Abstract
Captive breeding and rearing are central elements in conservation, management, and recovery planning for many endangered species including Rio Grande Silvery Minnow, a North American freshwater cyprinid. Traditionally, the sole purpose of hatcheries was to produce as many fish as feasible for stocking and harvest. Production quotas are also an important consideration in hatchery programs for endangered species, but they must also maintain and maximize genetic diversity of fish produced through implementation of best breeding practices. Here, we assessed genetic outcomes and measures of productivity (number of eggs and larval viability) for three replicates of three mating designs that are used for this small, pelagic-spawning fish. These were 1) monogamous mating, 2) hormone-induced communal spawning, and 3) environmentally cued communal spawning. A total of 180 broodstock and 450 progeny were genotyped. Genetic diversity and egg productivity did not differ significantly among spawning designs (H e: F = 0.52, P = 0.67; H o: F = 0.12, P = 0.89; number of eggs: F = 3.59, P = 0.09), and there was evidence for variance in reproductive success among individuals in all three designs. Allelic richness declined from the broodstock to progeny generation in all breeding designs. There was no significant difference in the genetic effective size (regardless of the method used) among designs. Significantly more viable eggs were produced in environmentally cued communal spawn compared to the alternative strategies (F = 5.72, P = 0.04), but this strategy is the most difficult to implement.
Key words: captive spawning, effective population size, genetic diversity, Rio Grande Silvery Minnow
Hatcheries are now commonly used as conservation and management tools for threatened and endangered fish (e.g., Flagg et al. 2004; O’Reilly and Doyle 2007; Fraser 2008). Most importantly, hatcheries can provide vital supplementary stock to bolster census sizes of wild populations and ostensibly enhance species’ recovery. Supplemental stocking is necessary when the natural environment lacks sufficient resources to fully support reproduction, recruitment, and growth of wild fish at self-sustaining densities (e.g., US Fish and Wildlife Service 2009), or when natural or anthropogenic disturbance extirpates wild fish completely from an otherwise suitable natural habitat. Hatchery stocks are somewhat immune to vagaries and fluctuations of the natural environment and so provide a measure of insurance against extinction from catastrophic disturbance in the wild for critically endangered fish. On the down side, supplemental stocking from hatcheries can severely decrease genetic diversity of wild stocks under certain circumstances (Ryman and Laikre 1991; Ryman 1994; Tessier et al. 1997; Christie et al. 2012; but see Gow et al. 2011). Moreover, fish produced in hatcheries can suffer reduced survival and reproduction compared to wild fish (e.g., Dowling et al. 1996; Araki et al. 2007; Thériault et al. 2011) because of hatchery-imposed (domestication) selection and/or relaxation of natural selection pressures in the wild (e.g., Bryant and Reed 1999; Waples 1999). Concerns about hatcheries as a recovery tool thus revolve around their potential to diminish chances of long-term persistence by negatively impacting genetic diversity and mean fitness of the species.
To mitigate negative genetic effects, conservation hatchery goals and benchmarks have evolved considerably from initial focus on simply meeting production quotas to development of breeding protocols that maximize genetic diversity in progeny (e.g., Ballou 1984; Koljonen et al. 2002; Russello and Amato 2004). Some hatchery programs have even adopted measures such as naturalized spawning and rearing so as to attempt to ameliorate domestication selection and other selective effects (e.g., Gruenthal et al. 2012). Many of these developments are motivated by genetic theory that can, in principle, be universally applied across species (Hedrick 1992). However, breeding and rearing protocols must also be based on the life history and reproductive biology of the focal species in order to be successful. Most practical knowledge of endangered fish breeding and rearing is based on extensive data that have been gathered for trout and salmon (family Salmonidae) for well over a century. Comparatively little data are available for other fish, including members of the family Cyprinidae. Fish in this group have a broad range of life histories and reproductive features. This family is the most evolutionarily diverse and among the most imperiled groups of freshwater fish in North America (Jelks et al. 2008). There are captive breeding programs for some of these species, including the Rio Grande Silvery Minnow, Hybognathus amarus.
Unlike salmonids, most cyprinids, including Rio Grande Silvery Minnow, mature rapidly at small body size. Small body size (<100mm total length) imposes limits to the kinds of captive breeding protocols that can be effectively implemented in the hatchery. For example, stripping males and female of gametes is very difficult to do successfully (Moore 1944; Bottrell et al. 1964; Platania and Altenbach 1998; but see Fisch et al. 2013), which limits the potential to employ designs that are frequently implemented for larger-bodied species (e.g., a complete factorial design). On the other hand, the reproductive biology of Rio Grande Silvery Minnow permits effective communal spawning in the hatchery, but it is unknown whether this breeding strategy leads to unacceptably high levels of variance in reproductive success among spawners that could limit diversity of progeny (e.g., Frost et al. 2006; Herlin et al. 2008). This underscores the premise that knowledge of life history and reproductive biology of the focal species is essential to choose hatchery practices that simultaneously meet production and conservation goals.
The focal species in this study, Rio Grande Silvery Minnow, was listed as endangered in 1994 due to dramatic declines in abundance and extirpation from the majority of its former range (Bestgen and Platania 1991; US Department of the Interior 1994). Historically, the Rio Grande Silvery Minnow was found in the Pecos River in New Mexico and Texas, the Rio Grande in New Mexico and Texas, and the Federal Republic of Mexico. The species is now restricted to a ~280-km stretch of the Rio Grande mainstem from downstream of Cochiti Dam to the headwaters of Elephant Butte reservoir, New Mexico. Six years after species’ listing in the Federal Register, refuge populations and an experimental captive propagation was established. Beginning in 2003, a full-scale supportive breeding program was implemented that, to date, has released over a million fish to the Middle Rio Grande in New Mexico (US Fish and Wildlife Service 2009). In 2008, the species was reintroduced into the Rio Grande at Big Bend National Park with progeny from captive stocks. Several hundred thousand fish have been released annually since then.
