Abstract
Background The present study investigated whether single nucleotide polymorphisms (SNPs) in the alpha-protein kinase 1 (ALPK1) gene are associated with gout in aboriginal and Han Chinese Taiwanese.
Methods A total of 1351 aborigines from the community (511 cases and 840 controls) and 511 Han people from hospital (104 cases and 407 controls) were recruited. SNPs in potentially functional regions of the 38 genes within 4q25 were identified and genotypes determined by direct sequencing. Quantitation of blood ALPK1 mRNA expression levels and luciferase assay of gout-associated rs231253 pGL3-SNP constructs cotransfected with hsa-miR-519e were examined.
Results We found that ALPK1 gene was the most determinant of gout. Three SNPs of rs11726117 M861T [C], rs231247 [G] and rs231253 [G] were most associated with gout risk [odd ratios (OR) ≥1.44, P ≤ 3.78 × 10−6) in aborigines. A replication set using Han people had risk at rs11726117 and rs231247 (OR ≥1.72, P ≤ 4.08 × 10−3). From pooled analysis (Breslow-Day test, P > 0.33) assuming an additive model, each increasing copy of the risk allele of rs11726117 [C], rs231247 [G] and rs231253 [G] showed significantly elevated OR for gout ≥1.42 (P ≥ 1.53 × 10−6). Consistently, the composite homozygous of linked 3 SNPs (versus wild-type, OR = 1.83, P = 8.21 × 10−4) had strong associations with ALPK1 mRNA expression. Luciferase showed reduced hybridization between hsa-miR-519e and construct carrying gout-associated rs231253 [G] than the wild-type [C] (P = 6.19 × 10−4).
Conclusions Our study found that a newly identified ALPK1 gene can effectively interfere with microRNA target recognition and modulates the mRNA expression; and the varying distribution of the implicated SNPs among cases and controls in the two studied populations suggests a significant role in gout susceptibility.
Keywords: Gout, ALPK1 gene, SNP, microRNA, mRNA
Introduction
Gout is characterized by monosodium urate (MSU) deposits in the soft tissues of hyperuricaemic patients whose symptoms include recurrent flares of acute gout arthritis, tophaceous deposits of MSU crystals in joints and other tissues, chronic arthropathy, uric acid urolithiasis and renal impairment.1,2 Gout patients experience very painful attacks due to the precipitation of MSU crystals in joints, along the Achilles tendon and the first metatarsophalangeal joint, as well as the triggering of inflammatory cytokines, chemokines, proteases and oxidants that lead to damage such as chronic synovitis, cartilage loss and bone erosion.1–3 Hyperuricaemia is present in almost all gout patients, however only a minority (20%) of hyperuricaemic individuals experience gout symptoms.4 Tophus is a severe form of crystal arthropathy that can occur anywhere in the body. Without treatment, tophi occur in 30% of patients within 5 years of onset of gout,3 whereas others may continue to have flares of acute arthritis without the tophus occurrence. Therefore, studying a possible role for these familial hyperuricaemia- and gout-causing genes in common gout is justified.5
For the past two decades, public health researches have identified the Taiwanese aboriginals as having a high gout prevalence of 12%6,7 especially male aboriginals, with as high as 64%8 of gout cases exhibiting familial clustering of chronic tophaceous gout. Linguistic and archaeological evidence suggests that the aboriginals were originally one group of expanding Neolithic Setaria farmers who first settled in Taiwan as early as 5500 YBP from the southern coast of China.9 Their prevalence markedly differs from Taiwanese Han (who immigrated in 300 YBP in large numbers to Taiwan from Guangzhou and Fujian provinces) at 3%6 and tophi rates that have declined from 34%10 to 9%11 of cases during 1948–1984 to 1992–1999.
Following the genome-wide linkage of aboriginal gout multiplex at marker D4S2623 (at 114cM) on chr4q25 (logarithm of odds [LOD] = 4.29), the Gout Susceptibility1 (GOUT1, ID %138900) was a region hypothesized to influence Taiwanese aboriginal gout.12 The aborigines are likely to have been isolated in small groups for thousands of years, subjected to higher genetic drift and increased homogeneity. Today, the indigenous population is a minority comprising <3% of entire population of Taiwan. This is an opportunity to study gout-related gene variant differences among ethnicities with special demographic histories, adding to the existing suite of major gout genes discovered.
Methods
Study participants
This study recruited from a community-based Taiwanese aboriginal health cohort study and selected 1351 subjects from 2235 aboriginal highlanders residing in the north central mountains and examined at Township Health Stations since 1994, whom we closely followed up on 511 cases (38.6% had tophi) with chronic gout consistently observed for mean duration of 10 years.12,13 The general physicians based at health stations and visiting rheumatology medical specialists carefully diagnosed and followed gout in each aborigine using clinical history, physical exam and blood tests performed 2–4 times annually. Because of limited resources of clinical treatment in the area, only a small proportion (<5%) of aboriginal gout patients were treated with uric acid-lowering agents. Additionally, a replication set, 511 hospital-based Taiwanese Han (104 gout and 407 controls), of which cases have chronic gout, were consistently observed for mean duration of 8 years.14 All gout cases were strictly diagnosed by American College of Rheumatology criteria.15 Among cases, a high proportion had tophaceous gout, 39% aboriginals and 49% Han; these patients must have demonstrated two or more tophi, clearly visible and palpable from arms, legs, ears, or articular cartilage from other sites, and hyperuricaemia (>8 mg/dl) when their blood was collected. The controls were 840 healthy aboriginals from the same communities as the aboriginal cases, and 407 gout-free Han from the same hospitals as the Han cases, all ascertained to be unrelated, with no history of gout and not taking hypouricaemic agents for other medical conditions. Trained nurses recorded demographic data by standardized questionnaires. Blood specimens were analyzed using standardized hospital protocols. Hyperuricaemia was defined as uric acid exceeding 7.0 mg/dl in males and 6.0 mg/dl in females. Institutional review boards and ethics committees from Kaohsiung Medical University and National Health Research Institutes approved our study design. All participants gave their written informed consent.
