Abstract
Survivin is an oncogene that functions in cancer cell cytoprotection and mitosis. Here we report that differential expression in cancer cells of a C-terminal splice variant of survivin, termed survivin-ΔEx3, is tightly associated with aggressive disease and markers of unfavorable prognosis. In contrast to other survivin variants, survivin-ΔEx3 localized exclusively to nuclei in tumor cells and was phosphorylated at multiple residues by the checkpoint kinase Chk2 during DNA damage. Mutagenesis of the Chk2 phosphorylation sites enhanced the stability of survivin-ΔEx3 in tumor cells, inhibited the expression of phosphorylated H2AX (γH2AX) in response to double strand DNA breaks, and impaired growth after DNA damage. DNA damage-induced Chk2 phosphorylation, stabilization of p53, induction of the cyclin-dependent kinase inhibitor p21, and homologous recombination-induced repair were not affected. In vivo, active Chk2 was detected at the earliest stages of the colorectal adenoma-to-carcinoma transition, persisted in advanced tumors, and correlated with increased survivin expression. Together, our findings suggest that Chk2-mediated phosphorylation of survivin-ΔEx3 contributes to a DNA damage-sensing checkpoint that may affect cancer cell sensitivity to genotoxic therapies.
Keywords: Survivin, ΔEx3, alternative splicing, Chk2, DNA damage
INTRODUCTION
As a unique member of the Inhibitor of Apoptosis (IAP) gene family, survivin has attracted attention for its multiple roles in cell proliferation and cell death (1), and its abundant distribution in every human tumor, compared to normal tissues (2). Disparate oncogenic pathways control the differential expression of survivin in cancer (3), and influence its localization to multiple subcellular compartments (4, 5). Accordingly, transcription of the survivin locus is complex, giving rise to at least five mRNA species that encode, in addition to wild type (WT) survivin, the variants survivin-2B, -3B, -2α and -ΔEx3 (6, 7). Structurally, survivin-2α and -3B are generated by “read-through” into intron 2 (8), or via inclusion of an alternative exon 3B (9), whereas survivin-2B and -ΔEx3 originate from the insertion of an alternative exon 2B (10), or the skipping of exon 3 (11), respectively.
Elucidating the function(s) of the survivin spliced variants has been challenging, given their low level of expression in most cells, and the limited availability of isoform-specific reagents. For instance, survivin-2B has been reported to promote apoptosis, in vitro (10, 12). However, low levels of survivin-2B correlate with better survival in acute myeloid leukemia (13), and its silencing in ovarian cancer has been linked to greater sensitivity to taxanes (14). A role of the survivin isoforms in mitosis has been equally controversial, as this function has been proposed in some reports (9, 10), but negated in others (15).
In this study, we took a multidisciplinary approach of genome-wide bioinformatics, analysis of the DNA damage response, and evaluation of primary patient samples to dissect a potential role of survivin-ΔEx3 in cancer (6, 7). We found that survivin-ΔEx3 is a nuclear substrate of the checkpoint kinase, Chk2 (16) in its unique –COOH terminus (6, 7), and that this pathway contributes to a DNA damage-sensing checkpoint in tumor cells (17).
MATERIALS AND METHODS
Bioinformatics analysis
Fourteen cancer-related datasets with a total of 702 samples assayed on GPL5188 (Affymetrix Human Exon 1.0 ST) arrays were examined for expression of survivin-ΔEx3 (6, 7). Of the 14 datasets, 9 compared cancer or cancer-related cells with normal controls, and 5 compared either different cancers or the same cancer at different stages (Supplementary Table 1).
The HuEx-1_0-st Affymetrix microarray platform contains 22 probesets designed to detect sequences derived from three isoforms of the survivin locus (Figure 1A): NM_001168 (survivin), NM_001012270 (survivin-ΔEx3) and NM_001012271 (survivin-2B). Of the 22 probesets, 9 were retained in all 14 datasets, and 13 were removed due to expression below background. Probesets 3 and 16 were also removed as their expression profiles were the same as 6 other probesets that targeted the same isoforms. Of the remaining probesets (Figure 1A), 8 of the 9 probes targeted regions that were common to all three survivin isoforms. Probeset 9 specifically targets exon 3, which is deleted in survivin-ΔEx3 (6, 7). Specific expression of survivin-ΔEx3 was calculated as the difference between the average expression of the 8 common survivin probesets and probeset 9.
Figure 1. Genome-wide bioinformatics analysis of survivin-ΔEx3 in cancer.
A. Schematic diagram of the genomic organization of WT survivin, survivin-2B and -ΔEx3 (top), and distribution of probesets and datasets (bottom). B. Ratio between probeset 9 targeting WT survivin and survivin-2B, and average expression of probesets targeting all survivin isoforms (expected ratio is <1). The different colors indicate samples from 14 different datasets, incremental from left to right. C. Fold upregulation of survivin-ΔEx3 in tumors relative to matched normal tissues. The number of cases in normal (N) or tumor (T) groups is: ovary (N, n=31; T, n=8); stomach (N, n=25; T, n=25); brain (N, n=6; T, n=49); lung (N, n=20; T, n=20); prostate (N, n=29; T, n=150). Mean±SEM. The statistical analysis for differential expression of WT survivin in tumors versus normal samples is: ovary, p=3.10−7; stomach, p=3×10−5; brain, p=3.10−7; lung, p=0.02; prostate, p=1×10−5. D. Increased expression of survivin-ΔEx3 during tumor progression. OD, oligodendroglioma; GBM, grade IV glioblastoma; PrCa, prostate cancer; NB, neuroblastoma. The number of cases per each condition is: OD, n=2; GBM, n=28; localized PrCa, n=131; metastatic PrCa, n=19; stage (S) 1 NB, n=10; stage (S) 4 NB, n=37. Mean±SEM. The statistical analysis for differential expression of WT survivin in tumors of advanced versus early stage is: GBM versus OD, p=0.09; metastatic versus localized prostate cancer, p=2×10–11; NB S4 versus NB S1, p=0.002. E. Primary samples of non-small cell lung cancer (NSCLC, left) or prostate cancer (PrCa, right), and matched normal tissues (NSCLC, n=18; PrCa, n=2) were analyzed for differential expression of survivin-ΔEx3 mRNA, by quantitative RT-PCR. Mean±SEM.