When Rio Grande Silvery Minnow spawn, both sexes broadcast gametes into the water column. Spawning is synchronous and apparently cued by high-flow events associated with spring snowmelt runoff and rainstorm events, which increase turbidity in the river. High turbidity (which results in low visibility) makes direct observations of breeding behavior in the wild difficult; however, laboratory observations have been conducted (Platania and Altenbach 1998). Briefly, breeding for this species involves pursuit of a female by a male. The male nudges her abdominal region and, when she is ready to spawn, the male wraps himself around the female’s midsection. Eggs and sperm are then simultaneously released. The non-adhesive, semi-buoyant eggs absorb water and expand. Multiple spawning episodes with an interval of at least 10 minutes have been reported (Platania and Altenbach 1998). Based on the knowledge of breeding behavior in other broadcast spawners in which temporal and spatial aggregations of adults form during spawning, it is likely that groups of Rio Grande Silvery Minnow adults come together in the wild during these high-flow events (Osborne et al. 2006). Aggregation facilitates concentration of gametes in the water column and hence increases fertilization rates (Pennington 1985; Oliver and Babcock 1992; Hedgecock 1994). In captivity, Rio Grande Silvery Minnow have been induced to spawn by increasing both flow and turbidity, rather than by injection of carp pituitary extract (CPE).
The aim of this study was to evaluate levels of genetic diversity of progeny from three breeding strategies used at the Albuquerque Biopark including 1) hormone-induced communal spawning, 2) environmentally cued (flow, temperature, and turbidity) communal spawning, and 3) pairwise (monogamous) mating. Levels of genetic diversity and productivity (number of eggs and viability) among breeding designs were compared to help establish the most practical (in terms of the number of crosses) and beneficial hatchery practices that could simultaneously meet production goals and maintain target levels of genetic diversity in progeny.
Materials and Methods
Breeding trials
Broodstock for this study was collected as young-of-the-year (YOY) from the Isleta reach of the Middle Rio Grande, New Mexico, in 2005 as part of ongoing recovery efforts for the species (details regarding the study area are provided in Osborne et al. 2012). These fish were reared to adulthood at the Albuquerque Biological Park. Ten males and ten females were selected at random from this pool of potential breeders for each of three trials and three spawning treatments per trial, for a total of 180 adults used in the entire experiment (Figure 1). Each trial consisted of three breeding strategies: monogamous mating (MM), hormone-induced communal spawn (HICS), and environmentally cued communal spawn (ECCS), and three replicates of each strategy were conducted. Breeding strategies within a trial were conducted on the same day (Trial 1: 27 May 2006; Trial 2: 31 May 2006; Trial 3: 2 June 2006) at the Albuquerque Biological Park. For each MM and HICS, fish were anesthetized in a bath of MS-222 (0.1g per 3L of water). Anesthetized fish received an intraperitoneal injection of CPE (0.01g/5mL) at the base of one pelvic fin using a 1-cc insulin syringe. Once injected, the fish were returned to a freshwater bath to recover from the anesthesia and were then placed into spawning aquaria. Percent viability of each trial and replicate was estimated by randomly netting a total of 100 eggs from the hatching tank 24 hours post-fertilization. These were placed in a petri dish and the number of fertilized and unfertilized eggs determined based on visual inspection of the eggs. The fertilized eggs have a clear chorion and a distinct white blastula, whereas unfertilized eggs or unviable eggs are cloudy throughout and have no distinct nucleus.
Figure 1.
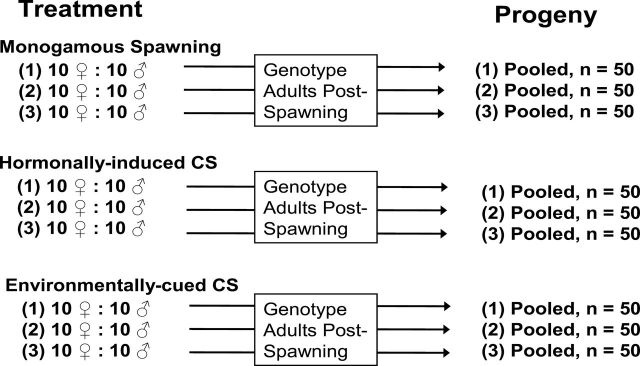
Experimental design and sampling strategy for broodstock and progeny across three breeding designs: MM, HICS, and ECCS.
MM
For each MM trial, 10 pairs of Rio Grande Silvery Minnow were placed in separate 378.5-L rectangular aquaria after injection with CPE. Adults and eggs were removed from the spawning aquaria after spawning was complete. The number of eggs was estimated volumetrically (Hunter et al. 1985) and were then moved to a 378.5-L hatching aquaria.
HICS
For each HICS trial, 10 males and 10 females were injected with CPE and placed in 4269-L circular tanks. All other methods were the same as the MM. MM and HICS trials were conducted indoors.
ECCS
For each trial, 10 males and 10 females were moved from a holding system to an 847-cm diameter outdoor raceway with a depth of 30cm, an average width of 58cm, and a volume of 5440L. Initial turbidity was 7 NTU (nephelometric turbidity units). To induce spawning, this was increased to 231 NTU. Over the course of the trial, turbidity declined to 120 NTU and averaged 155 NTU. Average flow in the raceway was 0.339 meters per second. Eggs were collected using a specially constructed screened collection box (8″ × 8″ × 36″) that was designed to permit eggs but exclude fish.
Sampling and Molecular Methods
Parents for each trial and replicate were fin-clipped after spawning. For MM, progeny of 10 pairs were pooled prior to sampling. Fifty larval fish from each treatment and replicate were collected approximately 1 week after hatching. All samples were stored in 95% ethanol. DNA was extracted from air-dried fin clips using proteinase-K digestion and organic extraction methods (Hillis et al. 1996). Each whole larval fish was placed in 1.5mL Eppendorf tube with 50µL of sterile water and pulverized using a plastic pestle. One in 10µL dilutions was made using this mixture. This extract was used as a template for PCR. Individuals were genotyped at nine microsatellite loci using the methods described in Osborne et al. (2012). Electrophoresis was conducted on an ABI377 automated sequencer and fragments sized using Genscan software.