DNA resequencing and SNP genotyping
Genomic DNA was extracted from peripheral whole blood by wizard genomic DNA purification kit (QIAGEN-Gentra Puregene Blood Kit) followed standard laboratory protocols. DNA resequencing used Dye Terminator kit (Applied Biosystems) and ABI 3730 DNA sequencers. ALPK1 gene SNPs were genotyped by TaqMan SNP allelic discrimination assay (Applied Biosystems). TaqMan reactions were based on manufacturer's protocol and samples ran on the ABI7900HT Real-Time polymerase chain reaction (PCR) platform (Applied Biosystems). Allelic discrimination was performed, and analysed by SDS software (v. 2.3) (Applied Biosystems).
mRNA
RNA analysis of 62 aborigines (23 gout and 39 controls) was performed using real-time PCR as described previously.16 Total RNA from human peripheral blood leukocytes was isolated by PAXgene Blood RNA Kit and generated cDNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). The expression experiment of ALPK1 mRNA was performed using pre-designed gene-specific TaqMan probes and primer pairs (Assays-ID: Hs00228473_m1), in triplicate per sample, and a control without template included in each plate. Reference housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified in same plate and equivalent to all samples.
Luciferase assay
The rs231253 allelic differences in promoter activity in the human HEK293 line were confirmed with use of the pGL3-basic vector. Cells were transfected with these reporter constructs and with pRLTK renilla (Promega) luciferase vector as a normalization control. We constructed HEK293 cells (2 × 104 cells per 96-well) were cotransfected with 0.1μg of pGL3-SNP (firefly luciferase), 0.02μg of pGL4.73 (renilla luciferase) (Promega) and 0.1μg of the hsa-miR-519e (Applied Biosystems) plasmid. Cells were lysed 72 h after transfection, in 50 μl of lysis buffer according to the Dual-Glo Luciferase Assay System protocol (Promega) and luciferase activity measured with TopCount NXT luminometer (Packard). Relative luciferase activity of ALPK1 reporter constructs was calculated as the ratio of firefly luciferase activity to renilla luciferase. Reporter assays shown were the average of four independent experiments in triplicates and were analysed by repeated measures analysis.
Statistical analysis
Multipoint analysis was performed using conditional-logistic model implemented in the S.A.G.E. v6.0.1 program for linkage fine-map. We used a one-parameter model and default value that constrained relative risks, λ2 = 3.634λ1–2.364, assuming the expression fix for mode of inheritance was a value approximately halfway between a dominant and a recessive model, corresponding to the Whittemore and Tu minmax model mode of inheritance parameter.17 The likelihood-ratio statistic was computed by multiplying the LOD (logarithm of odds) score by 4.6. Linkage disequilibrium (LD) coefficients (D’ and r2) implemented by Haploview v4.218 and PLINK v1.07.19 Genotype frequencies of the controls of ethnic groups regarding investigated SNPs conformed to Hardy-Weinberg equilibrium and were assessed by χ2 goodness-of-fit test. The means or proportions for baseline gout risk factors were calculated for cases and controls. Significance of associated risk factors was tested with the χ2 statistic for categorical variables and the generalized linear regression models for continuous variables. We analysed the association between SNPs and risk of gout using logistic regression analysis. Additive genetic effects were modelled by defining continuous variables with levels 0, 1 and 2 corresponding to genotypes (i.e. rs11726117 was coded as 0 for TT, 1 for CT and 2 for CC). The Breslow-Day test examined the odds ratio (OR) homogeneity between family-based substructures and population-based aboriginal gout subjects (Breslow-Day test, P = 0.22) were obtained. OR with 95% confidence intervals (CI) was adjusted for age, gender, body mass index, hypertension, alcohol use, hyperuricaemia, total cholesterol, log (triglycerides) and creatinine using a logistic regression model. Pooled analysis was performed by Cochran-Mantel-Haenszel test method using the PLINK v1.07. Case/control traits can be analysed for haplotype associations using PLINK v1.07. We categorized composite genotypes into three categories: risk-homozygous (rs11726117 [CC] + rs231247 [GG] + rs231253 [GG]), reference-homozygous ([TT] + [AA] + [CC]) and other-type. ALPK1 mRNA expression by composite genotypes was calculated.
Bioinformatics analysis
Binding site of 3’ untranslated region (UTR) polymorphism to miRNA was indicated by MicroCosm Targets version 5 at EMBL-EBI. RNAhybrid20 calculated miRNA::SNP interaction induced allele-induced minimum free energy (MFE) changes. Signal peaks of NF-κB (p65) within 100-10 kb upstream of the transcription initiation site of ALPK1 was shown to be present in GM12878 (lymphoblastoid) cell-line of ENCODE Yale TFBS by ChIP-seq Yale/UC-Davis/Harvard (release 3; May 2010). MOTIF Search explored all putative transcription binding sites in the ALPK1 promoter region.
