Abstract
Objectives
The mechanism explaining the strong association between IL28B rs12979860 polymorphisms and treatment outcome in chronic hepatitis C remains unclear. We explore whether IL28B protein [interferon (IFN)-λ3] plasma levels may vary according to IL28B genotype and/or following pegylated IFN-α/ribavirin therapy.
Patients and methods
A total of 112 HIV/hepatitis C virus (HCV)-coinfected patients who completed a course of pegylated IFN-α/ribavirin therapy were examined. Sustained virological response (SVR) was achieved by 56% of patients. IL28B rs12979860 alleles were genotyped using the 5′ nuclease assay with specific TaqMan probes. A specific enzyme immunoassay was used to measure IFN-λ3 plasma levels before initiating anti-HCV therapy and at week 4 of treatment.
Results
No significant differences between CC and non-CC IL28B carriers were found at baseline, either in the proportion of patients with detectable IFN-λ3 plasma levels or in their median values. In contrast, median IFN-λ3 plasma levels at week 4 of therapy significantly increased with respect to baseline in CC carriers [34.3 (16.7–56.3) versus 15.6 (15.6–30.3) pg/mL, respectively; P < 0.0001], but not in CT/TT carriers. Unexpectedly, increases in IFN-λ3 at week 4 of therapy did not predict SVR.
Conclusions
The exogenous administration of IFN-α may induce IFN-λ3 release in IL28B CC carriers, but not in CT/TT carriers. However, this finding does not account for the link between IL28B polymorphisms and treatment outcome.
Keywords: HIV/HCV coinfection, anti-HCV treatment, predictors of response
Introduction
The single nucleotide polymorphism rs12979860 upstream of the IL28B gene coding for interferon (IFN)-λ3 has recently been shown to strongly predict response to pegylated IFN-α/ribavirin therapy in patients with chronic hepatitis C.1 Accordingly, a role for innate immunity mediated by IFN-λ3 has been postulated in the control of hepatitis C virus (HCV) infection. However, the precise biological mechanism explaining why IL28B gene polymorphisms exert such a strong influence on hepatitis C treatment outcomes is unknown.
IFN-λ3 belongs to the family of type III IFNs, which share functional characteristics with type I IFNs (IFN-α and IFN-β). All are produced in response to viral infections and mediate their effects throughout the JAK-STAT signal transduction pathway.2,3 Moreover, the production of IFN-λ in vitro has been shown to be modulated by IFN-α.4,5 As IFN-λ3 is active against HCV in vitro following interaction with its receptor,6,7 IL28B gene polymorphisms might influence viral control modulating the expression and/or functionality of IFN-λ3. It is currently unclear to what extent the exogenous administration of IFN-α might exert its antiviral effect directly or through IFN-λ3. Therefore, it is of paramount importance to assess the in vivo mechanisms underlying the relationship between the two IFNs and how this interaction is modulated by IL28B gene polymorphisms. In this regard, the measurement of IFN-λ3 in the bloodstream of patients with chronic hepatitis C who are treated with pegylated IFN-α/ribavirin may provide some clues.
The strong association between IL28B gene polymorphisms and hepatitis C treatment outcomes has been reproduced in chronic hepatitis C patients coinfected with HIV.8 This is important because treatment response rates are lower in the HIV population and, hypothetically, HIV-associated immunodeficiency might help to unveil any role of the innate immunity in the clearance of HCV infection.
Patients and methods
All HIV/HCV patients that had completed a first course of pegylated IFN-α/ribavirin therapy and had validated outcomes at Hospital Carlos III, Madrid, Spain were identified. Treatment regimens included pegylated IFN-α 2a or 2b at standard doses (180 μg/week or 1.5 μg/kg/week, respectively) plus weight-adjusted ribavirin (1000 mg/day for patients weighing <75 kg and 1200 mg/day for patients weighing ≥75 kg). Following international guidelines, patients with HCV genotypes 1 or 4 received either 48 or 72 weeks of treatment and those with HCV genotype 3 received 24 or 48 weeks of treatment, according to the virological response at week 4. Based on the level of haemoglobin at week 4 of treatment, no dose reduction of ribavirin was necessary in any patient.
A control group of HCV/HIV-seronegative individuals matched by age and gender was similarly examined. Only subjects with both plasma and peripheral blood mononuclear cells properly stored in frozen conditions were selected for further studies. Written informed consent was obtained from all individuals and the study protocol was approved by the hospital's Ethics Committee.