Data Analysis—Genetic Diversity
Microsatellite data were checked for errors using Microsatellite Toolkit (add-in for Microsoft Excel, written by S. Park, available at http://animalgenomics.ucd.ie/sdepark/ms-toolkit/). Nei’s unbiased genetic diversity (H e) (Nei 1987), observed heterozygosity (H o), and allele frequencies were obtained using this program. The computer program MicroChecker (van Oosterhout et al. 2004) was used to examine the data for scoring errors due to stuttering, large allele dropout, and the presence of null alleles. We used the EM algorithm to estimate the frequency of null alleles at each locus as implemented in GENEPOP (Dempster et al. 1977; Hartl and Clark 1989; Kalinowski and Taper 2006). For each microsatellite locus and population, allelic richness (A R), total number of alleles, and inbreeding coefficients (F IS) were obtained using FSTAT version 2.9.3.1 (Goudet 1995). Global tests for linkage disequilibrium (non-random association of loci) were conducted for all pairs of loci using this program. Allelic richness was calculated using the methods described in Petit et al. (1998). The computer package ARLEQUIN (Schneider et al. 2000) was used to assess whether there were significant departures from Hardy–Weinberg equilibrium using the procedure of Guo and Thompson (1992). Bonferroni (Rice 1989) correction was applied to account for multiple simultaneous tests. We compared measures of genetic diversity (including genetic effective size) and productivity (number of eggs and viability) for the broodstock and progeny of each design using a one-way ANOVA as implemented in SigmaPlot version 11.0. Two-tailed t-tests were conducted to ascertain differences in diversity metrics between broodstock and progeny within each breeding design. Diversity metrics were also recalculated with the removal of two loci with consistent departures from HWE and relatively high frequencies of null alleles (Supplementary Table S1). Likewise, comparisons of diversity metrics between broodstock and progeny (via t-test as above) and among breeding designs (AMOVA) were also conducted after dropping these loci.
Parentage/Sibship Analysis
The computer program COLONY version 2.0 (Jones and Wang 2009) was employed to jointly infer parentage and sibship for each replicate and breeding trial. COLONY implements full-pedigree likelihood methods and considers the likelihood over the entire pedigree configuration, rather than for pairs of individuals (dyads) (Jones and Wang 2009). Three runs were conducted for each replicate of each mating design to determine the number of mates per individual, number of offspring per parent, and to estimate effective size (N e(SA)) (Wang 2009). For this analysis, input parameters were: both sexes polygamous (for HICS and ECCS) or monogamous (MM), no inbreeding, marker error rate of 0.05 (rather than the default 0.01), and full-likelihood analysis. No sibship prior was used, and allele frequencies were updated by accounting for inferred relationships. We determined that short runs were sufficient to converge on the most likely ML configuration by conducting replicate three runs but specifying different random number seeds. Replicates produced almost identical results, indicating convergence even with short runs (Wang 2009). The best configuration (as indicated by the highest likelihood score) was used to determine the number of offspring produced by broodstock and the number of mates. Using the COLONY results, we examined whether there was a correlation between the number of mates and reproductive success using ordinary least-squares linear regression conducted in SigmaPlot version 11.0 (Systat Software).
Genetic Effective Size
Genetic effective size was estimated using two methods: 1) the temporal method (N eV) and 2) sibship assignment method (N e(SA)) (Wang 2009). N eV and 95% confidence intervals (CIs) were estimated from temporal changes in microsatellite allele frequencies at nine microsatellite loci across year classes (parental to progeny generation), assuming sampling plan I according to Waples (1989; Equation 12). Confidence intervals were calculated using Equation 15 in Waples (1989). We also used the reduced microsatellite dataset (described above); however, this resulted in estimates of infinity in all cases. Hence we only report the estimates from the full dataset. Previously, we have shown that excluding loci that departed consistently from HWE when estimating N e has little effect on the estimates (Osborne et al. 2012). N e(SA) and 95% CIs were estimated using COLONY version 2.0 (Jones and Wang 2009), specifying a polygamous mating strategy.
Pedigree-based estimates of N e were also calculated using the estimates of family size obtained from the data using the program COLONY (Wang 2009). Hence N e was estimated, where N e = (Nk − 2)/[k + (V k/k) − 1] with N = total number of parents, k = average family size, and V k = variance of family size (Crow and Kimura 1970; Herbinger et al. 2006).
Results
Genetic Diversity
Microsatellite genotyping was conducted for 180 parents and 450 progeny across nine loci and three breeding designs. There were 73 departures from Hardy–Weinberg expectations from 162 comparisons (44 remained significant following Bonferroni correction). Twelve departures involved parental samples with the remainder occurring in the progeny. The loci involved were Ppro118, Ca6, Ca8, Lco1, Lco7, and Lco8, with two loci (Lco7 and Lco8) explaining more than half of the violations. Analysis with MicroChecker suggested that null alleles may be present in 17 instances involving broodstock samples and 35 instances involving progeny samples. Null allele frequencies are reported in Supplementary Table S1. There were 10 cases of linkage disequilibrium that were significant after Bonferroni correction for multiple comparisons. There were 19 instances of missing data for some microsatellites among broodstock despite repeated attempts at amplification. Seven microsatellites were involved: Ppro118 (n = 1), Ca6 (n = 1), Lco1 (n = 4), Lco6 (n = 3), Lco7 (n = 2), Lco8 (n = 6), and Ppro126 (n = 2). On average, 10–13% of progeny exhibited missing data across replicates of each spawning design.
There was considerable variation in the number of eggs and viability across replicates and breeding designs (Table 1). One-way ANOVA indicated that the number of eggs produced (P = 0.094) did not differ significantly; however, percent viability was significantly different (P = 0.041) across designs, with the monogamous design having lower viability than either HICS (P = 0.048) or ECCS (P = 0.017). Viability did not differ between the communal designs (0.479). Measures of genetic diversity for broodstock and progeny did not differ significantly between designs (Table 2). In all designs there was a significant decline in A R from the parental to progeny generation and H o also declined in the HICS and ECCS designs (Table 2).
Table 1.
Details of spawning strategy (MM, HICS, and ECCS) and the approximate number of eggs and viability per spawning event
| Treatment | Replicate | Estimate of number of eggs | Viability |
|---|---|---|---|
| MM | 1 | 5,310 | 65% |
| 2 | 7,110 | 20% | |
| 3 | 8,580 | 61% | |
| HICS | 1 | 9,000 | 85% |
| 2 | 7,500 | 99% | |
| 3 | 6,750 | 67% | |
| ECCS | 1 | 450 | 90% |
| 2 | 6,660 | 99% | |
| 3 | 3,000 | 94% |
Table 2.