Results
Clinical characteristics of study participants are presented in Table 1. Gout cases had a mean onset age of 40.8 years for aborigines and 45.2 years for Han. Cases showed high urate levels (≥8.9 mg/dl), ≥85.5% hyperuricaemia and ≥38.5% tophaceous gout, and higher mean total cholesterol, triglycerides and creatinine levels (P < 0.01). Gout cases also had significantly increased alcohol intake (P < 0.001). Aboriginal controls showed borderline-high uric acid (≥7 mg/dl) and triglycerides (199 mg/dl), and 31% were hypertensive. Type 2 diabetes, often comorbid with gout in Caucasians, was insignificant in the aborigines.
Table 1.
Characteristics of the study participants
| Taiwanese aborigines |
Taiwanese Han |
|||||
|---|---|---|---|---|---|---|
| Characteristic | Cases | Controls | P | Cases | Controls | P |
| Number | 511 | 840 | 104 | 407 | ||
| Age (SD), years | 51.1 (14.4) | 53.5 (16.5) | 0.0065 | 52.8 (13.7) | 55.2 (14.5) | 0.1308 |
| Men, n (%) | 392 (76.7) | 419 (49.9) | <0.0001 | 104 (100.0) | 405 (99.5) | 0.4738 |
| Age of onset (SD), years | 40.8 (15.0) | 45.2 (12.3) | ||||
| Duration of gout (SD), years | 9.9 (7.9) | 8.2 (6.3) | ||||
| Tophi, n (%) | 197 (38.6) | 51 (49.0) | ||||
| Systolic pressure (SD), mmHg | 139.1 (21.7) | 131.5 (20.7) | <0.0001 | 136.0 (18.0) | 131.7 (19.4) | 0.0535 |
| Diastolic pressure (SD), mmHg | 88.9 (14.1) | 83.3 (12.5) | <0.0001 | 85.6 (13.3) | 83.4 (11.7) | 0.1344 |
| Body mass index (SD), kg/m2 | 26.4 (4.2) | 26.5 (4.1) | 0.9322 | 26.0 (4.0) | 24.6 (3.4) | 0.0002 |
| Hypertension, n (%) | 225 (44.0) | 262 (31.2) | <0.0001 | 32 (30.8) | 71 (17.4) | 0.0025 |
| Type 2 diabetes mellitus, n (%) | 41 (8.0) | 62 (7.4) | 0.6661 | 2 (1.9) | 13 (3.2) | 0.4931 |
| Alcohol use, n (%) | 393 (76.9) | 446 (53.1) | <0.0001 | 35 (33.7) | 98 (24.1) | 0.0470 |
| Total cholesterol (SD), mg/dl | 186.9 (48.4) | 183.8 (46.3) | 0.2381 | 210.7 (48.1) | 190.9 (38.1) | <0.0001 |
| Triglycerides (SD), mg/dl | 268.5 (275.4) | 192.2 (245.3) | <0.0001 | 225.3 (121.4) | 144.2 (136.8) | <0.0001 |
| Log(triglycerides) (SD), mg/dl | 5.3 (0.7) | 4.9 (0.7) | <0.0001 | 5.3 (0.5) | 4.7 (0.6) | <0.0001 |
| Creatinine (SD), mg/dl | 1.2 (0.6) | 1.0 (0.2) | <0.0001 | 1.4 (0.4) | 1.2 (0.2) | <0.0001 |
| Uric acid (SD), mg/dl | 9.3 (2.4) | 7.0 (2.0) | <0.0001 | 8.9 (1.8) | 6.1 (1.3) | <0.0001 |
| Hyperurcaemia, n (%) | 437 (85.5) | 490 (58.3) | <0.0001 | 91 (87.5) | 99 (24.3) | <0.0001 |
SD, standard deviation. P-values from generalized linear regression models for continuous variables and from chi-square tests for categorical variables.