Plasma HCV-RNA was measured using a real-time PCR assay (COBAS TaqMan, Roche, Barcelona, Spain), which has a lower detection limit of 10 IU/mL. HCV genotyping was performed using a commercial hybridization assay (Versant HCV Genotype v2.0 LiPA, Siemens, Barcelona, Spain). Plasma HIV-RNA was measured using a commercial assay (Versant HIV-RNA v3.0, Siemens, Barcelona, Spain).
The IL28B rs12979860 single nucleotide polymorphism was genotyped using the 5′ nuclease assay with allele-specific TaqMan probes, as reported previously.1 A specific enzyme immunoassay was used to measure IFN-λ3 plasma levels (Interleukin 28B Cat.E2028Hu, USCN Life Science Inc., Wuhan, China). The capture antibody used in this assay is specific for IFN-λ3 (IL28B protein), with no significant cross-reactivity or interference with other molecules (<5% with IL28A and <1% with IL29). The lower limit of detection of the assay is 15.6 pg/mL. The intra- and inter-assay variations were determined to be 8% and 15%, respectively.
Statistical analysis
For continuous variables, data are given as median (IQR). Non-parametric tests were used to compare IFN-λ3 plasma levels in different groups (Mann–Whitney U-test) and at distinct timepoints (Wilcoxon signed-rank test). The association between categorical variables was assessed using χ2 or Fisher's exact test, as appropriate. All statistical analyses were performed using SPSS v13 software (SPSS Inc., Chicago, IL, USA). All P values were two-tailed and considered as significant only when <0.05.
Results
We identified 112 HIV/HCV patients at Hospital Carlos III, Madrid, Spain who had completed a first course of pegylated IFN-α/ribavirin therapy, had validated outcomes and had frozen specimens available for testing. Overall, sustained virological response (SVR) was achieved by 63 of them (56%), with the highest rate of SVR observed in HCV-3 as compared with HCV-1/4 patients (90% versus 44%, respectively; P < 0.0001). A rapid virological response was present in 31% of the overall population, with a higher rate in HCV-3 patients as compared with HCV-1/4 patients (60% versus 19%; P < 0.0001). Table 1 records the main characteristics of the study population. A control group of 30 HCV/HIV-seronegative individuals matched by age and gender was similarly examined. All persons included in the study were Caucasians.
Table 1.
Baseline characteristics of the study population
|
IL28B rs12979860 genotype |
||||
|---|---|---|---|---|
| Characteristics | Patients (n = 112) | CC (n = 51) | non-CC (n = 61) | P |
| Age (years), median (IQR) | 42 (39–46) | 42 (39–46) | 42 (39–46) | 0.91 |
| Male gender, n (%) | 84 (75) | 35 (69) | 49 (80) | 0.15 |
| CD4 count (cells/mm3), median (IQR) | 519 (391–692) | 517 (391–624) | 533 (389–778) | 0.44 |
| Patients on antiretroviral therapy, n (%) | 84 (75) | 38 (75) | 46 (75) | 0.91 |
| Plasma HIV-RNA in patients with no antiretroviral therapy (log10 copies/mL), median (IQR) | 3.4 (2.1–3.9) | 3.4 (2.3–3.9) | 3.0 (2.0–3.9) | 0.82 |
| Serum HCV-RNA (log10 IU/mL), median (IQR) | 6.6 (6.0–7.0) | 6.6 (6.0–7.0) | 6.7 (6.0–7.0) | 0.65 |
| Advanced liver fibrosis (kPa > 9.5), n (%) | 30 (27) | 13 (26) | 17 (28) | 0.83 |
| HCV genotype, n (%) | ||||
| 1 | 69 (62) | 27 (53) | 42 (69) | |
| 3 | 30 (27) | 20 (39) | 10 (16) | 0.021 |
| 4 | 13 (11) | 4 (8) | 9 (15) | |
| Patients with SVR to pegylated IFN-α/ribavirin therapy, n (%) | 63 (56) | 36 (71) | 27 (44) | 0.005 |
The overall frequency of the IL28B CC genotype was 45% in chronic hepatitis C patients and 43% in healthy controls. The CC genotype was recognized in 36/63 (57.1%) of chronic hepatitis C patients who achieved SVR versus 15/49 (30.6%) of those who did not respond to therapy (P = 0.005). Overall, IFN-λ3 plasma levels were detectable in 50% of healthy controls and in 46% of chronic hepatitis C patients at baseline, with no significant differences in median values [17 (15.6–52) and 15.6 (15.6–31) pg/mL, respectively; P = 0.42]. The same was true when comparing only individuals showing detectable IFN-λ3 plasma levels [50 (26–287) and 33 (26–71) pg/mL in healthy controls and patients, respectively; P = 0.3].