Average diversity measures (Nei’s unbiased gene diversity [H e], heterozygosity [H o], allelic richness [A R], and average inbreeding coefficient [F IS]) across three replicates of each breeding design, MM, HICS, and ECCS, for all loci and with Lco7 and Lco8 excluded. Total sample size (n) is also given for each treatment (broodstock: 3 replicates × 20 broodstock; progeny: 3 replicates × 50 progeny). P values for two-sided t-tests are given for comparisons between broodstock and progeny within treatments and for one-way ANOVA among breeding strategies
| n | N loci | H e | H o | A R | F IS | |
|---|---|---|---|---|---|---|
| Broodstock—MM | 60 | 9 | 0.843 | 0.760 | 11.648 | 0.099 |
| 7 | 0.840 | 0.797 | 12.464 | 0.051 | ||
| Progeny—MM | 150 | 9 | 0.773 | 0.677 | 9.068 | 0.126 |
| 7 | 0.767 | 0.699 | 9.695 | 0.089 | ||
| P value | 9 | 0.05 | 0.112 | <0.001 | 0.598 | |
| 7 | 0.057 | 0.123 | 0.031 | 0.385 | ||
| Broodstock—HICS | 60 | 9 | 0.834 | 0.744 | 11.596 | 0.111 |
| 7 | 0.832 | 0.778 | 12.078 | 0.065 | ||
| Progeny—HICS | 150 | 9 | 0.783 | 0.663 | 9.266 | 0.155 |
| 7 | 0.770 | 0.704 | 9.553 | 0.086 | ||
| P value | 9 | 0.071 | 0.010 | 0.007 | 0.072 | |
| 7 | 0.046 | 0.100 | <0.001 | 0.327 | ||
| Broodstock—ECCS | 60 | 9 | 0.854 | 0.764 | 12.081 | 0.108 |
| 7 | 0.835 | 0.809 | 12.257 | 0.052 | ||
| Progeny—ECCS | 150 | 9 | 0.798 | 0.679 | 9.389 | 0.150 |
| 7 | 0.792 | 0.721 | 9.436 | 0.089 | ||
| P value | 9 | 0.031 | 0.028 | 0.027 | 0.229 | |
| 7 | 0.034 | 0.020 | <0.001 | 0.494 | ||
| Broodstock—P values | 9 | 0.789 | 0.856 | 0.970 | 0.919 | |
| 7 | 0.524 | 0.684 | 0.796 | 0.928 | ||
| Progeny—P values | 9 | 0.891 | 0.619 | 0.801 | 0.493 | |
| 7 | 0.619 | 0.852 | 0.885 | 0.994 |
Parentage/Sibship Analysis
For HICS the average number of mates per individual was 3.358 (σ = 1.570). This value was marginally lower for ECCS (2.833; σ = 1.124). From the sample of 50 eggs, the average number of offspring per individual was 6.140 (σ = 3.371) for MM, 6.583 (σ = 3.870) for HICS, and 6.619 (σ = 3.230) for ECCS (Figure 2). Inclusion probabilities for full-sib families were high in almost all cases (except three instances where probabilities were less than 80% across all replicates and designs). For parentage assignments, probabilities were typically high in communal spawning trails (HICS: one case of a probability less than 80%; ECCS: 13 progeny with probabilities less than 80% for determination of parentage, the majority involving a single broodstock individual, and all probabilities greatly exceeded the alternatives). Probabilities of maternity and paternity for the monogamous design were lower, but inferred relationships were fairly consistent across multiple runs (MM1: 13 progeny could only be unambiguously assigned one parent; MM2: probabilities of parentage assignment for 15 progeny were <70% involving four broodstock; MM3: 18 progeny accuracy of parentage assignment was <70% involving four broodstock). There was a positive relationship between the number of mates and the number of offspring produced (HICS: r 2 = 0.676, P < 0.001; ECCS: r 2 = 0.454, P < 0.001).
Figure 2.

Number of offspring per parent and number of mates per parent across three breeding designs: (a) MM, (b) HICS, and (c) ECCS.
Effective Population Size
Variance effective size (N eV) was estimated between the parent and progeny generations. Effective size measured in this way gives an estimate of N e for the parental population (Waples 2005). For MM, N eV (harmonic mean across replicates) was 15; for HICS, average N eV was 23; and for ECCS, N eV was 19 (Table 3). Estimates of N e obtained using sibship assignment ranged from 13 to 17. The harmonic mean of effective size estimated using the pedigree-based estimate was 17 for all breeding designs. One-way ANOVA revealed that estimates of N e did not differ significantly among mating designs (pedigree-based N e: P = 0.983; N eV: P = 0.295; N e(SA): P = 0.264).
Table 3.
Average estimates of demographic N e, N eV and N e(SA) across replicates and breeding designs: MM, HICS, and ECCS. k is the mean family size, and V k is the variance in family size based on results from COLONY. P-values for ANOVA testing whether effective size differs between breeding designs are also given
| Spawning strategy | k | V k | N e | N eV | 95% CIs | 95% CIs | N e(SA) | −95% CIs | +95% CIs |
|---|---|---|---|---|---|---|---|---|---|
| MM 1 | 6.91 | 8.91 | 18.9 | 14.7 | 7.2 | 20.4 | 14 | 7 | 30 |
| MM 2 | 6.27 | 20.20 | 14.5 | 14.9 | 7.3 | 20.5 | 12 | 6 | 26 |
| MM 3 | 5.92 | 4.92 | 20.2 | 16.9 | 8.8 | 22.4 | 16 | 8 | 34 |
| Harmonic mean | 17.5 | 15.4 | 7.7 | 21.1 | 13.8 | 6.9 | 29.6 | ||
| HICS 1 | 5 | 5.05 | 19.6 | 20.8 | 12.1 | 25.6 | 13 | 7 | 30 |
| HICS 2 | 5.50 | 15.44 | 14.8 | 27.7 | 21.0 | 30.3 | 19 | 11 | 38 |
| HICS 3 | 6.67 | 8.95 | 18.7 | 20.7 | 12.0 | 25.5 | 20 | 11 | 42 |
| Harmonic mean | 17.4 | 22.6 | 14.1 | 26.9 | 16.7 | 9.2 | 35.9 | ||
| ECCS 1 | 6 | 6.27 | 19.5 | 16.2 | 8.3 | 21.8 | 16 | 9 | 34 |
| ECCS 2 | 7.14 | 10.44 | 18.5 | 30.3 | 25.9 | 31.7 | 13 | 7 | 30 |
| ECCS 3 | 6.83 | 17.06 | 16.2 | 15.3 | 7.5 | 20.9 | 11 | 6 | 28 |
| Harmonic mean | 18.0 | 18.7 | 10.3 | 24.0 | 13.0 | 7.1 | 30.5 | ||
| P values | 0.983 | 0.295 | 0.264 |
Discussion
Captive breeding, rearing, and release of hatchery fish to supplement wild stocks are critical components of efforts to prevent extinction and recover many species with diverse breeding and rearing ecologies. The Rio Grande Silvery Minnow, for example, is a small-bodied (<100mm standard length), pelagic spawner with different breeding behavior than salmonids and other commonly cultured fish. The hatchery program for this species has two goals: 1) production of sufficient numbers of minnows to effectively supplement the wild population in the Rio Grande in New Mexico and to support the reintroduced population in Big Bend (Texas), and 2) to maximize genetic diversity of captive stocks such that it is reflective of the wild population (US Fish and Wildlife Service 2009). This study addresses the latter goal (although data on progeny production and viability were also collected) and was designed to examine whether genetic composition of progeny differed among the three captive spawning designs employed for this species: MM, HICS, and ECCS.