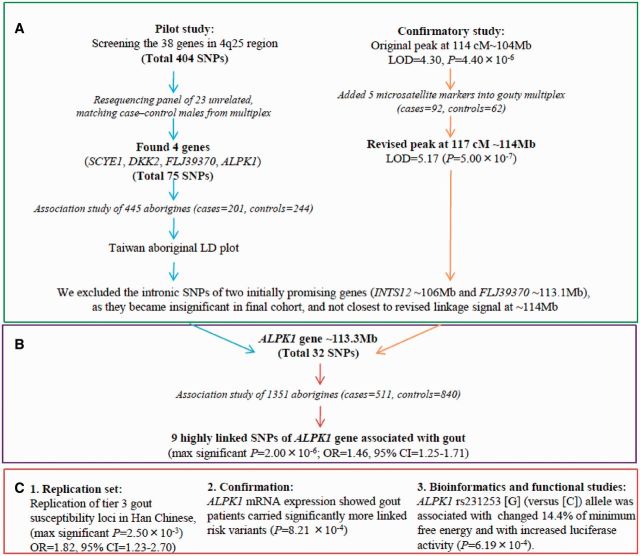
Our study scheme is summarized in Figure 1. A pilot study involved gene-centrically resequenced 666 polymerase-chain-reaction (PCR) amplicons including 38 gene exons, and flanking intron sequences of genes between D4S1647 and D4S2937 region in GOUT1 were typed and 404 SNPs exposed, using 23 unrelated male aboriginal gout case-control pairs to produce the association SNPs (Supplementary Table 1, available as Supplementary data at IJE online). Although, as for ALPK1, only one SNP is reported as significant (P = 0.041), we cannot rule out the possibility that ALPK1 variants contribute to the gout risk. Increased cohort size had narrowed these SNPs to those belonging to four genes (SCYE1, DKK2, FLJ39370, ALPK1; Supplementary Table 2, available as Supplementary data at IJE online). Separately, a confirmatory study by adding five dense microsatellite markers into the linkage peak at 114cM (P = 4.40 × 10−6, LOD = 4.29) moved the maximal signal to a new peak at 117cM (P = 5.00 × 10−7, LOD = 5.17; Supplementary Table 3 available as Supplementary data at IJE online). After re-examining many of previous genes that had been associated with smaller cohort sizes (e.g. INTS12 and FLJ39370), only ALPK1 SNPs remained consistently associated in all cohorts and at ∼113.3 Mb were closest to revised signal at ∼114 Mb on the physical map. The candidates that were not significant in the final cohort or closest to revised signal, such as intronic SNPs of INTS12 and FLJ39370, were discontinued.
Figure 1.
Region-wide association fine-map of chr4q25 to identify ALPK1. (A) We commenced with a pilot study that gene-centrically resequenced 38 genes (total 404 SNPs) in the 4q25 region using 23 unrelated male aboriginal gout case-control pairs that produced the association SNPs; a confirmatory study was made by adding five dense microsatellite markers into linkage peak at 114cM, then maximal signal migrated to a new peak at 117cM (P = 5.00 × 10−7, LOD = 5.17); raising the cohort size had narrowed down SNPs further to those within four genes (SCYE1, DKK2, FLJ39370 and ALPK1). (B) Using the final 1351 cohort, which consisted 511 gout cases and 840 controls, we isolated nine SNPs which were significantly associated with gout. (C) We replicated independently similar risk in the Han Chinese people. Interestingly, the linked variant at 3’UTR is predicted to be a binding site polymorphism of hsa-miR-519e, suggesting loss of gene regulation among carriers of the affected, a result consistent with luciferase activity in vitro and ALPK1 mRNA expression from blood samples findings
Thus, ALPK1 was selected as a Taiwanese aboriginal gout susceptibility gene and a linkage disequilibrium plot (44 SNPs) for 445 aborigines was constructed (Supplementary Figure 1 available as Supplementary data at IJE online). Furthermore, nine SNPs [located one in intron 7-8; four nonsynonymous in exon 11; two synonymous in exon 13 and 14; and two in 3’untranslated region (3’UTR)] were found to be significant with gout (Table 2) and in strong linkage disequilibrium (LD: D’≥0.85 and r2 ≥ 0.75 in controls). Three loci particularly, a nonsynonymous rs11726117 M861T [C], a synonymous rs231247 [G] and 3’UTR rs231253 [G] showed most significance with gout (P = 3.78 × 10−6, 2.00 × 10−6, 3.48 × 10−6, respectively) with odds ratios for gout after adjustment for hyperuricaemia and other covariates being 1.45 (95% CI = 1.21–1.73), 1.48 (95% CI = 1.24–1.77) and 1.43 (95% CI = 1.20–1.72), respectively. Independently using these three SNPs, we replicated a similar risk in Taiwanese Han (adjusted OR = 2.41 in rs11726117, adjusted OR = 2.15 in rs231247 and adjusted OR = 1.36 in rs231253). The haplotype-block showed high gout risk in two studied ethnicities (Supplementary Table 4 available as Supplementary data at IJE online).
Table 2.
