Abstract
DR (dietary restriction), or reduced food intake without malnutrition, is associated with extended longevity, improved metabolic fitness and increased stress resistance in a wide range of organisms. DR is often referred to as calorie restriction, implying that reduced energy intake is responsible for its widespread and evolutionarily conserved benefits. However, recent data indicate dietary amino acid restriction as a key mediator of DR benefits. In fruitflies, an imbalance in essential amino acid intake is thought to underlie longevity benefits of DR. In mammals, reduced dietary protein or essential amino acid intake can extend longevity, improve metabolic fitness and increase stress resistance. In the present paper we review two evolutionarily conserved signal transduction pathways responsible for sensing amino acid levels. The eIF2α (eukaryotic initiation factor 2α) kinase GCN2 (general amino acid control non-derepressible 2) senses the absence of one or more amino acids by virtue of direct binding to uncharged cognate tRNAs. The presence of certain amino acids, such as leucine, permits activation of the master growth regulating kinase TOR (target of rapamycin). These two signal transduction pathways react to amino acid deprivation by inhibiting general protein translation while at the same time increasing translation of specific mRNAs involved in restoring homoeostasis. Together, these pathways may contribute to the regulation of longevity, metabolic fitness and stress resistance.
Keywords: amino acid, dietary restriction (DR), eukaryotic initiation factor 2α (eIF2α), general amino acid control non-derepressible 2 (GCN2), target of rapamycin (TOR)
INTRODUCTION
Coding amino acids are necessary for protein synthesis, but are also involved in a number of other essential processes. For example, methionine is required for one-carbon transfer reactions, tryptophan is a precursor for NAD and serotonin biosynthesis, glutamate acts as a neurotransmitter, and a number of amino acids can serve as intermediate metabolites in a variety of processes ranging from gluconeogenesis to anaplerosis. Although lower organisms can synthesize all of the coding (and non-coding) amino acids from carbon skeletons and nitrogen, some of these biosynthetic pathways were lost during evolution of multicellular organisms. Thus humans (as well as mice and fruitflies) must acquire the EAAs (essential amino acids) through dietary means.
DR (dietary restriction), defined loosely as reduced food intake without malnutrition, was first shown to extend longevity in rats by Clive McKay in the 1930s [1]. In the ensuing decades, lifespan extension by DR has been demonstrated in multiple species, along with a number of other benefits, including extended healthspan, improved metabolicfitness and increased stress resistance.A large number of changes have been documented upon DR, including energy metabolism, gene expression, protein turnover, immune function and oxidative stress to name just a few [2]. However, which of these changes are evolutionarily conserved requirements for the benefits of DR remains unclear.
DR is also known as CR (calorie restriction), based, in part, on data in rodents suggesting that the reduction in total calorie intake is more important than the source of those calories (e.g. carbohydrates compared with protein) [3]. Previous data in fruitflies indicate that the source of the calories does matter, and that reduction of protein calories in the form of yeast contributes more to longevity extension than carbohydrate calories [4]. Furthermore, adding back EAAs erases the benefits of DR on longevity extension in fruitflies [5]. In rodents, it has long been known that reduced dietary intake of protein or certain amino acids, namely methionine and tryptophan, can also extend longevity [6,7]. What this has to do with lifespan extension by DR remains unclear, as there are many similarities, but also differences, between rodents on DR and those on methionine restriction [8].
How do cells sense the availability of amino acids, and what role might these mechanisms play in conveying benefits of DR? Cells have evolved different mechanisms to sense both the absence of individual amino acids as well as the presence of some others. Lack of amino acids is generally sensed by a surrogate marker, an uncharged cognate tRNA. Uncharged tRNAs then trigger adaptive responses through a variety of downstream mechanisms to rectify the situation. In bacteria, these involve direct effects on transcriptional control, whereas in eukaryotes uncharged tRNAs trigger a signal transduction pathway via direct interaction with the eIF (eukaryotic initiation factor) 2α kinase GCN(general amino acid control non-derepressible) 2. The presence of certain amino acids, such as leucine, can also be sensed, activating signal transduction pathways that integrate responses to environmental cues, including nutrients, through the TOR (target of rapamycin) kinase. Less is known about upstream amino-acidsensing mechanisms in this pathway. A common output of amino acid deficiency sensed by either mechanism is repressed general translation, but enhanced (or derepressed) translation of particular mRNAs involved in restoring homoeostasis. The mechanisms by which GCN2 activation and TOR repression affect translation are also distinct. In the present review, we will discuss both GCN2-and TOR-based amino-acid-sensing mechanisms, downstream effects on translation and how these pathways may contribute to the benefits of DR, including stress resistance and longevity.
SENSING AMINO ACID DEFICIENCY: FROM UNCHARGED tRNAs TO THE GCN2 KINASE
Uncharged tRNAs activate the stringent response to amino acid deprivation in bacteria
Protein synthesis is universally important to life as we know it, and mechanisms to sense the building blocks of proteins, amino acids, are conserved across evolution. In bacteria, nutrient deficiency activates the stringent response, so-called because of the stringent inhibition of transcription of stable RNAs including rRNA and tRNAs [9]. The stringent response is dependent on unusual nucleotides, ppGpp (guanosine 5′,3′ bispyrophosphate) or pppGpp (guanosine pentaphosphate), collectively referred to here as ppGpp [10,11]. ppGpp binds to the transcriptional regulator RelA and modulates RNA polymerase activity in a promoter-specific fashion, down-regulating some genes and up-regulating others [12]. Although the stringent response can occur upon deprivation of phosphate, carbon, iron or fatty acids, it is best understood in response to amino acid deprivation. When intracellular amino acids are low, cognate uncharged tRNAs accumulate and can compete with binding of charged tRNAs to the A-site of ribosomes. When an uncharged tRNA occupies an A-site [13], translation slows and the ribosome-associated ppGpp synthase RelA is activated [11]. ppGpp, in complex with the transcriptional regulator DksA, can then repress stable RNA transcription by binding to RNA polymerase near the active site and inhibiting its activity [14]. At the same time, ppGpp/DksA can activate transcription of amino acid biosynthetic genes.