Genetic Diversity
Genetic diversity in progeny and effective size did not differ significantly among breeding treatments, likely because variance in reproductive success occurred in all designs (see below). For all breeding designs, genetic diversity (H e, H O, and A R) declined between the parental and progeny generations. Allelic richness declined the most in all designs, although this may be a sampling artifact whereby rare alleles may not have been represented in our sample of 50 progeny. Specifically, although this collection of progeny was a subsample of a much large number of larvae produced (in thousands), it is possible that not all parental contributions are represented in this sample. Hence, it is not unexpected that A R is lower in the progeny sample. Results from COLONY suggested variance in reproductive success among individuals regardless of the breeding design. Three replicates had high variance in reproductive output, one in each different spawning design. For example, in the HICS, there was one instance of an individual contributing to 32% of the sampled offspring. Similarly, for the ECCS, a single parent contributed to 28% of offspring. We expected that variance in reproductive success would be minimal in the MM design, thereby maximizing allelic diversity and heterozygosity. However, even in MM design, variance in the proportion of eggs that are fertilized can be high due to factors including sperm availability, which may be determined by the distance to the female (when released), abundance of sperm, and behavior of the male. Variance may also be affected by egg and sperm traits and other traits that affect compatibility of mates. The presence of variance in reproductive success in all mating designs likely explains why genetic diversity does not differ between mating designs and why diversity is reduced compared to that of the parental broodstock. In fact, estimates of N e(SA) were less than the total number of broodstock used in the matings.
Viability and Fecundity
Our results suggest a tradeoff of productivity and quality of fish produced using the monogamous design. Viability of eggs produced using this design was significantly lower than in the communal spawning experiments. Reduced viability of offspring may result because MM precludes active mate choice. Mating in the wild is probably not random, as individuals compete for access to mating partners (Andersson 1994). Although we do not have specific information on mate choice in Rio Grande Silvery Minnow, it has been shown to occur in other broadcast spawning species, such as cod (Hutchings et al. 1999). Mate choice plays an important role in maximizing genetic diversity in genes involved with immunity (e.g., Milinski and Bakker 1990; Pen and Potts 1999; Reusch et al. 2001; Milinski 2003; Milinski et al. 2005). Additionally, breeding designs that facilitate mate choice can positively affect growth and viability of offspring. For example, offspring growth rates are higher in guppy (Poecilia reticulata) and pipefish (Syngnathus typhle) when produced by mating of females with preferred males (Reynolds and Gross 1992; Sandvik et al. 2000). For other pelagic spawners, offspring survival and escape behavior are increased compared to those produced by random mating. For example, in whitefish and cod, offspring survival may increase from 12% to 74%, respectively, when compared to mating situations where mate choice is precluded or limited (Wedekind et al. 2001; Rudolfsen et al. 2005). Taken together, these studies and our results suggest that elimination of mate choice in captive populations of Rio Grande Silvery Minnow (as would occur in the monogamous design) could have adverse consequences for growth rates and viability of progeny. Recently, Gruenthal and Drawbridge (2012) showed that communal spawning in the hatchery could be used effectively in larger-bodied pelagic broadcast spawning species. Differences in fecundity between monogamous and communal spawning could also be caused by disruptions to behaviors during the pre-spawning period leading to physiological disruptions during the final stages of gamete maturation and during spawning where, for example, gametes from one male may be insufficient to fertilize all eggs released by a female. The results presented here suggest that communal spawning can also be used effectively (in terms of achieving high viability and equivalent levels of diversity to a MM) in small-bodied, pelagic broadcast spawning species. However, if the tight link between the wild and captive populations were broken, for example in the case of a catastrophic population crash in the wild leading to a supportive breeding program that is more hatchery focused, then a marker-assisted breeding program (e.g., Doyle et al. 2001; Kozfkay et al. 2008; Fisch et al. 2012) would likely be warranted to minimize relatedness among breeding individuals.
Breeding Strategy and Variance in Reproductive Success in Rio Grande Silvery Minnow
Results of parentage/sibship analysis indicated that most individuals spawned with multiple partners and further suggested that the most likely breeding system in Rio Grande Silvery Minnow is polygamy. If groups of individuals come together to spawn, it is plausible that eggs are fertilized by sperm from multiple males that are in close proximity to the female. Specifically, likelihood analysis of genetic data (via COLONY) indicated that Rio Grande Silvery Minnow mated with an average of three individuals. Polygamy is also present in other broadcast spawning species, such as cod and lake sturgeon (e.g., Bruch and Binkowski 2002). In cod, mates pair up and release gametes into the water column, but the couple is often joined by satellite males, which swim among the eggs where they release sperm (Rowe et al. 2004). Multiple matings of this type increase the number of different genetic combinations in the offspring and may affect N e if one male/female fails to contribute. In a monogamous situation, the contribution of his/her partner is also nullified. In contrast, with multiple matings, the consequences are spread more evenly. Interestingly, we found, on average, that multiple mated individuals produced a greater number of offspring.