Result of the gout-associated ALPK1 SNPs
| Cases |
Controls |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | R/NR | 11 | 12 | 22 | 1 | 11 | 12 | 22 | 1 | HWE P | P (trend) | OR (95% CI) | Adjusted OR (95% CI)a |
| Taiwan aborigines | |||||||||||||
| rs9994944 | G/A | 229 | 221 | 61 | 0.66 | 324 | 392 | 124 | 0.62 | 0.76 | 1.87 × 10−2 | 1.21 (1.03–1.43) | 1.26(1.05–1.52) |
| rs2074388 G565D | A/G | 180 | 243 | 88 | 0.59 | 214 | 414 | 212 | 0.50 | 0.68 | 9.03 × 10−6 | 1.43 (1.22–1.67) | 1.41(1.18–1.69) |
| rs13148353 H642R | G/A | 191 | 240 | 80 | 0.61 | 244 | 404 | 192 | 0.53 | 0.32 | 1.04 × 10−5 | 1.36 (1.17–1.59) | 1.39(1.16–1.66) |
| rs2074379 M732I | A/G | 184 | 246 | 81 | 0.60 | 237 | 405 | 198 | 0.52 | 0.33 | 1.03 × 10−4 | 1.36 (1.17–1.59) | 1.37(1.14–1.63) |
| rs11726117 M861T | C/T | 198 | 237 | 76 | 0.62 | 244 | 396 | 200 | 0.53 | 0.11 | 3.78 × 10−6 | 1.44 (1.23–1.69) | 1.45(1.21–1.73) |
| rs231247 R1084R | G/A | 195 | 233 | 83 | 0.61 | 225 | 414 | 201 | 0.51 | 0.70 | 2.00 × 10−6 | 1.46 (1.25–1.71) | 1.48(1.24–1.77) |
| rs55840220 T1145T | A/G | 23 | 131 | 357 | 0.17 | 21 | 125 | 694 | 0.10 | <0.01 | 1.89 × 10−7 | 1.76 (1.42–2.18) | 1.71 (1.33–2.18) |
| rs231253, 3' UTR | G/C | 190 | 239 | 82 | 0.61 | 223 | 416 | 201 | 0.51 | 0.80 | 3.48 × 10−6 | 1.45 (1.24–1.70) | 1.43(1.20–1.71) |
| rs960583, 3' UTR | A/G | 25 | 159 | 327 | 0.20 | 19 | 224 | 597 | 0.16 | 0.71 | 1.37 × 10−3 | 1.39 (1.13–1.69) | 1.33(1.06–1.66) |
| Taiwanese Han | |||||||||||||
| rs11726117 M861T | C/T | 70 | 31 | 3 | 0.82 | 209 | 168 | 30 | 0.72 | 0.64 | 2.50 × 10−3 | 1.82 (1.23–2.70) | 2.41 (1.41–4.12) |
| rs231247 R1084R | G/A | 70 | 28 | 6 | 0.81 | 204 | 167 | 36 | 0.71 | 0.83 | 4.08 × 10−3 | 1.72 (1.18–2.50) | 2.15 (1.31–3.50) |
| rs231253, 3' UTR | G/C | 63 | 35 | 6 | 0.77 | 215 | 164 | 28 | 0.73 | 0.66 | 1.92 × 10−1 | 1.27 (0.89–1.83) | 1.36 (0.84–2.20) |
| Pooled analysisb | |||||||||||||
| rs11726117 M861T | C/T | 268 | 268 | 79 | 0.65 | 453 | 564 | 230 | 0.59 | 2.89 × 10−8 | 1.51 (1.31–1.75) | 1.53 (1.30–1.81) | |
| rs231247 R1084R | G/A | 265 | 261 | 89 | 0.64 | 429 | 581 | 237 | 0.58 | 2.05 × 10−8 | 1.52 (1.31–1.75) | 1.55 (1.31–1.83) | |
| rs231253, 3' UTR | G/C | 253 | 274 | 88 | 0.63 | 438 | 580 | 229 | 0.58 | 1.53 × 10−6 | 1.42 (1.23–1.65) | 1.42 (1.21–1.68) | |
R, risk allele; NR, non-risk allele; 11 indicates at-risk homozygote, 12 indicates heterozygote; 22 indicates wild-type homozygote; HWE, Hardy-Weinberg equilibrium; 3' UTR, three prime untranslated region; OR, odd ratios; CI, confidence interval.
ORs and P-values were for each case-control study separately under an additive model of inheritance.
aOR with 95% CI in parentheses was adjusted for age, gender, body mass index, hypertension, alcohol use, total cholesterol, log (triglycerides), creatinine and hyperuricaemia by a logistic regression model.
bPooled analysis was performed by the Cochran-Mantel-Haenszel test method using the PLINK v1.07. Three SNPs revealed homogeneity in the association with gout (Breslow-Day test, P > 0.33). OR with 95% CI in parentheses was adjusted for ethnicity, hyperuricaemia and other covariates using a logistic regression model.
For aborigines, those aforementioned three most important SNPs were within 10.2 k of each other (113571846-113582071 base pair), having an r2 > 0.74. Model comparison test was significant for at-risk haplotype [CGG] versus reference haplotype [TAC] (OR = 1.43, 95% CI = 1.22–1.67) and the likelihood ratio test (χ2 = 20.5, degree of freedom [df] = 1, P = 5.86 × 10−6). For Han Chinese people, of those three SNPs generated, four were common haplotypes (rs11726117/rs231247, r2 = 0.88; rs11726117 or rs231247/rs231253, r2 > 0.41). Model comparison test was significant for at-risk haplotype [CGG] versus reference haplotype [TAC] (OR = 2.05, 95% CI = 1.29–3.26) and the likelihood ratio test (χ2 = 28.9, df = 3, P = 2.34 × 10−6). SNPs of rs11726117, rs231247 and rs231253 showed homogeneity in associating with gout (Ethnic Breslow-Day test, P > 0.33). Therefore, pooled analysis of the three SNPs by Cochran-Mantel-Haenszel assuming an additive model was used to test for associations (Table 2). When individuals were combined into a pooled analysis, each increasing copy of the [C] allele at rs11726117, [G] allele at rs231247 and [G] allele at rs231253 conferred a significantly elevated OR for gout of 1.51 (P = 2.89×10−8), 1.52 (P = 2.05×10−8) and 1.43 (P = 1.53 × 10−6) after controlling for ethnicity only. The above results were similar even after controlling for ethnicity and hyperuricaemia and other covariates concerning the risk estimate for rs11726117 [C] (OR = 1.53, 95% CI = 1.30–1.81), rs231247 [G] (OR = 1.55, 95% CI = 1.31–1.83) and rs231253 (OR = 1.42, 95% CI = 1.21–1.68). From a 2-df model, the results were consistent with an underlying additive model. For instance, compared with reference allele homozygotes, the OR for heterozygotes in the full multivariable model was 1.68 (95% CI = 1.19–2.36) for rs11726117, and the risk allele homozygotes had the highest risk (OR = 2.43, 95% CI = 1.70–3.47), again remaining consistent with an additive genetic model.