Cell culture and antibodies
Human lung adenocarcinoma H460, breast adenocarcinoma MDA-431 and MCF-7, glioblastoma LN229, and colorectal adenocarcinoma HCT116 and SW480 cells were obtained from the American Type Culture Collection. HCT116-DR-GFP cells were kindly provided by Dr. S. Powell (Memorial Sloan Kettering Cancer Center, New York, NY). Consistent with editorial guidelines, all cell lines were used within six months of receipt from the cell bank. The following antibodies to Chk2 (Santa Cruz), Thr68 phosphorylated Chk2 (Cell Signaling), survivin (Novus Biologicals), p53 (Calbiochem), Ser15-phosphorylated p53 (Cell Signaling), p21 (Calbiochem), Ser139-phosphorylated histone H2AX, i.e γH2AX (Millipore), Aurora B (Bethyl Laboratories), Alexa Fluor® 488 (Invitrogen), FLAG (Sigma-Aldrich), β-actin (Sigma-Aldrich), COX-IV (Cell Signaling), and RCC1 (Santa Cruz) were used.
Mutagenesis
Substitution of predicted Chk2 phosphorylation sites Thr79→Ala, Thr127→Ala, and Ser98→Ala in the unique –COOH terminus of survivin-ΔEx3 was carried out using QuikChange Site-Directed Mutagenesis Kit (Stratagene) with oligonucleotides (mutated sequences underlined): 5′-ATGCAAAGGAAACCAGCAATAAGAAGAAAGAAT-3′ (Thr79, ACA→GCA), 5′-TTATTCCCTGGTGCCGCCAGCCTTCCTGTGGGC-3′ (Thr127, ACC→GCC), and 5′-AATCCATGGCAGCCAGGCGCTCGATGGCACGGC-3′ (Ser98, AGC→GCC). Mutant constructs were confirmed by DNA sequencing.
Transfections
Tumor cell types (105/well) were transfected with FLAG-tagged cDNAs in the presence of lipofectamine 2000 (Invitrogen) and 250 μl Opti-MEM I (Invitrogen) per well (18). In some experiments, HCT116 transfectants were treated with or without etoposide (2.5μM), immunoprecipitated with an antibody to FLAG (2 μg) plus protein G-Sepharose, and immune complexes were analyzed with an antibody to phosphorylated Ser residues (Upstate-Millipore), by Western blotting. Gene silencing by small interfering RNA (siRNA) was carried out with non-targeting or SMART pool siRNA to Chk2 (Dharmacon). Two survivin-ΔEx3-specific siRNA sequences, 5′-GAAGAAAGAAUUUGAGGAAUU-3′ and 5′-GCAAAGGAAACCAACAAUAUU-3′, were used.
Protein expression and purification
Recombinant GST fusion proteins were expressed in E. coli, as described (19).
Protein analysis and turnover
Tumor cell types (6–7×107) were fractionated in mitochondrial and cytosolic extracts as described (20). In some experiments, extracts (5 μg) from HCT116 transfectants were separated on 13% acrylamide:bis-acrylamide, 29:1 gels in the presence of 20 μM Phos-tag™ Acrylamide AAL-107M (Wako) containing 3% MeOH and 40 μM MnCl2.Pull down experiments with recombinant proteins (10 μg) were carried out as described (19). For protein stability, HCT116 transfectants were treated with 0.1 mg/ml cycloheximide, harvested after 0, 2, 4, and 6 h, and analyzed by Western blotting.
Kinase assays
Recombinant proteins were incubated with human active Chk2 (R&D Systems, 0.1 μg/μl) in kinase buffer (pH 7.2) containing 25 mM MOPS, 12.5 mM β-glycerolphosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT, and 0.1 mM ATP in the presence or absence of 32P-γATP (Amersham) for 20 min at 30°C, and detection of phosphorylated bands by autoradiography. Recombinant Cdc25 was a control Chk2 substrate.
γH2AX reactivity
For analysis of DNA damage, tumor cell types were treated with etoposide (2.5 μM) or camptothecin (CPT, 2.5 μM) for 16 h at 37°C, followed by 4 h or 8 h recovery. Cells (1×106) were fixed in 1% paraformaldehyde for 15 min and 70% ethanol for 30 min, permeabilized in 0.2% Triton X-100 plus 1% BSA for 10 min, and incubated with an antibody to γH2AX for 90 min. After addition of Alexa Fluor® 488, cells were stained with propidium iodied (PI), and analyzed by multiparametric flow cytometry. Alternatively, cells grown on glass coverslips were processed as above and analyzed by fluorescence microscopy for γH2AX-containing foci formation using a Nikon E600 upright microscope. Fluorescence intensity and number of foci was quantified using Image J software.
Homologous double strand DNA break repair assay
A pDR-GFP plasmid that utilizes two modified GFP genes (I-Sce1-GFP and iGFP) to create a recombination reporter was used (21). Stable HCT116-pDR-GFP clones were co-transfected with pCMV-I-SceI alone or with various cDNAs, and analyzed after 3 d by multiparametric flow cytometry (22).