In Atlantic salmon, a positive correlation was identified between numbers of mates and realized reproductive success of females (Garant et al. 2001). Several genetic benefits of multiple mating by females have been suggested, including acquisition of good genes and maximization of genetic diversity within clutches (reviewed in Jennions and Petrie 2000). Petrie and Kempenaers (1998) hypothesized that where environmental conditions are unstable, good genes for the next generation may not persist, so females may increase their chances of acquiring good genes by mating with multiple males. In addition, maximizing diversity within a clutch may reduce the potential costs of inbreeding (Stockley et al. 1993) and may compensate for deleterious effects of genetic incompatibility between mates (Tregenza and Wedell 1998).
Caveats
In several cases, alleles were identified in parents that were not detected in progeny. The reverse was also true, whereby alleles were seen in the progeny but were not detected in the broodstock. The presence of alleles in broodstock but not progeny may be caused by sampling error in which rare alleles in the broodstock can be difficult to detect by sampling only 50 individuals. For multiplex PCRs that amplify both small and large microsatellites, dropout of the largest allele can occur. In this study, PCR and screening were repeated for individuals that were homozygous at locus Ppro118 to reduce the possible impact of dropout. Samples with anomalous alleles were also rescreened to eliminate the possibility of PCR and electrophoresis errors, scoring or recording errors. As all broodstock were sampled, possible explanations for presence of alleles in the progeny but failure to be detected in the broodstock include 1) missing parental genotypes, 2) null alleles in the broodstock, 3) allelic dropout, 4) scoring errors, 5) de novo mutations. Missing parental genotypes explain eight observations compared to 469 cases of missing genotypes in the progeny generation. Microsatellites have high mutation rates, so de novo mutations may explain a few of the remaining cases. In a study of genetic parentage in the catfish, two de novo mutations were observed from 2,450 gametes (Tatarenkov et al. 2006). For some of the microsatellites we employed, particularly Lco6 and Lco7, there was a high PCR failure rate, especially for MM replicate number two. The high rate of missing data for this group of progeny suggests that these fish may have been very small when they were sampled and thus had small quantities of DNA, making amplification difficult. Interestingly this sample (monogamous replicate 1) also had a high number of unviable eggs (~80%).
Goals of the Rio Grande Silvery Minnow Captive Propagation Program
One of the main goals of the captive propagation program for Rio Grande Silvery Minnow is to maximize diversity in the captive stocks and to prevent losses of diversity in the wild population through augmentation (e.g., via a Ryman–Laikre effect; Ryman and Laikre 1991). We have shown previously that the program is achieving these goals on a broad scale. Despite severe fluctuations in wild population sizes, genetic diversity has been retained and even increased in the wild over the last decade (Osborne et al. 2012). Maintenance of a strong connection between the wild population and the captive stock has been a high priority (US Fish and Wildlife Service 2009). For example, fish reared from wild-caught eggs are a priority for stocking the wild population and for incorporation into the captive broodstock. Also, there has been a focus on attempting to design breeding protocols that recognize the mode of reproduction of Rio Grande Silvery Minnow, particularly at the Albuquerque Biological Park. The results presented above suggest that, under the tested conditions, genetic diversity was maintained in the progeny produced using communal spawning designs (which permits mate choice and multiple matings) at levels similar to those achieved with a monogamous design. Given this result, we suggest that communal spawning be incorporated into the breeding plan for Rio Grande Silvery Minnow.
Conclusions
Although this study was limited to three replicates of each breeding design and our progeny sample sizes were relatively small, it does provide some insight into genetic and other potential impacts of the different breeding designs. When designing captive breeding programs for endangered species, it is important to consider the life history and the reproductive biology of the species. Here, we have shown that Rio Grande Silvery Minnow is likely a polygamous communal spawning species. We have also shown that spawning groups of individuals (hormone-induced and environmentally-cued) does not cause a reduction in diversity when compared to the monogamous designs, at least over a single generation, nor are there significant difference in effective size between designs (although we are cognizant that our estimates of N e apply to enumeration of offspring at a very early life stage, so they are not necessarily indicative of results across an entire life cycle). Communal spawning also appears to maximize viability of eggs. However, communal spawning (with equalized sex ratios) should still be used with caution as observations suggest that as the breeding season progresses fewer individuals contribute (Ulibarri M, personal communication). For this reason, it may be advisable to use communal spawning early in the breeding season to maximize contribution of individuals and switch to paired matings as the breeding season progresses. Other benefits of communal spawning schemes include the ability to include more breeders as compared to the number of paired matings that can realistically be conducted in the short breeding period of Rio Grande Silvery Minnow and in the space available.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
This study was supported by the Middle Rio Grande Endangered Species Collaborative Program administered through the US Bureau of Reclamation.
Supplementary Material
Acknowledgments
We gratefully acknowledge Christine Cooper, Tamara Max, Tracy Diver, and Ashleigh Gallion for vital laboratory assistance during this project, and Kim Ward from the Albuquerque Biological Park for assistance with the captive breeding portion of the study, and the technical support of the University of New Mexico’s Molecular Biology Facility, which is supported by National Institutes of Health grant number P20GM103452. We also thank Robin Waples and two anonymous reviewers for helpful comments on a previous version of the manuscript.