Uncharged tRNAs trigger the amino acid starvation response in eukaryotes by activating an eIF2α kinase
As in bacteria, intracellular uncharged tRNAs signal amino acid depletion in eukaryotic cells. Unlike bacteria, yeast have a dedicated uncharged tRNA-sensing molecule, the signaltransducing kinase GCN2 [15]. GCN2 has a domain with homology with HisRS (histidyl-tRNA synthetase) that binds to uncharged tRNAs [16,17], resulting in kinase activation through homodimerization and autophosphorylation [15,18]. Besides itself, activated GCN2 has only one other known target, eIF2α. Phosphorylation of eIF2α prevents efficient translational initiation at the starting methionine codon. This slows translation of most mRNAs while at the same time favouring the translation of select mRNAs with specific regulatory elements in their 5′-UTRs (untranslated regions). This phenomenon of selective translational up-regulation of specific mRNAs in the face of overall reduced global translation, known as translational derepression, is described in more detail later in the present review. One such derepressed mRNA encodes GCN4, the main effector of the GAAC (general amino acid control) pathway responsible for activating the transcription of amino acid biosynthetic and transport genes [19,20]. In mammals, the GCN4 orthologue controlling the transcriptional response to amino acid starvation, ATF4 (activating transcription factor 4), is similarly stabilized by translational derepression.
Conditions leading to GCN2 activation
In theory, any uncharged tRNA can bind the HisRS-like domain of GCN2. In yeast, a handful of conditions leading to accumulation of uncharged tRNAs have been experimentally verified to activate GCN2. These conditions include amino acid depletion, mutation of aminoacyl-tRNA synthetase genes or chemical inhibition leading to the accumulation of tRNAs for histidine, tryptophan, lysine, arginine, serine and the BCAAs (branched-chain amino acids) leucine, isoleucine and valine [17]. Interestingly, deprivation of a single amino acid can result in deacylation of non-cognate tRNAs as well. For example, leucine starvation of auxotrophic yeast results in accumulation of uncharged serine and threonine tRNAs in addition to leucine tRNAs [21].
In mammals, GCN2 can be rapidly activated in brain and liver (but apparently not kidney) upon ingestion of a meal lacking EAAs including leucine, histidine, tryptophan or lysine [22–24]. GCN2 can also be activated by non-dietary depletion of amino acids [25]. For example, in the context of an acute stress such as trauma or sepsis, increased NO (nitric oxide) production by NOS (NO synthase) can rapidly deplete the conditional EAA arginine from the blood. Local depletion of the EAA tryptophan in the placenta by the tryptophan catabolizing enzyme IDO (indoleamine 2,3-dioxygenase) can anergize T-cells that might otherwise react against the fetus. This purposeful immunosuppression is dependent on GCN2 expression in T-cells [26]. Other amino-acid-catabolizing enzymes, such as asparaginase, have been co-opted as chemotherapeutic agents against blood-borne cancers; the response to this agent in mice is dependent on GCN2 [27]. Simulated amino acid starvation can also be achieved by blocking the charging of tRNAs with their cognate amino acid. Halofuginone, for example, is a competitive inhibitor of the prolyl-tRNA synthetase [28] that can activate the amino acid starvation response in vitro and in vivo [29,30]. It is used to treat a variety of maladies ranging from psoriasis (an autoimmune disorder) to cancer.
Thus depletion of essential, non-essential or conditionally essential amino acids by dietary, enzymatic or pharmacological means can activate the amino acid starvation response by increasing the concentration of uncharged tRNA species and activating GCN2. Because GCN2 regulates adaptive changes to perceived amino acid deficiency, mice lacking this protein appear normal in the absence of such a challenge. This is not the case for one of the prime effectors of the GCN2 response, ATF4, which is required not only for the response to amino acid insufficiency, but also for the normal anabolic response to insulin, mediated, at least in part, through mTORC [mTOR (mammalian TOR) complex] 1 [31,32]. Cells lacking ATF4 require excess non-EAAs including cysteine (or antioxidants such as glutathione or N-acetylcysteine) [33], and ATF4-knockout mice have multiple developmental abnormalities and are smaller than control littermates [34].