Soft agar colony formation
HCT116 transfectants (5×105) treated with or without etoposide (1.25 μM) were analyzed for colony formation in semi-solid medium, as described (20). Colony size (≤100 μm or 100–200 μm) was quantified using NIH Image J software.
Patient samples and RT-PCR
All patient-related investigations were approved by an Institutional Review Board. Histologically confirmed tumor and matched normal tissue samples from chemo-naive patients with diagnosis of non-small cell lung cancer (n=18), or prostate cancer (n=2) were analyzed for differential expression of survivin-ΔEx3 mRNA by RT-PCR using primers 5′-TGGAAGGCTGGGAGCCA (forward) and 5′-AGAAAGAATTTGAGGAAACTGCGGA-3′ (reverse). Relative levels of survivin-ΔEx3 mRNA were quantified using β2-microglobulin (β-2M, Assay-on-Demand chemistry, Hs 99999907_m1) as a reference gene and the 2^-DDCt formula.
Histology
Anonymous primary human tissue samples representative of different stages of the colorectal adenoma-to-carcinoma transition (hyperplasia, moderate or severe dysplasia, or adenocarcinoma) were analyzed for expression of survivin, Chk2, Thr68-phosphorylated Chk2 or p53, by immunohistochemistry (20). Twenty cases per condition were scored, and a cutoff of 5% positively stained cells was used.
Statistical analysis
Data were analyzed using the unpaired t test on a GraphPad software package for Windows (Prism 4.0). Differences in survivin-ΔEx3 gene expression in datasets with paired cancer/control samples (datasets 3 and 4) were analyzed using the two-tail t-test. Additional comparisons were done using the Spearman correlation test. The level of significance was set at p<0.05.
RESULTS
Differential expression of survivin-γEx3 in human cancer
We used a genome-wide bioinformatics approach to examine the expression of survivin-ΔEx3 in cancer-associated GEO datasets (Supplementary Table 1) A schematic diagram of the genomic organization of the survivin gene, and its relationship to the probesets on the arrays are shown in Figure 1A. By principal component analysis (Supplemental Figure 1A), the expression of probeset 9 (deleted in survivin-ΔEx3) differed from that of other 8 probesets common to other survivin isoforms, validating the ability of the microarray platform to distinguish between survivin isoforms that either contain or lack exon 3 (Supplementary Figure 1A). A hierarchical clustering based on differential probeset expression is shown in Supplementary Figure 1B. Given the average expression of probesets 4, 5, 13, 14, 15 and 17 (X = cumulative expression of all three isoforms) and probeset 9 (Y = expression of survivin and survivin-2B, but not survivin-ΔEx3), we determine that Y<X for the majority of samples (Figure 1B). Calculated as the difference X–Y, and then log2 transformed, survivin-ΔEx3 mRNA was expressed at significantly higher levels in tumors of the ovary, stomach, brain, lung and prostate, compared to matched normal tissues (Figure 1C). WT survivin was also differentially expressed in tumor versus normal tissues in the same datasets (Figure 1, legend) (2).
Survivin-ΔEx3 was expressed at higher levels in grade IV glioblastoma compared to low grade oligodendrogliomas, metastatic compared to localized prostate cancer, and stage 4 compared to stage 1 neuroblastoma (Figure 1D and Supplementary Table 2). Survivin-ΔEx3 expression correlated with hallmarks of disease progression, including poor survival (p=0.01, spearman correlation), recurrence (p=6×10−5, spearman correlation) and advanced Gleason grade (p=0.0004) in prostate cancer, N-myc amplification in neuroblastoma, H.pylori infection in gastric cancer, and chromosome 11q13 amplification in head and neck cancer (Supplementary Table 2).
To validate these bioinformatics predictions, quantitative PCR analysis of primary tumor samples and matched normal tissues was next carried out. In these experiments, survivin-ΔEx3 mRNA was consistently over-expressed in cases of non-small cell lung cancer (Figure 1E, left) and prostate cancer (Figure 1E, right), compared to matched normal tissues.
Subcellular localization of survivin-γEx3
Consistent with previous observations (6, 7), endogenous survivin-ΔEx3 mRNA was expressed at lower abundance in MCF-7 cells, compared to WT survivin (Supplementary Figure 2A). When transfected in HCT116, H460 or MDA-431 tumor cells, FLAG-tagged survivin-ΔEx3 was exclusively localized to nuclei, whereas WT survivin was distributed in nuclear and cytosolic fractions (Figure 2A) (23). In addition, survivin-ΔEx3 did not localize to mitochondria in transfected tumor cells, as opposed to an abundant pool of WT survivin associated with tumor mitochondria (Figure 2B). Functionally, treatment of H460 cells with the apoptosis inducer, staurosporine (STS), caused loss of mitochondrial transmembrane potential, in a reaction unaffected by transfection of control plasmid or survivin-ΔEx3 (Figure 2C). Conversely, a ~50% knockdown of endogenous survivin-ΔEx3 expression using isoform-specific siRNA (Supplementary Figure 2A) produced nuclear events, with accumulation of tumor cells with G2/M DNA content, and appearance of a cell population with >4N DNA content (Supplementary Figure 2B). In contrast, survivin-ΔEx3- specific siRNA did not reduce WT survivin mRNA levels, and a non-targeting siRNA had no effect (Supplementary Figure 2A, B).
Figure 2. Nuclear localization of survivin-ΔEx3 in cancer.