Given that our population of HIV/HCV-coinfected individuals was heterogeneous in terms of plasma HIV-RNA and CD4 count, as well as with respect to HCV characteristics (genotype or HCV-RNA), we tested the effect of these variables on baseline IFN-λ3 plasma levels. Whereas there was no significant association between IFN-λ3 plasma level and CD4 count, patients with detectable plasma HIV-RNA harboured significantly higher levels of IFN-λ3 than those with suppressed HIV replication [27 (15.6–38) and 15.6 (15.6–27) pg/mL, respectively; P = 0.007]. Moreover, HIV viraemic patients were those more frequently displaying detectable IFN-λ3 levels (72% versus 37%, respectively; P = 0.001). Differences according to HCV genotype were also noticed. HIV/HCV-coinfected patients with HCV-1 tended to show detectable IFN-λ3 more frequently than subjects harbouring other HCV genotypes (51% versus 37%, respectively; P = 0.1) as well as higher median IFN-λ3 plasma levels [24 (15.6–38) versus 15.6 (15.6–26) pg/mL, respectively; P = 0.05].
When HIV/HCV-coinfected patients were split into four groups according to status of HIV viraemia and HCV genotype, there were significant differences in baseline median levels of IFN-λ3 (Kruskal–Wallis test, P = 0.003) and in the proportion of patients with detectable IFN-λ3 (χ2 test, P = 0.003). The highest IFN-λ3 plasma levels [35 (25–110) pg/mL] and the highest proportion of IFN-λ3 detectability (86%) were observed in patients with detectable HIV viraemia and HCV-1 genotype, whereas the lowest IFN-λ3 plasma levels [15.6 (15.6–23) pg/mL] and proportion of detectable IFN-λ3 (28%) were seen in patients with non-HCV-1 genotypes and undetectable plasma HIV-RNA. Of note, IFN-λ3 plasma levels in patients with HCV-1 genotype and detectable plasma HIV-RNA were higher than in seronegative controls (P = 0.06). There was no association between IFN-λ3 plasma level and serum HCV-RNA, either in the whole study population or only in the HCV-1 genotype patients.
In chronic hepatitis C patients, no significant differences between IL28B CC and non-CC carriers were found at baseline, either in the proportion of patients with detectable IFN-λ3 plasma levels (45% versus 46%, respectively) or in their median values (Figure 1a). Median IFN-λ3 values were also similar in CC and non-CC carriers in the subset of individuals with detectable levels [31 (26–41) versus 34 (26–195) pg/mL, respectively; P = 0.34]. Similar results were observed in healthy controls when comparing IL28B CC and non-CC carriers [19 (15.6–190) versus 15.6 (15.6–47) pg/mL, respectively; P = 0.62]. In a multivariate logistic regression analysis, only HCV-1 genotype (OR 2.3; 95% CI 0.98–5.5; P = 0.05) and detectable plasma HIV-RNA (OR 5.2; 95% CI 1.94–14; P = 0.001) remained as independent predictors of detectable plasma IFN-λ3 levels.
Figure 1.
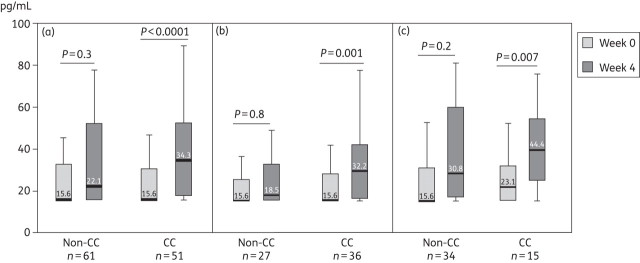
Median IFN-λ3 plasma levels during pegylated interferon-α/ribavirin therapy according to IL28B rs12979860 genotype. In the whole population of patients (a); in patients with SVR (b); and in non-responders (c).
At week 4 of pegylated IFN-α/ribavirin therapy, the proportion of patients with detectable IFN-λ3 levels significantly increased in both CC and non-CC carriers (from 45% to 82% and from 46% to 70%, respectively; P < 0.01 in both groups). However, the increase was more pronounced in CC carriers and, as a consequence, the proportion of IFN-λ3 detectability at week 4 tended to be higher in CC than in non-CC patients (82% versus 70%, respectively; P = 0.1). Likewise, median values of IFN-λ3 plasma levels increased at week 4 of therapy, although variations from baseline reached statistical significance only in CC carriers (Figure 1a). Accordingly, the percentage of variation in IFN-λ3 plasma levels at week 4 with respect to baseline was significantly higher in CC than in non-CC carriers [24% (0–151) and 0% (−11–70), respectively; P = 0.02]. Although non-HCV-1 genotype was also significantly associated with increased IFN-λ3 plasma levels at week 4 in univariate analysis, the multivariate logistic regression analysis showed that the only variable significantly associated with increased IFN-λ3 plasma levels at week 4 of therapy was the IL28B CC genotype (OR 2.6; 95% CI 1.2–5.8; P = 0.02).