Specificity of the GCN2-dependent amino acid starvation response
GCN2 is one of a family of four kinases that share a common target, Ser51 of the translation initiation factor eIF2α. In addition to amino acid deficiency, GCN2 can also be activated by glucose deprivation and UV irradiation [35]. The other three eIF2α kinases are activated by diverse signals in different tissues, ranging from haem deficiency in erythrocytes [HRI (haem-regulated eIF2α kinase)] to endoplasmic reticulum stress in pancreatic β-cells {PERK [PKR (double-stranded-RNA-dependent protein kinase]-like endoplasmic reticulum kinase} to infectious or metabolic stress in a variety of tissues (PKR). Global translational suppression and translational derepression of targets such as ATF4 are not unique to GCN2-mediated eIF2α phosphorylation upon amino acid starvation. Nonetheless, some specificity of each response to the appropriate stimulus is maintained, possibly as a result of additional, unique targets [36]. PKR, for example, has other direct targets besides eIF2α, including protein phosphatase 2A [37]. Although GCN2 has no known direct targets besides eIF2α, DNA-PK (DNA-dependent protein kinase) can be phosphorylated in a GCN2-dependent manner upon exposure of cells to UV [38].
SENSING AMINO ACID SUFFICIENCY: ROLE OF THE TOR SIGNAL TRANSDUCTION PATHWAY
Amino acid, energy and growth factor signalling integrated through TOR
Our present understanding of the ability of cells to sense the presence, rather than absence, of specific amino acids is based on genetic and pharmacological perturbation of the signal transduction cascade involving the TOR serine/threonine kinase. TOR is best known for controlling cell size and proliferation in response to the presence of adequate energy, nutrients and growth factors. TOR was first identified as the cellular target of the growth-inhibiting bacterial compound rapamycin, and is conserved from yeast to humans. Yeast has two TOR isoforms: TOR1 and TOR2, which are functionally distinct. Mammalian cells have a single TOR kinase, mTOR, which exists in two structurally and functionally distinct multi-protein complexes: mTORC1 and mTORC2. mTORC1 is acutely inhibited by rapamycin and is sensitive to nutrient levels, whereas mTORC2 is neither readily sensitive to nutrients nor acutely inhibited by rapamycin [39].
The kinase activity of mTORC1 is regulated by its association with the Ras-related GTPase Rheb (Ras homologue enriched in brain). In its GTP-bound form, Rheb directly activates mTORC1. The Rheb-specific GAP (GTPase-activating protein) is the TSC (tuberous sclerosis complex) 2 protein, which functions as a heterotrimer with its binding partners TSC1 and TBC1D7 [40,41]. The TSC1–TSC2–TBC1D7 complex is the critical negative regulator of mTORC1 signalling with respect to most of its upstream regulators, such as growth factors, insulin and energy levels [42]. The best-characterized downstream effectors of mTORC1 signalling include 4E-BP1 and 4E-BP2 (eIF4E-Binding proteins 1 and 2), and the p70 ribosomal protein S6K1 and S6K2 (S6 kinases 1 and 2). These proteins are directly phosphorylated by mTORC1, and are thus commonly used as markers of TOR activity [43].
Select amino acids regulate TORC1 signalling
Amino acid levels regulate both protein synthesis and autophagic proteolysis. Evidence supporting the role of mTORC1 in this regulation came from cell culture studies in which phosphorylation of mTORC1 substrates S6K and 4E-BP were associated with translational control and inhibition of autophagy upon amino acid stimulation [44–48]. In rat adipocytes, addition of amino acids to the medium stimulates S6K phosphorylation, which is sensitive to rapamycin treatment [46]. In CHO (Chinesehamster ovary) cells, amino acid withdrawal results in a rapid dephosphorylation of S6K and 4E-BP1 and abolishes the ability of growth factors to stimulate S6K activity. Re-addition of single amino acid dropout mixtures lacking leucine or arginine reduces S6K activity by 90% or 70% respectively [45]. Interestingly, re-addition of either leucine or arginine individually is unable to stimulate S6K activity. Similar studies carried out in different contexts and cell types, including H4IIE hepatoma, L6 muscle and pancreatic β-cells, revealed important details about the regulatory properties of individual amino acids on TOR activity. All agreed on the prominent role of leucine [47,49–51]. However, in certain contexts, arginine stimulates S6K activity [52], whereas in others each of the BCAAs (leucine, isoleucine and valine) has similar potency in stimulating 4E-BP1 phosphorylation [47]. The non-EAA glutamine can also modulate leucine- and argininestimulated mTORC1 signalling, although in different directions in different cell types. In rat intestinal epithelial cells and myogenic C2C12 cells, glutamine antagonizes arginine- and leucinemediated mTORC1 activation respectively [52–54]. In isolated rat hepatocytes, leucine and glutamine work additively to increase mTORC1 signaling [50]. In HeLa cells, the interaction between glutamine and leucine has been ascribed to a heterodimeric bidirectional amino acid antiporter consisting of SLC7A5 (solute carrier family 7 member 5)/SLC3A2 (solute carrier family 3 member 2) that regulates the simultaneous efflux of glutamine and influx of leucine and other EAAs [55]. Interestingly, in a previous study, carried out in a variety of cell types, it was shown that leucine and glutamine co-operate to activate mTORC1 signalling through enhancing glutaminolysis and 2-oxoglutarate (α-ketoglutarate) production [56].
In yeast auxotrophs, deprivation of leucine (and to a lesser degree lysine and histidine) reversibly inhibits TORC1-dependent phosphorylation of Sch9 (the yeast orthologue of S6K). Inhibition of protein synthesis with cyclohexamide results in accumulation of intracellular free amino acids, with leucine accumulating more than others, and increases Sch9 phosphorylation [57]. Taken together these observations are consistent with leucine primacy in TOR activation crossing evolutionary boundaries.