A. The indicated tumor cells were transfected with vector, FLAG-WT survivin (SVV) or FLAG-survivin-ΔEx3, and cytosolic (C) or nuclear (N) fractions were analyzed by Western blotting. RCC1 was a nuclear marker. B. The indicated tumor cells were transfected as in A, and cytosolic (C) or mitochondrial (M) fractions were analyzed by Western blotting. COX-IV was a mitochondrial marker. For panels A and B, the position of survivin (top) or survivin-ΔEx3 (bottom) bands is indicated. C. MDA-431 cells were transfected with vector or survivin-ΔEx3, treated with staurosporine (STS), and analyzed for mitochondrial membrane potential by JC-1 staining and multiparametric flow cytometry. Data are expressed as ratio between green (FL1)/red (FL2) fluorescence. The percentage of cells in each quadrant is indicated.
Chk2 phosphorylation of survivin-ΔEx3
By sequence analysis, three potential phosphorylation sites matching the consensus for the DNA damage kinase, Chk2 (16) were identified at Thr79, Ser98 and Thr127in the unique –COOH terminus of survivin-ΔEx3 (Figure 3A) (6, 7). In contrast, no Chk2 phosphorylation sites were predicted in WT survivin. Accordingly, active Chk2 readily phosphorylated recombinant survivin-ΔEx3, but not WT survivin, in a kinase assay (Figure 3B). Ala substitution of Thr127 in survivin-ΔEx3 (ΔEx3-Mut1), or double mutation of Thr79 and Ser98 (ΔEx3-Mut2) partially reduced phosphorylation by Chk2 (Figure 3B). In addition, Ala mutagenesis of all three predicted Chk2 phosphorylation sites at Thr127, Thr79 and Ser98 (ΔEx3-Mut3) abolished Chk2 phosphorylation of survivin-ΔEx3 (Figure 3B).
Figure 3. Chk2 phosphorylation of survivin-ΔEx3.
A. The position of three putative Chk2 phosphorylation sites at Thr79, Ser98 and Thr127 in survivin-ΔEx3 is indicated. B. Active Chk2 was incubated with recombinant survivin (SVV), survivin-Ex3 or survivin-ΔEx3 Thr127→Ala (Mut1), Thr79/Ser98→Ala (Mut2) or Thr127/Thr79/Ser98→Ala (Mut3) in a kinase assay, and radioactive bands were visualized by autoradiography. Cdc25 was a control Chk2 substrate. Bottom, Coomassie blue staining of recombinant proteins. C. HCT116 cells were transfected with the indicated FLAG-tagged cDNA constructs, treated with (+) or without (−) etoposide and analyzed for changes in electrophoretic migration of phospho-tagged protein bands, by Western blotting. A white dotted line is drawn across the center of each band. D. HCT116 transfected with FLAG-survivin-ΔEx3 or FLAG-survivin-ΔEx3-Mut3 were immunoprecipitated (IP) with an antibody to FLAG before or after etoposide treatment (top), and immune complexes were analyzed with an antibody to phosphorylated Ser residues, by Western blotting (bottom). TCE, total cell extracts. E. HCT116 cells transfected as indicated were treated with cycloheximide (CHX), harvested at the indicated time intervals after release and analyzed by Western blotting. Right, densitometric quantification of protein bands. Similar results were obtained in an independent experiment: survivin-ΔEx3, 2 h, 61.2%; 4 h, 40.6%; 6 h, 3.6%; survivin-ΔEx3-Mut3, 2 h 58.1%; 4 h, 48.7;6 h, 25.7%.
We next asked whether Chk2 phosphorylated survivin-ΔEx3, in vivo, potentially as part of a DNA damage response in tumor cells (17). First, treatment of HCT116 cells with the DNA damaging agent, etoposide, did not result in electrophoretic mobility changes of transfected FLAG-WT survivin (Figure 3C). In contrast, etoposide-induced DNA damage caused gel retardation of FLAG-survivin-ΔEx3 in HCT116 transfectants (Figure 3C). A Chk2 phosphorylation-defective FLAG-survivin-ΔEx3-Mut3 (Figure 3B) transfected in HCT116 cells did not exhibit gel shift retardation after DNA damage (Figure 3C), and no changes in electrophoretic mobility were observed for survivin-ΔEx3 or survivin-ΔEx3-Mut3 in untreated cultures (Figure 3C). Second, survivin-ΔEx3 immune complexes precipitated from etoposide-treated HCT116 cells (Figure 3D, top) reacted with an antibody to phosphorylated Ser residues, compared with untreated cultures (Figure 3D, bottom). In contrast, survivin-ΔEx3-Mut3 immunoprecipitates (Figure 3D, top) did not exhibit changes in phosphorylated Ser reactivity, with or without etoposide (Figure 3D, bottom).
A potential regulation of survivin-ΔEx3 by Chk2 phosphorylation was next investigated. WT survivin transfected in HCT116 cells remained stable over a 6-h period (Figure 3E). In contrast, survivin-ΔEx3 exhibited a much accelerated turnover, with an estimated half-life of ~1 h (Figure 3E). Ablation of Chk2 phosphorylation in survivin-ΔEx3-Mut3 improved protein stability, prolonging its half-life to ~2 h (Figure 3E). Expression of survivin-ΔEx3 or survivin-ΔEx3-Mut3 did not affect endogenous Chk2 levels in HCT116 transfectants (Figure 3E). In addition, recombinant survivin-ΔEx3 or survivin-ΔEx3-Mut3 did not associate with endogenous Chk2 in pull down experiments (Supplementary Figure 3A). As control, recombinant survivin, but not survivin-ΔEx3, associated with the mitochondrial chaperone, Hsp60 in tumor cells (Supplementary Figure 3B) (19).