Finally, we tested whether increases in IFN-λ3 plasma levels influenced the chance of subsequent SVR. It was not the case. Unexpectedly, similar increases in IFN-λ3 plasma levels at week 4 of treatment occurred in CC carriers who achieved SVR (Figure 1b) and in those who failed therapy (Figure 1c).
Discussion
This study was designed to study the relationship between IL28B polymorphisms and IFN-λ3 plasma levels, as well as how it could be influenced following the exogenous administration of IFN-α. We show that IFN-λ3 plasma levels at baseline were not modulated by IL28B genotypes in HIV/HCV-coinfected patients. This is in contrast to findings from another study that reported higher levels of IL28A/B in CC carriers than in non-CC carriers.9 However, that study included subjects with acute and self-limited hepatitis C besides chronically infected individuals. More importantly, the assay used was specific for IL28A, with partial cross-reactivity for IL28B (IFN-λ3). Despite this lower specificity of the assay, detection of IL28A/B only occurred in 27% of the subjects, while in our study using an assay specific for IFN-λ3, half of our patients had measurable levels of this protein.
An interesting finding of our study was the association between baseline IFN-λ3 plasma levels and both HIV-RNA and HCV-1 genotype. Although the correlation between serum HCV-RNA and IFN-λ3 levels did not reach statistical significance, patients with HCV-1 genotype had significantly higher serum HCV-RNA. Altogether, these findings suggest that there is an up-regulation of the IFN-λ3 system driven by HCV replication. A similar up-regulation of IFN-α production has been shown in uncontrolled HIV infection.10
On longitudinal follow-up, our results show that IFN-λ3 is up-regulated after exogenous administration of IFN-α and that this up-regulation is influenced by the IL28B genotype. It seems to be a tight in vivo relationship between the two IFNs, IFN-α and IFN-λ3, and IL28B polymorphisms modulate their interaction. However, the better response to pegylated IFN-α/ribavirin therapy in chronic hepatitis C patients harbouring the IL28B CC genotype does not seem to be mediated by an increased production of IFN-λ3 during therapy.
In summary, IFN-λ3 plasma levels significantly increase after 4 weeks of treatment with pegylated IFN-α/ribavirin in HIV patients with chronic hepatitis C, particularly in those with favourable IL28B alleles. However, the lack of association between IFN-λ3 plasma levels and SVR is against an up-regulation of IFN-λ3 by IFN-α as the biological mechanism responsible for the link between IL28B polymorphisms and treatment outcome.
Funding
This work was supported in part by grants from Fundación Investigación y Educación en SIDA (IES), Red de Investigación en SIDA (RIS, FIS-RD06/0006), Agencia Lain Entralgo and the European Union 6th Framework Programme (NEAT, No. LSHP-CT-2006-037570).
Transparency declarations
None to declare.
Acknowledgements
We would like to thank all of the patients who participated in the study.
References
- 1.Ge D, Fellay J, Thompson A, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. doi:10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 2.Kotenko S, Gallager G, Baurin V, et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. doi:10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 3.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. doi:10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 4.Sirén J, Pirhonen J, Julkunen I, et al. IFN-α regulates TLR-dependent gene expression of IFN-α, IFN-β, IL-28, and IL-29. J Immunol. 2005;174:1932–7. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 5.Ank N, West H, Bartholdy C, et al. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–9. doi: 10.1128/JVI.80.9.4501-4509.2006. doi:10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–4. doi: 10.1128/JVI.79.6.3851-3854.2005. doi:10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons α and λ inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–98. doi: 10.1053/j.gastro.2006.09.052. doi:10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 8.Rallón NI, Naggie S, Benito JM, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–9. doi: 10.1097/QAD.0b013e3283391d6d. doi:10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langhans B, Kupfer B, Braunschweiger I, et al. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–65. doi: 10.1016/j.jhep.2010.08.020. doi:10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Benlahrech A, Patterson S. HIV-1 infection and induction of interferon alpha in plasmacytoid dendritic cells. Curr Opin HIV AIDS. 2011;5:373–8. doi: 10.1097/COH.0b013e328349592a. doi:10.1097/COH.0b013e328349592a. [DOI] [PubMed] [Google Scholar]