References
- Andersson M. 1994. Sexual selection. Princeton (NJ): Princeton University Press; [Google Scholar]
- Araki H, Cooper B, Blouin MS. 2007. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 318: 100–103 [DOI] [PubMed] [Google Scholar]
- Ballou JD. 1984. Strategies for maintaining genetic diversity in captive populations through reproductive technology. Zoo Biol. 3: 311–323 [Google Scholar]
- Bestgen KR, Platania SP. 1991. Status and conservation of the Rio Grande Silvery Minnow, Hybognathus amarus . Southwest Nat. 36: 225–232 [Google Scholar]
- Bottrell CE, Ingersol RH, Jones RW. 1964. Notes on the embryology, early development, and behavior of Hybopsis aestivalis tetranemus (Gilbert). T Am Microsc Soc. 83: 391–399 [Google Scholar]
- Bruch RM, Binkowski FP. 2002. Spawning behavior of lake sturgeon (Acipenser fulvescens). J Appl Ichthyol. 18: 570–579 [Google Scholar]
- Bryant EH, Reed DH. 1999. Fitness2009 decline under relaxed selection in captive populations. Cons Biol. 1: 665–669 [Google Scholar]
- Christie MR, Marine ML, French RA, Waples RS, Blouin MS. 2012. Effective size of a wild salmonid population is greatly reduced by hatchery supplementation. Heredity. 109: 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 1970. An introduction to population genetics theory. New York: Harper and Row; [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. 1977. Maximum Likelihood from incomplete data via the EM algorithm (with discussion). J R Stat Soc Series B. 39: 1–38 [Google Scholar]
- Dowling TE, Minckley WL, Marsh PC, Goldstein E. 1996. Mitochondrial DNA diversity in the endangered razorback sucker (Xyrauchen texanus): analysis of hatchery stocks and implications for captive propagation. Cons Biol. 10: 120–127 [Google Scholar]
- Doyle RW, Perez-Enriquez R, Takagi M, Taniguchi N. 2001. Selective recovery of founder genetic diversity in aquacultural broodstocks and captive, endangered fish populations. Genetica 111:291–304 [DOI] [PubMed] [Google Scholar]
- Fisch KM, Ivy JA, Burton RS, May B. 2013. Evaluating the performance of captive breeding techniques for conservation hatcheries: a case study of the Delta Smelt captive breeding program. J Hered. 104:92–104 [DOI] [PubMed] [Google Scholar]
- Flagg TA, Mahnken CVW, Iwamoto RN. 2004. Conservation hatchery protocols for Pacific salmon. Am Fish Soc Symp. 44: 603–619 [Google Scholar]
- Fraser DJ. 2008. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol Appl. 1: 535–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LA, Evans BS, Jerry DR. 2006. Loss of genetic diversity due to hatchery culture practices in barramundi (Lates calcarifer). Aquaculture. 261: 1056–1064 [Google Scholar]
- Garant D, Dodson JJ, Bernatchez L. 2001. A genetic evaluation of mating system and determinants of individual reproductive success in Atlantic salmon (Salmo salar L.). J Hered. 92: 137–145 [DOI] [PubMed] [Google Scholar]
- Gow JL, Tamkee P, Heggenes J, Wilson GA, Taylor EB. 2011. Little impact of hatchery supplementation that uses native broodstock on the genetic structure and diversity of steelhead trout revealed by a large-scale spatio-temporal microsatellite survey. Evol Appl. 4: 763–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 1995. FSTAT version 1.2: a computer program to calculate F-statistics. J Hered. 86: 485–486 [Google Scholar]
- Gruenthal KM, Drawbridge MA. 2012. Toward responsible stock enhancement: broadcast spawning dynamics and adaptive genetic management in white seabass aquaculture. Evol Appl. 5: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 48: 361–372 [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 1989. Principles of population genetics. 3rd edn Sunderland (MA): Sinauer Associates Inc; [Google Scholar]
- Hedgecock D. 1994. Does variance in reproductive success limit effective population sizes of marine organisms? Beaumont AR, editor. Genetics and evolution of aquatic organisms. London: Chapman and Hall; p. 122–134 [Google Scholar]
- Hedrick PW. 1992. Genetic conservation in captive populations and endangered species. In: Jain SK, Botsford LW, editors. Applied population biology. Dordrecht (The Netherlands): Kluwer; p. 45–68 [Google Scholar]
- Herbinger CM, O’reilly PT, Verspoor E. 2006. Unravelling first-generation pedigrees in wild endangered salmon populations using molecular genetic markers. Mol Ecol. 15: 2261–2275 [DOI] [PubMed] [Google Scholar]
- Herlin M, Delghandi M, Wesmajervi M, Taggart JB, McAndrew BJ, Penman DJ. 2008. Analysis of the parental contribution to a group of fry from a single day of spawning from a commercial Atlantic cod (Gadus morhua) breeding tank. Aquaculture. 274: 218–224 [Google Scholar]
- Hillis D, Moritz C, Mable C. 1996. Molecular systematics. Sunderland (MA): Sinauer; [Google Scholar]
- Hunter JR, Lo NCH, Leong HJ. 1985. Batch fecundity in multiple spawning fishes. Lasker R, editor. An egg production method for estimating spawning biomass of pelagic fish: application to the Northern Anchovy, Engraulis mordax. NOAA Technical Report NMFS. Springfield (VA): US Department of Commerce; p. 67–77 [Google Scholar]
- Hutchings JA, Bishop TD, McGregor-Shaw CR. 1999. Spawning behaviour of Atlantic cod, Gadus morhua: evidence of mate competition and mate choice in a broadcast spawner. Can J Fish Aquat Sci. 56: 97–104 [Google Scholar]
- Jelks HL, Walsh SJ, Burkhead NM, Contreras-Balderas S, Díaz-Pardo E, Hendrickson DA, Lyons J, Mandrak NE, McCormick F, Nelson JS, et al. 2008. Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries. 33: 327–407 [Google Scholar]
- Jennions MD, Petrie M. 2000. Why do females mate multiply? A review of the genetic benefits. Biol Rev Camb Philos Soc. 75: 21–64 [DOI] [PubMed] [Google Scholar]
- Jones OR, Wang J. 2010. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 10: 551–555 [DOI] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML. 2006. Maximum likelihood estimation of the frequency of null alleles at microsatellite loci. Conserv Biol. 7: 991–995 [Google Scholar]
- Kozfkay CC, Campbell MR, Heindel JA, Baker DJ, Kline P, Powell MS, Flagg T. 2008. A genetic evaluation of relatedness for broodstock management of captive, endangered Snake River sockeye salmon, Oncorhynchus nerka . Conserv Genet. 9:1421–1430 [Google Scholar]