Participation of survivin-γEx3 in a DNA damage-sensing checkpoint in tumor cells
Etoposide treatment of HCT116 cells resulted in the appearance of punctate nuclear foci containing Ser139-phosphorylated histone H2AX, or γH2AX (24), a marker of unrepaired DNA damage (17), by fluorescence microscopy (Figure 4A, B). Expression of WT survivin or survivin-ΔEx3 did not affect formation of γH2AX-containing foci in response to etoposide (Figure 4A, B). In contrast, transfection of survivin-ΔEx3-Mut3 inhibited the appearance of γH2AX-containing nuclear foci after DNA damage (Figure 4A, B). Similar results were obtained by quantification of γH2AX reactivity by multiparametric flow cytometry. Accordingly, HCT116 cells transfected with vector, WT survivin or survivin-ΔEx3 exhibited increased γH2AX reactivity after DNA damage, in a response that peaked 16 h after etoposide treatment (Supplementary Figure 4A), and returned to baseline during a 4- to 8-h recovery period (Figure 4C). In contrast, transfection of survivin-ΔEx3-Mut3 suppressed the induction of γH2AX reactivity after DNA damage (Supplementary Figure 4A, Figure 4C).
Figure 4. Survivin-ΔEx3 regulation of a DNA damage sensing checkpoint.
A. HCT116 cells were transfected with the indicated cDNAs, treated with etoposide and analyzed at the indicated time intervals for γH2AX-containing foci formation, by fluorescence microscopy. Magnification, x100. B. Quantification of γH2AX foci formation. FU, fluorescence intensity. C, D. HCT116 cells were transfected as indicated, exposed to etoposide (Etop, C), or camptothecin (CPT, D) for a 16 h treatment (16T), followed by 4 h and 8 h recovery (+4R, +8R) and analyzed for γH2AX expression by multiparametric flow cytometry. Normalized data from two independent experiments (Expt) are shown. E. HCT116 DR-GFP cells were transfected with pCMV-I-SceI without (None) or with the indicated cDNAs, and analyzed for changes in GFP expression, by multiparametric flow cytometry. The percentage of GFP+ cells under the various conditions tested is indicated. PE, phycoerythrin.
To test the specificity of γH2AX modulation by survivin-ΔEx3, we next treated HCT116 cells with the topoisomerase I inhibitor, camptothecin (CPT), which induces single strand DNA breaks, as opposed to the double strand breaks (DSB) caused by etoposide. CPT induced robust and time-dependent γH2AX reactivity in vector transfectants, by multiparametric flow cytometry (Supplementary Figure 4B, Figure 4D). Similarly, transfection with WT survivin, survivin-ΔEx3 or survivin-ΔEx3-Mut3 did not affect the magnitude or time-course of γH2AX induction by CPT (Supplementary Figure 4B, Figure 4D).
We next asked whether Chk2 phosphorylation of survivin-ΔEx3 affected homologous- directed DSB repair using HCT116 cells stably transfected with a GFP-DR recombination reporter (21, 22). Co-transfection of HCT-pDR-GFP cells with pCMV-I-SceI plus vector resulted in the appearance of GFP+ cells, indicative of homologous DSB repair (Figure 4D) (21,22). Addition of WT survivin, survivin-ΔEx3 or survivin-ΔEx3-Mut3 to these cells also resulted in quantitatively comparable degree of homologous DSB repair (Figure 4E). No GFP+ cells were detected in the absence of pCMV-I-SceI (Figure 4E), ruling out spontaneous intrachromosomal DNA conversion (21, 22).
Requirements of survivin-γEx3 for a DNA damage-sensing checkpoint
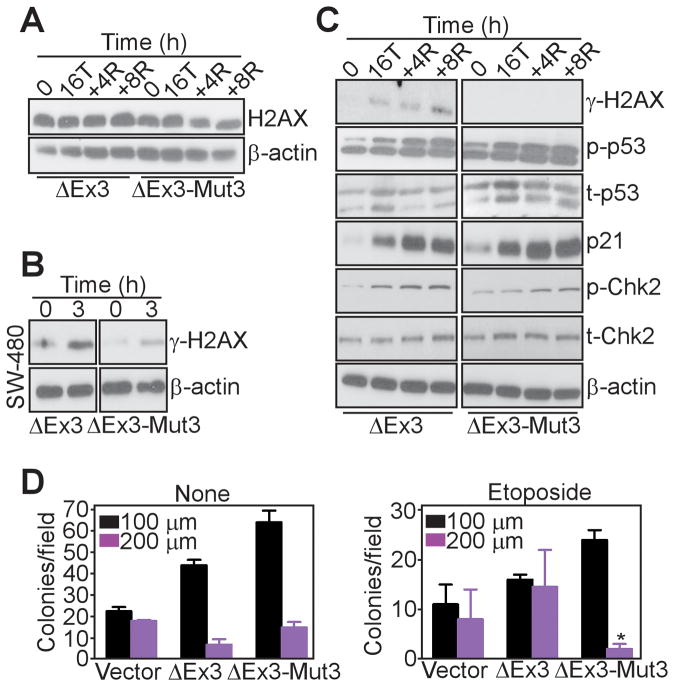
We next asked whether the effect of survivin-ΔEx3 on DSB foci formation reflected changes in total H2AX expression. HCT116 cells transfected with survivin-ΔEx3 or survivin-ΔEx3-Mut3 did not exhibit significant changes in endogenous H2AX levels, in the presence or absence of DNA damage (Figure 5A). Second, we tested whether this pathway required the DNA damage-sensing Mre11-Rad50-Nsb1 (MRN) complex, which is deficient in HCT116 cells (25). Transfection of survivin-ΔEx3 in colorectal adenocarcinoma SW480 cells, which express the MRN complex, resulted in robust γH2AX induction in response to etoposide, and this response was abolished by expression of survivin-ΔEx3-Mut3 (Figure 5B).