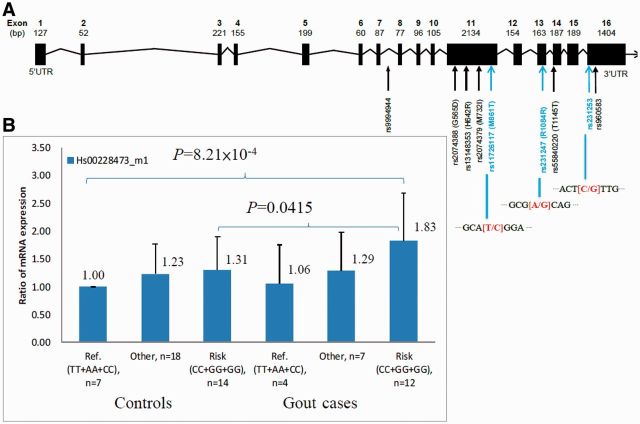
To quantitate the expression differences in genotype carriers of these three SNPs, a real-time PCR of ALPK1 mRNA was performed on peripheral blood leukocytes of 62 aborigines (Figure 2, and Supplementary Figure 2 available as supplementary data at IJE online). The rs11726117 [CC] and [CT] genotypes when compared with AA genotype were found to be 1.22 to 1.49 significantly higher (P-trend = 0.0007). Composite genotype analyses of the rs11726117 [CC], rs231247 [GG] and rs231253 [GG] showed association with gout. ALPK1 transcribed significantly higher in gout cases, specifically the homozygous of linked three SNPs [CC+GG+GG] versus wild-type [TT+AA+CC] (OR = 1.83, 95% CI = 1.24–2.68; P = 8.21 × 10−4) with post hoc compared Dunnett's test. At-risk homozygous [CC+GG+GG] was also significantly more highly expressed in cases than its composite genotype in controls (OR = 1.40, 95% CI = 1.01–1.93, P = 4.15 × 10−2).
Figure 2.
Genomic structure of ALPK1 with nine linked loci and mRNA expression in gout aboriginal cohort (A) Exons are indicated by black boxes. Most significant three loci are indicated by blue text and alleles in red text. SNP rs9994944 is located in intron 7-8, followed by four nonsynonymous variants in exon 11, two synonymous variants in exon 13 and exon 14 located within alpha-kinase domain, and rs231253 and rs960583 located in three prime untranslated region. SNP rs11726117 [C], rs231247 [G] and rs231253 [G] have most significant P values from association study. Therefore, three SNPs were entered into real-time PCR validation. (B) Real-time PCR result of ALPK1 mRNA (Hs00228473_m1) on peripheral blood leukocytes of 62 gout aborigines demonstrated elevated ALPK1 mRNA (at-risk [CC+GG+GG] in cases compared with wild-types [TT+AA+CC] in controls) with post hoc compared Dunnett's test (OR = 1.83, 95% CI = 1.24–2.68; P = 8.21 × 10−4). Error bar indicates 95% confidence interval; PCR, polymerase chain reaction
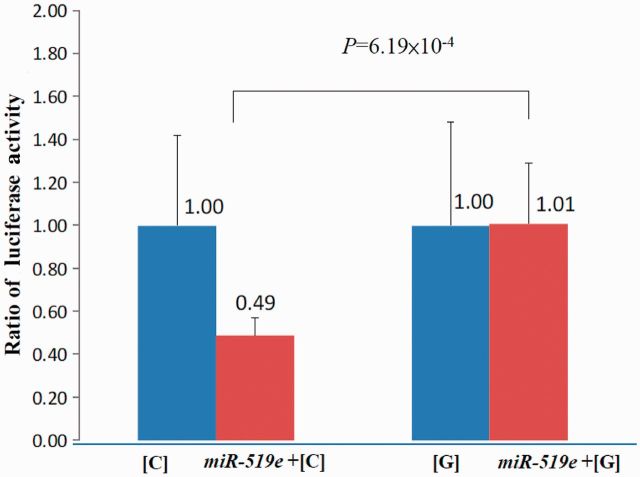
It was found that a microRNA, hsa-miR-519e (chr19q13.42), shared binding site complementarily with 3’UTR rs231253, and that gout-associated risk [G]-allele creates a slight kink in the mRNA structure with respect to [C]-allele thus becoming less negative in free energy state, denoting a less stable hybridization [MFE change: −25.4 kcal/mol to −22.2 kcal/mol, (Supplementary Figure 3 available as supplementary data at IJE online). There was a 2-fold increase in luciferase activity of rs231253 on the construct carrying [G] allele following an overexpression of the interacting hsa-miR-519e (P = 6.19 × 10−4; Figure 3].
Figure 3.