- Koljonen M-L, Tähtinen J, Säisä M, Koskiniemi J. 2002. Maintenance of genetic diversity of Atlantic salmon (Salmo salar) by captive breeding programmes and the geographic distribution of microsatellite variation. Aquaculture. 212: 69–92 [Google Scholar]
- Milinski M. 2003. The function of mate choice in sticklebacks: optimizing Mhc genetics. J Fish Biol. 63: 1–16 [Google Scholar]
- Milinski M, Bakker TCM. 1990. Female sticklebacks use male coloration in mate choice and hence avoid parasitized males. Nature. 344: 330–333 [Google Scholar]
- Milinski M, Griffiths S, Wegner KM, Reusch TB, Haas-Assenbaum A, Boehm T. 2005. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci U S A. 102: 4414–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore GA. 1944. Notes on the early life-history of Notropis girardi. Copeia. 1944: 209–214 [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press; [Google Scholar]
- Oliver JK, Babcock RC. 1992. Aspects of the fertilization ecology of broadcast spawning corals: sperm dilution effects and in situ measurements of fertilization. Biol Bull. 183: 409–417 [DOI] [PubMed] [Google Scholar]
- O’Reilly P, Doyle RW. 2007. Live gene banking of endangered populations of Atlantic salmon. Verspoor E, Stradmeyer L, Nielsen JL. The Atlantic salmon: genetics, conservation and management. Dordrecht (The Netherlands): Springer; . p. 346–380 [Google Scholar]
- Osborne MJ, Benavides MA, Aló D, Turner TF. 2006. Genetic effects of hatchery propagation and rearing in the endangered Rio Grande Silvery Minnow. Rev Fish Sci. 14: 127–138 [Google Scholar]
- Osborne MJ, Carson EW, Turner TF. 2012. Genetic monitoring and complex population dynamics: insights from a 12-year study of the Rio Grande silvery minnow. Evol Appl. 5: 553–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Potts WK. 1999. The evolution of mating preferences and major histocompatibility complex genes. Am Natl. 153: 145–164 [DOI] [PubMed] [Google Scholar]
- Pennington JT. 1985. The ecology of fertilization of echinoid eggs: the consequences of sperm dilution, adult aggregation, and synchronous spawning. Biol Bull. 169: 417–430 [DOI] [PubMed] [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. 1998. Identifying populations for conservation on the basis of genetic markers. Cons Biol. 12: 844–855 [Google Scholar]
- Petrie M, Kempenaers B. 1998. Extra-pair paternity in birds: explaining variation between species and populations. Trends Ecol Evol. 13: 52–58 [DOI] [PubMed] [Google Scholar]
- Platania SP, Altenbach CS. 1998. Reproductive strategies and egg types of seven Rio Grande basin cyprinids. Copeia. 3: 559–569 [Google Scholar]
- Reusch TB, Häberli MA, Aeschlimann PB, Milinski M. 2001. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 414: 300–302 [DOI] [PubMed] [Google Scholar]
- Reynolds JD, Gross MR. 1992. Female mate preference enhances offspring growth and reproduction in a fish, Poecilia reticulata . P Roy Soc B Biol Sci. 250: 57–62 [Google Scholar]
- Rice WR. 1989. Analyzing tables of statistical tests. Evolution. 43: 223–225 [DOI] [PubMed] [Google Scholar]
- Rowe S, Hutchings JA, Bekkevold D, Rakitin A. 2004. Depensation, probability of fertilization, and the mating system of Atlantic cod (Gadus morhua L.). ICES J Mar Sci. 61: 1144–1150 [Google Scholar]
- Rudolfsen G, Figenschou L, Folstad I, Nordeide JT, Søreng E. 2005. Potential fitness benefits from mate selection in the Atlantic cod (Gadus morhua). J Evol Biol. 18: 172–179 [DOI] [PubMed] [Google Scholar]
- Russello MA, Amato G. 2004. Ex situ population management in the absence of pedigree information. Mol Ecol. 13: 2829–2840 [DOI] [PubMed] [Google Scholar]
- Ryman N. 1994. Supportive breeding and effective population size: differences between inbreeding and variance effective numbers. Cons Biol. 8: 888–890 [Google Scholar]
- Ryman N, Laikre L. 1991. Effects of supportive breeding on the genetically effective population size. Cons Biol. 5: 325–329 [Google Scholar]
- Sandvik M, Rosenqvist G, Berglund A. 2000. Male and female mate choice affects offspring quality in a sex-role-reversed pipefish. Proc Biol Sci. 267: 2151–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. 2000. ARLEQUIN ver. 2.000: a software for population genetics data analysis. Switzerland: University of Geneva; [Google Scholar]
- Stockley P, Searle JB, Macdonald DW, Jones CS. 1993. Female multiple mating behaviour in the common shrew as a strategy to reduce inbreeding. Proc R Soc Lond B. 254:173–179 [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Barreto F, Winkleman DL, Avise J. 2006. Genetic monitoring in the channel catfish, Ictalurus punctatus, a species with uniparental nest guarding. Copiea. 4: 735–741 [Google Scholar]
- Tessier N, Bernatchez L, Wright JM. 1997. Population structure and impact of supportive breeding inferred from mitochondrial and microsatellite DNA analyses in land-locked Atlantic salmon Salmo salar L. Mol Ecol. 6: 735–750 [Google Scholar]
- Thériault V, Moyer GR, Jackson LS, Blouin MS, Banks MA. 2011. Reduced reproductive success of hatchery coho salmon in the wild: insights into most likely mechanisms. Mol Ecol. 20: 1860–1869 [DOI] [PubMed] [Google Scholar]
- Tregenza T, Wedell N. 1998. Benefits of multiple mates in the cricket Gryllus bimaculatus. Evolution. 52: 1726–1730 [DOI] [PubMed] [Google Scholar]
- US Department of the Interior 1994. Endangered and threatened wildlife and plants: final rule to list the Rio Grande silvery minnow as an endangered species. Federal Registar. 59: 36988–36995 [Google Scholar]
- US Fish and Wildlife Service 2009. Rio Grande Silvery Minnow Genetics Management and Propagation Plan. [cited 2011 July 1 ]. Available from: http://www.middleriogrande.com/LinkClick.aspx?fileticket=nAj3x8zOMgA%3dandtabid=455andmid=1041 [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 4: 535–538 [Google Scholar]
- Wang J. 2009. A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol Ecol. 18: 2148–2164 [DOI] [PubMed] [Google Scholar]
- Waples RS. 1989. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics. 121: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. 1999. Dispelling some myths about hatcheries. Fisheries. 24: 2–21 [Google Scholar]
- Waples RS. 2005. Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Mol Ecol. 14:3335–3352 [DOI] [PubMed] [Google Scholar]
- Wedekind C, Muller R, Spicher H. 2001. Potential genetic benefits of mate selection in whitefish. J Evol Biol. 16: 224–232 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