Figure 5. Requirements of survivin-ΔEx3 during the DNA damage response.
A. HCT116 cells were transfected as indicated, incubated with etoposide for a 16 h treatment (16T) followed by 4 h and 8 h recovery (+4R, +8R), and analyzed by Western blotting. B. SW480 cells transfected and treated as in A and analyzed by Western blotting. C. Transfected HCT116 cells were treated as in A, and analyzed by Western blotting. D. HCT116 cells were transfected as indicated, left untreated (None) or exposed to etoposide, and analyzed for colony formation in semisolid medium. Colonies were imaged by phase contrast microscopy (magnification, x50), and quantified for diameters of 50–100 μm or 100–200 μm. Mean±SEM of replicates from two independent experiments.
Finally, we looked at the activation of downstream pathway(s) of DNA damage upon expression of the various survivin isoforms. Transfection of survivin-ΔEx3-Mut3 suppressed γH2AX induction in etoposide-treated HCT116 cells (Figure 5C), consistent with the data above. However, stabilization of p53, phosphorylation of p53 on Ser15, or upregulation of the cyclin-dependent kinase inhibitor, p21, was indistinguishable in cells expressing survivin-ΔEx3 or survivin-ΔEx3-Mut3 (Figure 5C). Similarly, no differences were observed in etoposide-induced Thr68 phosphorylation of Chk2 in the presence of the various survivin isoforms (Figure 5C), and transfection of survivin-ΔEx3 or survivin-ΔEx3-Mut3 did not alter the expression of the chromosomal passenger protein, Aurora B during DNA damage, compared to WT survivin (Supplementary Figure 5).
Cellular implications for survivin-γEx3 participation in the DNA damage response
In the absence of DNA damage, transfection of survivin-ΔEx3 or survivin-ΔEx3-Mut3 significantly enhanced HCT116 colony formation in soft agar, compared to control cultures (Figure 5D, left), suggesting a cytoprotective role of both molecules irrespective of DNA damage or Chk2 phosphorylation. Expression of survivin-ΔEx3 still supported HCT116 colony formation after etoposide-induced DNA damage (Figure 5D, right). In contrast, transfection of survivin-ΔEx3-Mut3 impaired the ability of HCT116 cells to form large colonies (up to 200 μm in diameter) after etoposide treatment (Figure 5D, right).
As far as potential implications of this pathway for human tumorigenesis, Thr68-phosphorylated, i.e active Chk2 became abundantly expressed at the earliest, hyperplastic stages of the colorectal adenoma-to-carcinoma transition, and persisted in advanced disease stages (Supplementary Figure 6A, B). In contrast, active Chk2 was undetectable in normal epithelium, and total Chk2 levels remained unchanged throughout disease progression (Supplementary Figure 6A, B). The expression of active Chk2 correlated with aberrantly increased levels of survivin and p53, which rose in dysplastic lesions and remained elevated throughout all stages of colorectal transformation (Supplementary Figure 6A, B).
DISCUSSION
In this study, we have shown that survivin-ΔEx3 (6, 7) is a nuclear protein phosphorylated by the checkpoint kinase, Chk2 (16) during DNA damage on three novel amino acids in its unique –COOH terminus. Ablation of Chk2 phosphorylation enhanced survivin- ΔEx3 stability in vivo, suppressed γH2AX reactivity specifically in response to DSB, and impaired long-term clonogenic tumor cell survival. When analyzed in human tumors, survivin-ΔEx3 was differentially expressed in tumors compared to normal tissues, correlating with markers of unfavorable prognosis.
Previous studies had suggested a link between survivin and the DNA damage response. This was originally associated with the ability of survivin to counter radiation-induced apoptosis (26), but recent evidence suggested a more direct participation of the survivin network in DNA damage and repair, potentially independent of cytoprotection (27). Accordingly, interference with survivin expression or function resulted in reduced DNA repair, especially in gliomas (28), and therapeutic targeting of survivin with the small molecule transcriptional suppressor, YM155 (29), delayed DSB repair (30). Despite a potential suggestion that survivin may control the expression of the DNA repair protein, Ku70 (31), the molecular basis for a potential direct participation of the survivin network in DNA damage or repair has remained largely unexplored.
The data presented here add a new layer of complexity to these observations, and propose a role of the survivin spliced variant, ΔEx3 (6, 7) in sensing unrepaired DNA damage in tumor cells. This extends the panoply of functions previously associated with survivin-ΔEx3, including nucleolar (32), or mitochondrial control of apoptosis inhibition (33, 34), or endothelial cell migration and invasion (11), implicating Chk2 phosphorylation (16) as a novel regulator of survivin-ΔEx3. As shown here, Chk2 phosphorylation of survivin-ΔEx3 targeted its unique –COOH terminus introduced by the splicing event (6, 7), occurred during DNA damage, in vivo, and contributed to accelerated degradation of survivin-ΔEx3, compared to WT survivin. A role of survivin-ΔEx3 in the DNA damage response had not been previously proposed; however, this is consistent with the exclusive nuclear localization of this survivin isoform (35), which has been postulated to involve a bipartite nuclear localization signal, also introduced in its –COOH terminus by the splicing event (36).