Higher expression from miRNA hybridized with gout-associated ALPK1 rs231253 [G] allele. Approximately 2-fold differential luciferase activity (P = 6.19 × 10−4) between pGL3-SNP constructs of gout-associated rs231253 [G] allele and wild-type [C] allele cotransfected with hsa-miR-519e that shared complementary binding. Values represent mean ± SD of four independent experiments performed in triplicate. SD, standard deviation
Discussion
Region-wide association fine-map of 4q25 showed the ALPK1 gene to be most associated with gout in Taiwanese aboriginals and Han and closest to newly revised linkage signal at 117cM (LOD = 5.2) in this study. Particularly, three ALPK1 loci of the nonsynonymous rs11726117 M861T [C], synonymous rs231247 [G] and 3’UTR rs231253 [G] were most associated with gout risk. These could cause alterations to the normal physiological function of the ALPK1 of the alpha-kinase family although the functions of ALPK1 are less known. For example, the missense rs11726117 M861T is a threonine substitution located forward of the catalytic domain, and potentially a new phosphorylation site for ALPK1 with preferentially phosphorylate threonine residues.21,22 Synonymous rs231247 (R1084R) codes for an amino acid in the alpha catalytic domain, located adjacent to a conserved invariant glutamine, which structurally maintains the alpha-helix C of subdomain III and near a polar residue that binds H2O to help orientate the ATP γ-phosphate in the binding groove.21,23 We identified 3’UTR rs231253 [G] showing association with risk of gout (pooled analysis, OR = 1.42). Although the rs231253 [G] is marginal associated with gout (OR = 1.36) in Han people, we cannot rule out the possibility that rs231253 [G] contributes to the gout risk. One replication study is needed to clarify the genetic effects in the future. With functional testing, we found that the gout cases, carrying risk rs231253 [G], showed 83% increased ALPK1 mRNA expression (P = 8.21 × 10−4), and in vitro experimentation for verification of the rs231253 [G] showed increased expression in the luciferase assay (P = 6.19 × 10−4). With bioinformatic prediction, SNP rs231253 [G] presented a less stable hybridization of hsa-miR-519e/target duplex, as suggesting disruption in gene silencing (Supplementary Figure 3, available as supplementary data at IJE online).
Uric acid, generated by degradation of purines, is further cleared by degradation to allantoin catalyzed by hepatic uricase, or by excretion largely in the kidneys. Humans, unlike most mammals, lack uricase, and therefore rely on renal excretion as a critical determinant of systemic urate levels.2 Genome-wide association studies have identified the common polymorphisms in several genes involved in the renal urate transport that are associated with hyperuricaemia and gout, including SLC2A9, SCL22A12, SCL22A11, SLC17A3, ABCG2 and SLC17A1 genes.24,25 ABCG2 rs2231142 Q141K has been suggested as a major causative gene variant for gout.25 However, renal urate transporter gene variants explain only about 5.3% of the total variation of serum urate concentrations in people of Caucasian ancestry.24,25 Therefore, many other genes with partial effects and accompanied by high urate levels contribute to formation of MSU crystals and the clinical presentation of acute gout arthritis and chronic tophaceous disease.
We found in vitro that the ALPK1 and MSU may synergistically induce ERK1/2 and p38 phosphorylation, then regulate cytokine expressions through the activation of NF-κB pathway.16 The activated NF-κB complexes are known to translocate into the nucleus and bind to NF-κB DNA binding motifs to trigger transcription of genes that are critical to inflammation, such as cytokines, chemokines and cell adhesion molecules.1,26 The ENCODE ‘Yale TFBS’ track has revealed the entire 10 kb of ALPK1 promoter region to contain several signal peaks of NF-κB transcription factor binding motifs suggested in an overall positive feedback (Supplementary Figure 4, available as supplementary data at IJE online).
In conclusion, the ethnic replication of association findings between ALPK1 loci and gout has led us to consider ALPK1 as a significant new candidate susceptible gene for gout. Uniquely, a 3’UTR rs231253 variant on ALPK1 has been found to disrupt miRNA gene regulation and contribute to a higher mRNA expression among the patients affected.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This work was supported by the National Health Research Institutes (grant number NHRI-98A1-PDCO-0307101) and the National Science Council (grant numbers NSC97-2314-B-039-007-MY3 and NSC99-2628-B-037-039-MY3).
Author contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr Ko had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and design and final approval of the version: Ying-Chin Ko; drafting the article: Albert Min-Shan Ko, Hung-Pin Tu; acquisition of samples with information: Hung-Pin Tu, Shang-Lun Chiang, Shun-Jen Chang; performance of experiments: Tze-Tze Liu, Chung-Yee You, Jan-Gowth Chang; analysis and interpretation of the data: Ying-Chin Ko, Albert Min-Shan Ko, Jan-Gowth Chang, Hung-Pin Tu, Shun-Jen Chang, Allen Min-Jen Ko, Chien-Hung Lee, Chi-Pi Lee, Chung-Ming Chang and Shih-Feng Tsai; miRNA bioinformatics: Albert Min-Shan Ko, Yu-Fan Liu.
Supplementary Material
Acknowledgements
The authors thank the medical staff of the Jianshih and Wufong Township Health Station (Dr Xian-Min Qiu and nurses), Kaohsiung Medical University Hospital (Dr Gau-Tyan Lin, Dr Tsan-Teng Ou (rheumatologist) and Dr Su-Shin Lee) and Kaohsiung Chang-Gung Hospital (clinical rheumatologist, Dr Chung-Jen Chen and Han-Ming Lai) for ascertainment of clinical phenotypes and data collection. Some results of this paper were obtained using the software package S.A.G.E., supported by a U.S. Public Health Service Resource Grant (RR03655) from the National Centre for Research Resources.
