a small conserved sequence. Cell. 1988 May 6;53(3):441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- Postel E. H., Mango S. E., Flint S. J. A nuclease-hypersensitive element of the human c-myc promoter interacts with a transcription initiation factor. Mol Cell Biol. 1989 Nov;9(11):5123–5133. doi: 10.1128/mcb.9.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouer E., Van Huynh T., Lavareda de Souza S., Lang M. C., Fischer S., Benarous R. Structure of the human lck gene: differences in genomic organisation within src-related genes affect only N-terminal exons. Gene. 1989 Dec 7;84(1):105–113. doi: 10.1016/0378-1119(89)90144-3. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Schwab M., Bishop J. M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Traut W., Fanning E. Sequence-specific interactions between a cellular DNA-binding protein and the simian virus 40 origin of DNA replication. Mol Cell Biol. 1988 Feb;8(2):903–911. doi: 10.1128/mcb.8.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L., Johnson E. M. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol. 1990 Sep;10(9):4899–4904. doi: 10.1128/mcb.10.9.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L., Johnson E. M. Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res. 1989 Oct 11;17(19):7693–7705. doi: 10.1093/nar/17.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990 Jun 15;61(6):1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]