Bridging the GAP between leucine sensing and TOR activation
Unlike the GCN2 pathway, the most upstream mechanisms directly responsible for amino acid sensing permissive of TOR activation, and even the subcellular compartment in which this occurs, remain unresolved. TOR itself does not appear to bind amino acids or surrogates such as cognate tRNAs as GCN2 does. A number of candidates have been proposed as intermediates in TOR activation by amino acids. Some initial studies suggested a role for TSC1/TSC2 in amino acid sensing. However, budding yeast do not have TSC proteins, and mammalian TSC1- or TSC2-null cells are still sensitive to amino acid withdrawal, suggesting that amino acids signal to mTORC1 independently of TSC1/TSC2 [40,58]. hVps34 (human vacuolar protein sorting 34) has also been proposed to participate in amino acid sensing. This class III PI3K (phosphoinositide 3-kinase) participates in the regulation of mTORC1 by amino acids, possibly through its effects on vesicle trafficking and/or through generation of membrane compartments necessary for mTORC1 activation [59,60].
Two independent groups, using complementary biochemical and genetic approaches, showed that the Rag family of small GTPases is required for amino acid sensing by mTORC1 [61,62]. Rag GTPases are orthologues of the yeast Gtr1p and Gtr2p GTPases, which function as obligate heterodimers and regulate the Gap1 amino acid permease and microautophagy. The critical role of Rags in mTORC1 signalling and its regulation of cellular growth is further evidenced by in vivo experiments in Drosophila, where constitutively active RagA leads to flies with increased cell and wing size, whereas dominant-negative RagA leads to flies with decreased wing size [62]. Mammalian Rags also exist in heterodimers with one Gtr1p-like (RagA or RagB) and one Gtr2p-like (RagC or RagD) partner [63]. Unlike the Rheb GTPase, purified Rag GTPases are unable to stimulate mTORC1 kinase activity in vitro [62].
Instead, Rag-mediated mTORC1 activation involves recruitment of the complex to the outer lysosomal membrane, where mTORC1 can interact with its activator Rheb [61,64]. The Rag heterodimer is anchored to the lysosomal membrane via the protein complex called ‘Ragulator’ [64,65] and activated by the presence of amino acids. RagA/B in its GTP-bound form with RagC/D in its GDP-bound form constitutes the active heterodimer, which recruits mTORC1 to the lysosomal membrane through an interaction with Raptor (regulatoryassociated protein of mTOR). The Ragulator complex is functionally analogous to the yeast EGO complex due to its interaction with Gtr1p and Gtr2p, and its regulation of amino acid signalling to TOR at the vacuolar membrane [57,64]. Thus spatial regulation of the mTORC1 complex has emerged as an important aspect of amino-acid-mediated control [66].
Lysosomes/vacuoles are a major site of protein degradation and amino acid recycling with high concentrations of free amino acids. On the basis of mTORC1 localization and activation at the cytoplasmic face of the lysosomal membrane, the relevant amino-acid-sensing mechanism has been proposed to sense lysosomal rather than cytoplasmic free amino acid pools [64]. A Drosophila siRNA (small interfering RNA) screen targeting the lysosomal components that might be involved in amino acid signalling to TOR revealed the requirement of V-ATPase (vacuolar ATPase), and this was confirmed in mammalian cells [67]. Although the ATPase activity of V-ATPase is required for amino acid permissive signalling, V-ATPase does not function in transport of amino acids between the cytoplasm and the lysosome. V-ATPase does, however, appear to function upstream of the Rag–Ragulator interaction, in which amino acids stimulate GEF (guaninenucleotide-exchange factor) activity of the pentameric Ragulator complex towards RagA and RagB [68]. In further support of the importance of lysosomal free amino acid pools, stimulation of amino-acid-starved cells with radioactively labelled amino acids leads to their rapid appearance in the lysosome [67].
At face value, this lysosome-centred rather than cytoplasmiccentred view of amino acid sensing appears to be at odds with a previous study identifying the leucyl-tRNA synthetase as the intracellular leucine sensor responsible for mTORC1 activation [69]. siRNA directed at the leucyl-tRNA synthetase renders HEK (human embryonic kidney)-293T cells unable to phosphorylate S6K in response to leucine or isoleucine withdrawal and restimulation. Consistent with its requirement for leucine-permissive mTORC1 activation, the leucyl-tRNA synthetase also localizes to the cytoplasmic face of the lysosomal membrane, interacts with the Rag GTPase and has GAP activity specifically towards RagD, promoting the active GDP-bound form in the presence of leucine. Its ability to stimulate mTORC1 is dependent on leucine binding, but is independent of its ability to charge leucine tRNA. Nonetheless, the subcellular localization of the free leucine, and whether other TOR-permissive amino acids such as arginine function through similar mechanisms, remain to be seen.