Conflict of interest: None declared.
KEY MESSAGES
ALPK1 is a gout-associated gene in Taiwan aborigines and with replication in Han Chinese people.
ALPK1 mRNA transcription is transcribed more highly among gout cases with variants of this gene. Luciferase activity assay showed in vitro that the 3’UTR variant affected the complementary binding motif to a microRNA, leading to this increased expression.
ALPK1 recognizes phosphorylation sites related to regulation of cytokine expression, but the molecular mechanism and function need further study.
References
- 1.Choi HK, Mount DB, Reginato AM. Pathogenesis of gout. Ann Intern Med. 2005;143:499–516. doi: 10.7326/0003-4819-143-7-200510040-00009. [DOI] [PubMed] [Google Scholar]
- 2.Terkeltaub R. Update on gout: new therapeutic strategies and options. Nat Rev Rheumatol. 2010;6:30–38. doi: 10.1038/nrrheum.2009.236. [DOI] [PubMed] [Google Scholar]
- 3.Richette P, Bardin T. Gout. Lancet. 2010;375:318–28. doi: 10.1016/S0140-6736(09)60883-7. [DOI] [PubMed] [Google Scholar]
- 4.Hediger MA, Johnson RJ, Miyazaki H, Endou H. Molecular physiology of urate transport. Physiology (Bethesda) 2005;20:125–33. doi: 10.1152/physiol.00039.2004. [DOI] [PubMed] [Google Scholar]
- 5.Merriman TR, Dalbeth N. The genetic basis of hyperuricaemia and gout. Joint Bone Spine. 2011;78:35–40. doi: 10.1016/j.jbspin.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 6.Chang HY, Pan WH, Yeh WT, Tsai KS. Hyperuricemia and gout in Taiwan: results from the Nutritional and Health Survey in Taiwan (1993-96) J Rheumatol. 2001;28:1640–46. [PubMed] [Google Scholar]
- 7.Chang SJ, Ko YC, Wang TN, Chang FT, Cinkotai FF, Chen CJ. High prevalence of gout and related risk factors in Taiwan's Aborigines. J Rheumatol. 1997;24:1364–69. [PubMed] [Google Scholar]
- 8.Chang SJ, Chen CJ, Hung HP, Ou TT, Ko YC. Community-based study in Taiwan aborigines concerning renal dysfunction in gout patients. Scand J Rheumatol. 2004;33:233–38. doi: 10.1080/03009740310004919. [DOI] [PubMed] [Google Scholar]
- 9.Lee GA, Crawford GW, Liu L, Chen X. Plants and people from the Early Neolithic to Shang periods in North China. Proc Natl Acad Sci U S A. 2007;104:1087–92. doi: 10.1073/pnas.0609763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu TF. Diversity of clinical features in gouty arthritis. Semin Arthritis Rheum. 1984;13:360–68. doi: 10.1016/0049-0172(84)90016-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen SY, Chen CL, Shen ML, Kamatani N. Trends in the manifestations of gout in Taiwan. Rheumatology (Oxford) 2003;42:1529–33. doi: 10.1093/rheumatology/keg422. [DOI] [PubMed] [Google Scholar]
- 12.Cheng LS, Chiang SL, Tu HP, et al. Genomewide scan for gout in taiwanese aborigines reveals linkage to chromosome 4q25. Am J Hum Genet. 2004;75:498–503. doi: 10.1086/423429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu HP, Ko AM, Wang SJ, et al. Monoamine oxidase A gene polymorphisms and enzyme activity associated with risk of gout in Taiwan aborigines. Hum Genet. 2010;127:223–29. doi: 10.1007/s00439-009-0765-z. [DOI] [PubMed] [Google Scholar]
- 14.Tu HP, Chen CJ, Tovosia S, et al. Associations of a non-synonymous variant in SLC2A9 with gouty arthritis and uric acid levels in Han Chinese subjects and Solomon Islanders. Ann Rheum Dis. 2010;69:887–90. doi: 10.1136/ard.2009.113357. [DOI] [PubMed] [Google Scholar]
- 15.Wallace SL, Robinson H, Masi AT, Decker JL, McCarty DJ, Yu TF. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977;20:895–900. doi: 10.1002/art.1780200320. [DOI] [PubMed] [Google Scholar]
- 16.Wang SJ, Tu HP, Ko AM, et al. Lymphocyte alpha-kinase is a gout-susceptible gene involved in monosodium urate monohydrate-induced inflammatory responses. J Mol Med (Berl) 2011;89:1241–51. doi: 10.1007/s00109-011-0796-5. [DOI] [PubMed] [Google Scholar]
- 17.Whittemore AS, Tu IP. Simple, robust linkage tests for affected sibs. Am J Hum Genet. 1998;62:1228–42. doi: 10.1086/301820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–65. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–54. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drennan D, Ryazanov AG. Alpha-kinases: analysis of the family and comparison with conventional protein kinases. Prog Biophys Mol Biol. 2004;85:1–32. doi: 10.1016/S0079-6107(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 22.Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: a new class of protein kinases with a novel catalytic domain. Curr Biol. 1999;9:R43–45. doi: 10.1016/s0960-9822(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 23.Middelbeek J, Clark K, Venselaar H, Huynen MA, van Leeuwen FN. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. 2010;67:875–90. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dehghan A, Kottgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–61. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pare G, Ridker PM, Rose L, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.