Against this backdrop, Chk2 has long been recognized as a pivotal regulator of genomic integrity in response to DNA damage. Activated via phosphorylation on Thr68 by the DNA damage sensor kinase, ataxia telangiectasia mutated (ATM), active Chk2 in turn phosphorylates panoply of downstream targets coordinating checkpoint activation, cell cycle arrest, and apoptosis (16). Adding to this scenario, Chk2 phosphorylation of survivin-ΔEx3 was required to induce time-dependent induction of γH2AX, a marker of unrepaired DNA damage (24) important to recruit repair proteins to nuclear foci (37). Further studies are required to fully elucidate how a Chk2-survivin-ΔEx3 signaling axis contributes to a DNA damage response in tumor cells. However, the evidence presented here suggests that this pathway is specific for DSB, does not affect endogenous H2AX levels, is independent of the MRN complex (38), and is not required for homologous-directed DNA repair. Taken together, these findings suggest a model in which Chk2 phosphorylation of survivin-ΔEx3 provides a feedback amplification loop to enhance DSB sensing via optimal induction of γH2AX, which, in turn, may facilitate recruitment of repair proteins, including DNA-PK (39), or Ku70 (31), at sites of DNA damage (27). Given the rapid turnover of Chk2-phosphorylated survivin-ΔEx3, in vivo, this pathway may contribute to the earliest assembly of the repair machinery at DNA foci, and would be expected to confer a survival advantage for tumor cells exposed to genotoxic stimuli. Consistent with this hypothesis, survivin-ΔEx3 was preferentially observed in tumors compared to normal tissues, and correlated with markers of aggressive disease, in vivo (40, 41). Conversely, loss of Chk2 phosphorylation of survivin-ΔEx3 was detrimental for tumor growth, and impaired long-term clonogenic tumor cell survival after DSB.
Aside from its tumor suppressive properties, which are inactivated by mutagenesis in selected tumors (42), Chk2 has been intensely pursued for novel cancer therapeutics (43). Broadly defined as ‘checkpoint abrogation’, this strategy relies on the premise that pharmacologic inhibition of Chk2 (and Chk1) may prevent cell cycle arrest in response to DNA damage, and thus sensitize tumor cells to apoptosis (44). While validating Chk2 as an important cancer therapeutic target (45), the data presented also anticipate a broader role of this pathway in tumor progression, via exploitation of the survivin network. Mechanistically, this may involve Chk2-mediated release of cytoprotective survivin from its mitochondrial stores during DNA damage (18), coupled with efficient DSB sensing via phosphorylation of survivin-ΔEx3 (this study). More evidence is needed to further credential this model, in vivo, but it is intriguing that both active Chk2 and survivin become expressed at the earliest stages of malignant transformation in humans, for instance during the adenoma-to-carcinoma transition. The early appearance of Chk2 in comparable settings has been interpreted as a barrier to further malignant transformation (46, 47). However, it is also possible that Chk2 exploitation of the survivin network under these conditions may paradoxically contribute to accelerated tumor development, simultaneously promoting resistance to apoptosis (18), and faster control of genotoxic damage (this study). This scenario also reiterates the broad contribution of survivin family proteins to multiple aspects of tumorigenesis (1), and validate therapeutic strategies aimed at globally suppressing aberrant survivin gene transcription in tumors (30).
Supplementary Material
Acknowledgments
We thank Drs. Akiko Miyamoto and Hiroshi Kawasaki for help with the procurement of human tissue samples, and Simon Powell for HCT116-DR-GFP cells.
Footnotes
This work was supported by NIH grants CA140043, CA78810, CA118005. Support for Core Facilities utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
The authors declare that no conflict of interest exists.
References
- 1.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 3.Guha M, Altieri DC. Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle. 2009;8:2708–10. doi: 10.4161/cc.8.17.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knauer SK, Bier C, Habtemichael N, Stauber RH. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006;7:1259–65. doi: 10.1038/sj.embor.7400824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musacchio A. Surfing Chromosomes (and Survivin) Science. 2010;330:183–4. doi: 10.1126/science.1197261. [DOI] [PubMed] [Google Scholar]
- 6.Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005;92:212–6. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampath J, Pelus LM. Alternative splice variants of survivin as potential targets in cancer. Curr Drug Discov Technol. 2007;4:174–91. doi: 10.2174/157016307782109652. [DOI] [PubMed] [Google Scholar]
- 8.Caldas H, Honsey LE, Altura RA. Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Mol Cancer. 2005;4:11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knauer SK, Bier C, Schlag P, Fritzmann J, Dietmaier W, Rödel F, et al. The Survivin Isoform Survivin-3B is Cytoprotective and can Function as a Chromosomal Passenger Complex Protein. Cell Cycle. 2007;6:1501–8. [PubMed] [Google Scholar]
- 10.Ling X, Cheng Q, Black JD, Li F. Forced Expression of Survivin-2B Abrogates Mitotic Cells and Induces Mitochondria-dependent Apoptosis by Blockade of Tubulin Polymerization and Modulation of Bcl-2, Bax, and Survivin. J Biol Chem. 2007;282:27204–14. doi: 10.1074/jbc.M705161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldas H, Fangusaro JR, Boue’ DR, Holloway MP, Altura RA. Dissecting the role of endothelial survivin-DeltaEx-3 in angiogenesis. Blood. 2007;109:1479–89. doi: 10.1182/blood-2006-02-003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small TW, Pickering JG. Nuclear degradation of Wilms tumor 1-associating protein and survivin splice variant switching underlie IGF-1-mediated survival. J Biol Chem. 2009;284:24684–95. doi: 10.1074/jbc.M109.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner M, Schmelz K, Wuchter C, Ludwig WD, Dorken B, Tamm I. In vivo expression of survivin and its splice variant survivin-2B: Impact on clinical outcome in acute myeloid leukemia. Int J Cancer. 2006 doi: 10.1002/ijc.21995. [DOI] [PubMed] [Google Scholar]