MECHANISMS OF GCN2- COMPARED WITH TOR-BASED TRANSLATIONAL CONTROL
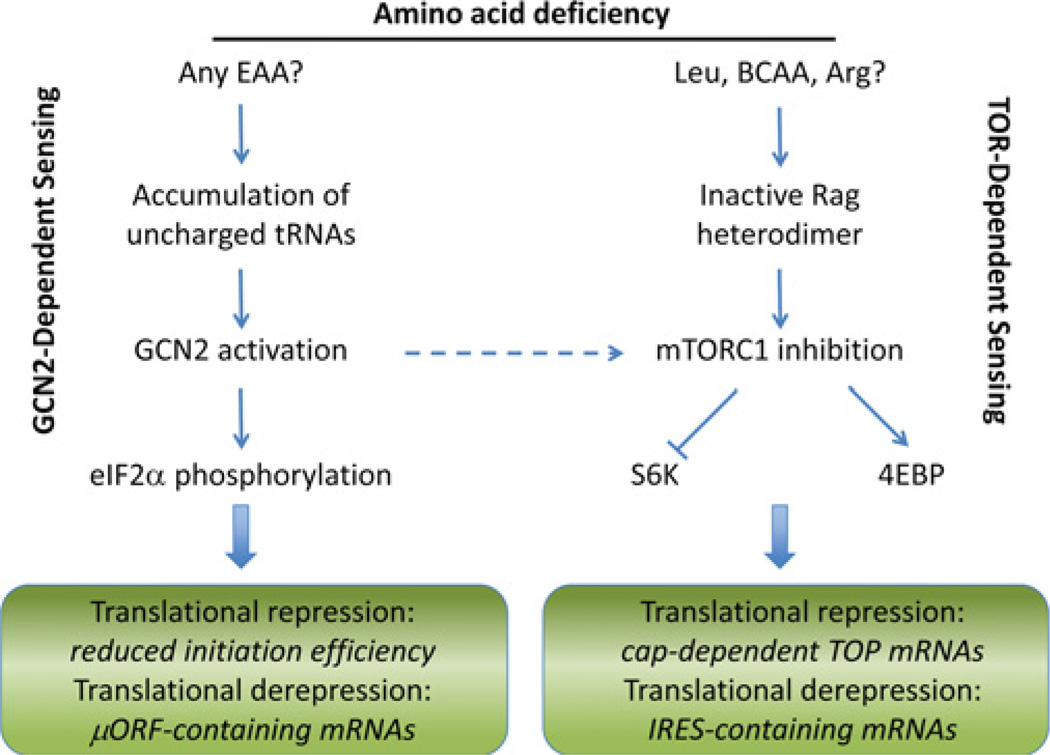
Translation of mRNA is one of the most energy requiring processes in the cell. It must therefore be coupled to the availability of cellular energy and coding amino acids. The immediate response to energy or amino acid insufficiency is to repress translation. At the same time, cells increase translation of specific messages involved in metabolic adaptation, for example transcription factors controlling the expression of amino acid biosynthetic and transport genes. This process is known as translational derepression. Just asGCN2 and TOR pathways sense amino acids by different mechanisms, so do they differ in the ways they accomplish translational repression and derepression (Figure 1).
Figure 1. Integration of amino acid sensing with translational control.
Two signal transduction mechanisms are involved in sensing intracellular amino acids: GCN2 senses the absence of many amino acids and the TOR pathway senses the presence of particular amino acids. Both repress general translation and derepress translation of specific messages through distinct mechanisms.
Translational regulation by GCN2
Translation initiation occurs upon pairing of the AUG start codon with Met-tRNAiMet (initiator methionyl-tRNA), which is part of the so-called ternary complex together with eIF2 and GTP. This is followed by hydrolysis of eIF2-bound GTP and release of eIF2-GDP from the 43S ribosome. Phosphorylation of the α subunit of eIF2 by GCN2 inhibits GDP/GTP exchange by eIF2B, reducing ternary complex availability and thus repressing initiation of translation of most mRNAs.
Some mRNAs, however, contain short upstream ORFs (open reading frames), or µORFs (microORFs), that influence translation of the downstream full-length ORF depending on the status of eIF2α phosphorylation. Reinitiation of translation following µORF translation requires reassociation of the ternary complex. This depends both on the concentration of ternary complex (and hence eIF2α phosphorylation status) and the scanning distance/ time to the next AUG codon. Increasing the distance between µORFs increases the chance of ternary complex association before reaching the next AUG start codon [70]. Under non-starvation conditions, reinitiation of translation after the first µORF is relatively efficient, decreasing the chance of translation of the downstream full-length ORF. Under amino-acid-starvation conditions, reinitiation after the first µORF is less efficient, increasing the chance that the ribosome will scan past subsequent µORFs and reinitiate translation further downstream at the full-length ORF. Thus the distance between µORFs and timing of ribosomal reinitiation are critical factors in translational derepression of mRNAs such as GCN4 upon intracellular amino acid starvation. Stabilization of ATF4 occurs by a similar mechanism [71], indicating conservation of this simple, yet elegant, translational control mechanisms from yeast to mammalian cells.
Translational regulation by mTOR
The mTORC1 pathway controls protein synthesis most directly by phosphorylating and inhibiting a repressor of cap-dependent mRNA translation, 4E-BP1. Translation of most cellular mRNAs is initiated via this common mechanism involving assembly of eIF4F (comprising eIF4E, eIF4G and eIF4A) on the 5′ 7-methyl-GTP cap. eIF4F is required for the loading of the small ribosomal subunit on the mRNA. 4E-BPs repress translation by inhibiting the formation of the eIF4F complex [72]. Phosphorylation of 4E-BPs by mTORC1 results in their dissociation from eIF4E, facilitating cap-dependent translation [40]. Acute inhibition of mTORC1 with rapamycin, or the more potent inhibitor Torin 1, reduces protein synthesis by approximately 30 % or 60 % respectively, without affecting the status of eIF2α phosphorylation [25]. Torin 1 inhibits translation of nearly all (99.8 %) cellular mRNAs, but the magnitude of repression is greatest for mRNAs containing 5′ TOP (terminal oligopyrimidine) motifs in their 5′-UTRs. Importantly, this class of mRNAs encodes all ribosomal proteins and major translation factors, therefore influencing the protein synthetic capacity of the cell. Torin-1-mediated translational repression requires 4E-BPs and is lost in cells lacking these proteins [25].
mTORC1 also regulates translation indirectly through phosphorylation of S6K. Phosphorylation on Thr389 activates S6K towards ribosomal protein S6, a component of the 40S ribosome important for translation in rapidly proliferating cells. It also phosphorylates a number of other downstream targets including eIF4B, eEF2K (eukaryotic translation elongation factor 2 kinase) and SKAR [72]. Phosphorylation of S6K targets can promote translation by a variety of direct and indirect mechanisms, including regulation of eIF4B recruitment, inhibition of PDCD4 (programmed cell death protein 4)-mediated inhibition of eIF4A, splicing-dependent translation of some mRNA messages via SKAR, and regulation of ribosome biogenesis [73,74]. The ability of amino acids to regulate phosphorylation of both S6K and 4E-BPs was shown in previous studies [47].
Although mTORC1 inhibition represses cap-dependent translation, it also selectively derepresses translation of select mRNAs that do not depend on cap-binding for ribosome entry. Conceptually, this parallels translational derepression upon GCN2 activation. However, instead of a µORF, it depends on an IRES (internal ribosome entry site) in the 5′-UTR for cap-independent ribosome recruitment. IRES-containing mRNAs are not only resistant to mTORC1 inhibition by Torin 1, but are in fact translated with greater efficiency under such conditions [25]. IRES-containing mRNAs are often involved in the response to stress, for example NRF2 (NF-E2-related factor 2), the master transcriptional regulator of genes in the Phase II response to oxidative/electrophilic stress. NRF2 is translationally up-regulated during oxidative stress despite the attenuation of global protein synthesis due to an IRES-dependent mechanism [75,76]. However, whether NRF2 is translationally derepressed upon amino acid starvation or mTORC1 inhibition remains to be elucidated.
Protein deficiency is expected to simultaneously repress mTOR and activate GCN2. How can efficient translational derepression occur when mRNA cap-binding and translation initiation are both inhibited by mTORC1 repression and GCN2 activation respectively? One possibility is that mRNAs subject to control by GCN2/eIF2α lack TOP motifs, limiting the effect of mTORC1/4E-BP-based repression on cap-binding. Another possibility is that IRES-containing mRNAs also contain µORFs; CAT1 (cationic amino acid transporter 1) is an example of such an mRNA [77]. IRES-mediated translational derepression can also occur despite phosphorylation of eIF2α. For example, translation of XIAP (X-linked inhibitor of apoptosis) occurs via an eIF2α-independent mechanism of translation initiation dependent on eIF5B [78]. Thus translational derepression probably depends on features within a particular 5′-UTR, as well as the status of both GCN2 and TOR activity.
CO-ORDINATION OF AMINO ACID STARVATION RESPONSES THROUGH GCN2 AND TOR
Why did separate amino-acid-sensing pathways evolve together in eukaryotes? Are they redundant or do they play separate roles? What is the connection between the two? Amino acid sensing through the TOR pathway may have evolved primarily to coordinate information on the presence of adequate nutrients (including amino acids), energy and other environmental conditions favourable for growth. Leucine and the other BCAAs are abundant in a variety of proteins. In mammals, they are also the only free amino acids to increase in peripheral blood after a meal in proportion to their levels in the diet (the remaining 17 are retained in the gut and liver and released in a controlled fashion) [79]. These qualities of BCAAs may be advantageous for a surrogate marker of availability of all amino acids.
GCN2-based sensing of individual amino acid deficiencies may have evolved for different reasons. Dietary amino acid deficiencies are common and must be detected rapidly in order to avoid negative effects ranging from reduced growth to fatty liver [80]. Protein sources with amino acid deficiencies include rice (low amounts of the EAA lysine), casein (limiting for both tryptophan and methionine) and gelatin (lacks tryptophan altogether). Xenobiotic anti-metabolites targeting individual amino acid metabolic pathways (for example, tRNA synthetase inhibitors) may also be widespread in Nature. Interestingly, yeast can make all 20 coding amino acids, and thus do not activate the GAAC response when amino acids are absent (for example in minimal growth media) [81]. However, they do activate the response when individual amino acids are deficient or in the presence of tRNA synthetase inhibitors. Thus the GCN2-based GAAC response may have evolved under different selective pressures than the TOR-based response, although both function to optimize growth potential and prioritize metabolic demands to fit environmental conditions.
Are responses to amino acid deprivation through GCN2 and TOR co-ordinated? In yeast, GCN4 translation is derepressed upon inhibition of TOR with rapamycin in a GCN2- and TOR1-dependent fashion [82]. Mechanistically, TOR inhibition may reduce inhibitory phosphorylation of GCN2, thus promoting eIF2α phosphorylation and GCN4 translational derepression. GCN4 is also a major effector of the transcriptional response to TOR inhibition by rapamycin on a scale equivalent to the canonical transcriptional activator and TOR substrate GLN3 [83,84]. Genes co-regulated by both GCN4 and GLN3 include those involved in nitrogen assimilation, amino acid transport, amino acid biosynthesis and other transcriptional activators.