- 14.Vivas-Mejia PE, Rodriguez-Aguayo C, Han H-D, Shahzad MMK, Valiyeva F, Shibayama M, et al. Silencing Survivin Splice Variant 2B Leads to Antitumor Activity in Taxane-Resistant Ovarian Cancer. Clin Cancer Res. 2011;17:3716–26. doi: 10.1158/1078-0432.CCR-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noton EA, Colnaghi R, Tate S, Starck C, Carvalho A, Ferrigno PK, et al. Molecular analysis of survivin isoforms: evidence that alternatively spliced variants do not play a role in mitosis. J Biol Chem. 2006;281:1286–95. doi: 10.1074/jbc.M508773200. [DOI] [PubMed] [Google Scholar]
- 16.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Op Cell Biol. 2009;21:245–55. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeggo PA, Lobrich M. DNA double-strand breaks: their cellular and clinical impact? Oncogene. 2007;26:7717–9. doi: 10.1038/sj.onc.1210868. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh JC, Dohi T, Raskett CM, Kowalik TF, Altieri DC. Activated checkpoint kinase 2 provides a survival signal for tumor cells. Cancer Res. 2006;66:11576–9. doi: 10.1158/0008-5472.CAN-06-3095. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–94. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 20.Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114:1117–27. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–50. [PubMed] [Google Scholar]
- 23.Fortugno P, Wall NR, Giodini A, O’Connor DS, Plescia J, Padgett KM, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–85. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 24.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, et al. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J Biol Chem. 2006;281:30814–23. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
- 26.Kanwar RK, Cheung CH, Chang JY, Kanwar JR. Recent advances in anti-survivin treatments for cancer. Curr Med Chem. 2010;17:1509–15. doi: 10.2174/092986710790979935. [DOI] [PubMed] [Google Scholar]
- 27.Rodel F, Reichert S, Sprenger T, Gaipl US, Mirsch J, Liersch T, et al. The role of survivin for radiation oncology: moving beyond apoptosis inhibition. Curr Med Chem. 2011;18:191–9. doi: 10.2174/092986711794088362. [DOI] [PubMed] [Google Scholar]
- 28.Chakravarti A, Zhai GG, Zhang M, Malhotra R, Latham DE, Delaney MA, et al. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene. 2004;23:7494–506. doi: 10.1038/sj.onc.1208049. [DOI] [PubMed] [Google Scholar]
- 29.Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, et al. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non small-cell lung cancer. J Clin Oncol. 2009;27:4481–6. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- 30.Iwasa T, Okamoto I, Suzuki M, Nakahara T, Yamanaka K, Hatashita E, et al. Radiosensitizing effect of YM155, a novel small-molecule survivin suppressant, in non small cell lung cancer cell lines. Clin Cancer Res. 2008;14:6496–504. doi: 10.1158/1078-0432.CCR-08-0468. [DOI] [PubMed] [Google Scholar]
- 31.Jiang G, Ren BO, Xu LEI, Song S, Zhu C, Ye F. Survivin may enhance DNA double-strand break repair capability by up-regulating Ku70 in human KB Cells. Anticancer Res. 2009;29:223–8. [PubMed] [Google Scholar]
- 32.Song Z, Wu M. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene. 2005 doi: 10.1038/sj.onc.1208097. [DOI] [PubMed] [Google Scholar]
- 33.Wang HW, Sharp TV, Koumi A, Koentges G, Boshoff C. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi’s sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J. 2002;21:2602–15. doi: 10.1093/emboj/21.11.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcles MH, Wang HW, Koumi A, Tsai YH, Yu M, Godfrey A, et al. Characterisation of the anti-apoptotic function of survivin-DeltaEx3 during TNFalpha-mediated cell death. Br J Cancer. 2007;96:1659–66. doi: 10.1038/sj.bjc.6603768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, et al. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ. 2002;9:1334–42. doi: 10.1038/sj.cdd.4401091. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez JA, Span SW, Ferreira CG, Kruyt FA, Giaccone G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein Survivin. Exp Cell Res. 2002;275:44–53. doi: 10.1006/excr.2002.5492. [DOI] [PubMed] [Google Scholar]
- 37.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 38.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–58. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 39.Reichert S, Rodel C, Mirsch J, Harter PN, Tomicic MT, Mittelbronn M, et al. Survivin inhibition and DNA double-strand break repair: A molecular mechanism to overcome radioresistance in glioblastoma. Radiother Oncol. 2011 doi: 10.1016/j.radonc.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 40.Koike H, Sekine Y, Kamiya M, Nakazato H, Suzuki K. Gene expression of survivin and its spliced isoforms associated with proliferation and aggressive phenotypes of prostate cancer. Urology. 2008;72:1229–33. doi: 10.1016/j.urology.2007.12.064. [DOI] [PubMed] [Google Scholar]
- 41.Nakano J, Huang C, Liu D, Masuya D, Yokomise H, Ueno M, et al. The clinical significance of splice variants and subcellular localisation of survivin in non-small cell lung cancers. Br J Cancer. 2008;98:1109–17. doi: 10.1038/sj.bjc.6604253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motoyama N, Naka K. DNA damage tumor suppressor genes and genomic instability. Curr Op Gen Dev. 2004;14:11–6. doi: 10.1016/j.gde.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Bolderson E, Richard DJ, Zhou B-BS, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15:6314–20. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Ghamrasni S, Pamidi A, Halaby MJ, Bohgaki M, Cardoso R, Li L, et al. Inactivation of chk2 and mus81 leads to impaired lymphocytes development, reduced genomic instability, and suppression of cancer. PLoS Genet. 2011;7:e1001385. doi: 10.1371/journal.pgen.1001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorgoulis VG, Vassiliou L-VF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 47.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.