Cross-talk between GCN2 and mTOR also exists in mammalian cell culture and animal models, although the direction of interaction appears to be different than in yeast. In human lymphocytic leukaemic cell lines, treatment with L-asparaginase, an enzyme that degrades asparagine in culture medium and activates GCN2, inhibits mTORC1 phosphorylation of its targets S6K and 4E-BP1 in a dose-dependent fashion [85]. Likewise, S6K activity is reduced in human T-lymphoblastoid cells exposed to various amino acid alcohols that selectively inhibit specific tRNA loading, including L-histidinol, L-leucinol, L-phenylalaninol and L-methioninol [86]. Both studies suggest cross-talk between GCN2 and mTORC1 signalling, with uncharged tRNAs initiating the changes in signal transduction.
Genetic evidence in mice is also consistent with GCN2 activation occurring upstream of mTORC1 repression. In response to dietary leucine deprivation or asparaginase treatment, phosphorylation of mTORC1 targets S6K and 4E-BP1 is reduced in the liver and pancreas, and this depends on the GCN2 kinase [24,27]. Leucine-deficient diets also improve insulin signalling in the liver as measured by increased phosphorylation of the insulin receptor, and whole-body insulin sensitivity as measured by an insulin tolerance test [87]. However, in GCN2-knockout mice on a leucine-deficient diet, mTOR signalling in the liver is increased and the improvement in insulin sensitivity is lost. Interestingly, GCN2-knockout mice on a leucine-deficient diet also develop hepatic steatosis due to increased expression of SREBP (sterol-regulatory-element-binding protein) 1c-dependent lipogenic genes including Fasn (fatty acid synthase), ApoC4 (apolipoprotein C-IV) and G6pd (glucose-6-phosphate dehydrogenase), which are under the control of mTORC1 in the liver. This suggests a model in which failure to repress mTORC1 allows inappropriate activation of SREPB1c targets [88,89]. Potentially complicating this model is the fact that leucine deprivation on its own would be predicted to reduce mTORC1 activity by GCN2-independent mechanisms. Nonetheless, taken together these findings are consistent with a model in which GCN2 activation by pharmacological or dietary means can suppress mTORC1 activity.
By what mechanisms could GCN2 inhibit mTORC1 in mammals? Three ATF4 transcriptional targets may contribute to reduced mTORC1 signalling downstream of GCN2 activation. GADD34 (growth-arrest and DNA-damage-inducible protein 34) is a phosphatase that removes an inhibitory phosphate on TSC2, thus repressing mTORC1, whileatthe same time turning down the GCN2 responsebyremoving the phosphate on Ser51 of eIF2α [90]. 4E-BP1[91] and the TSC activator/mTORC1 repressor REDD1 (regulated in development and DNA damage responses 1) [92], can also be up-regulated at the transcriptional level by ATF4 upon endoplasmic reticulum stress, although their ability to effect mTORC1 activity following eIF2α phosphorylation by GCN2 has not been demonstrated [27].
AMINO ACID SENSING IN DR BENEFITS
Dietary protein/amino acid modulation and longevity
DR has been intricately associated with aging research since the discovery in the 1930s that reduced food intake extends longevity of experimental rodents [1]. Today, we know that the benefits of DR occur in a variety of organisms and are pleiotropic in nature. These benefits include enhanced metabolic fitness and improved resistance to multiple forms of acute stress, ranging from heat shock to surgical ischaemia/reperfusion injury. Still, lifespan extension in a variety of model organisms is the most studied and perhaps best understood of its pleiotropic benefits.
Classically, DR has been described as reduced food intake without malnutrition. DR is not a single intervention, but it rather loosely describes a variety of interventions ranging widely in both dietary composition and timing of food intake [93]. Despite a number of studies aimed at dissecting the nutritional basis of DR, no consensus yet exists onthe relative contributions of overall reduced calorie intake compared with the restriction of particular macronutrients such as protein [4].
In yeast, chronological lifespan can be extended by restriction of asparagine, glutamate or methionine in the medium, in addition to deletion of TOR1 or Sch9, inhibition of glutamine synthetase and rapamycin treatment [94]. Unfortunately in worms, another well-characterized model organism in which the genetics of longevity extension and DR are particularly well characterized, the lack of purified diets and the ability of starvation to increase the lifespan of adults makes these questions difficult to address.
In fruitflies, reduction of the calories via titration of the sole source of protein, yeast extract, provides greater longevity extension than isocaloric reduction of sucrose [4]. Adding back purified EAAs to a restricted sucrose/yeast-based diet optimized for longevity abrogates lifespan extension, whereas adding back EAA minus methionine (or to a lesser degree tryptophan) does not [5]. Perhaps most unexpectedly, adding back methionine alone (but not other individual EAAs) on top of the DR regimen abrogated a well-known negative consequence of DR, reduced fecundity. Although the involvement of reduced insulin-like peptide signalling is implicated in longevity benefits [5], the status of GCN2 or TOR under conditions of DR and EAA add-back remains to be reported.
In rodents, a number of studies have reported moderate lifespan extension upon dietary protein restriction as with DR [95]. However, interpretation of DR and protein restriction studies are both complicated by the fact that protein and carbohydrates are readily interconverted in vivo. A different approach was to reduce individual dietary EAAs, namely tryptophan and methionine. It is not clear why only these two have been tested in rodents for longevity benefits. In the case of tryptophan, there is some evidenceofincreased maximal lifespan and a delay in aging-related phenotypes in at least a subset of tryptophan-restricted rats [7,96]. The underlying mechanistic hypothesis in these studies involved reduction of serotonin, a downstream metabolite of tryptophan. Likewise, methionine restriction has been shown to extend lifespan in male rats and mice [6,8].
Interestingly, tryptophan and methionine are the two least abundant EAAs by weight in casein, a protein commonly used in purified diets for experimental rodents. Thus DR regimens using casein as the sole source of protein would be expected, at some level of restriction, to become limited for these two EAAs. Indeed, there are many overlapping phenotypes shared both by DR and isolated methionine restriction, including reduced adiposity, extended maximal longevity, increased resistance to acetaminophen toxicity in the liver, reduced insulin and IGF1 (insulin growth factor 1) levels and reduced thyroid hormone [8]. Nonetheless, there are differences as well, and future studies will be necessary to address whether the benefits of each are derived from fundamentally distinct or overlapping mechanisms.
To the best of our knowledge, other amino-acid-restricted diets have not been tested for lifespan extension, but have been shown to induce a number of other benefits. For example, methionine restriction contributes to adiposity resistance by altering the lipogenic/lipolytic balance [97], leucine deprivation improves insulin sensitivity [87] and tryptophan deprivation protects against surgical ischaemia/reperfusion injury to both kidney and liver [29]. The latter two benefits are absent from mice lacking GCN2.
Role of GCN2 and TOR in protein/amino acid restriction benefits
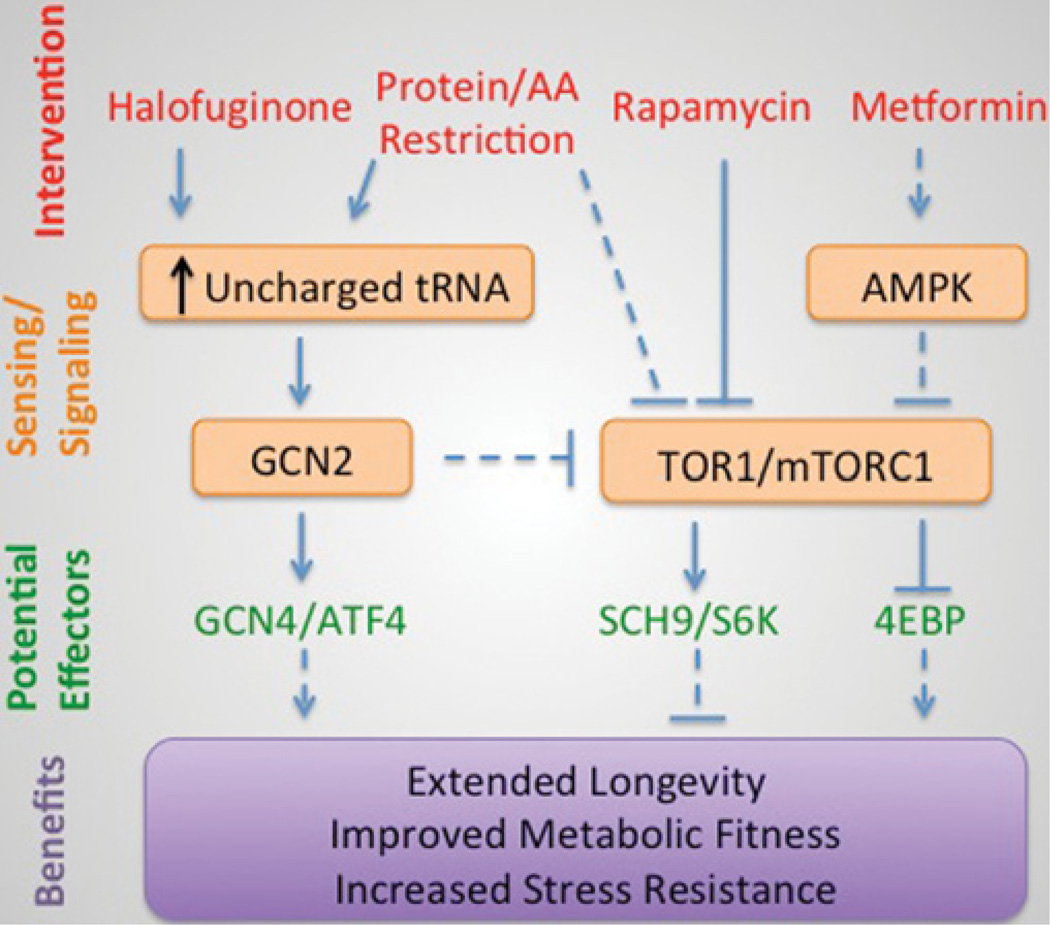
What roles do the TOR and/or GCN2 signal transduction pathways play in regulating longevity, metabolic fitness and stress resistance upon restriction of protein or specific amino acids? There is ample evidence that reduction of TOR signalling through genetic or pharmacological manipulation can extend longevity in a variety of organisms. For example, deletion of TOR1 extends lifespan in yeast and rapamycin treatment extends lifespan in rodents [98]. Genetic ablation of the downstream target of TOR, Sch9 in yeast and S6K in rodents, can also extend longevity [99,100]. In flies, 4E-BP is up-regulated upon DR and may contribute to translational derepression of nuclear-encoded mitochondrial electron transport chain components [101]. Because DR can reduce TOR signalling to downstream targets including S6K and 4E-BP, this could be a primary mechanism underlying its longevity effects (Figure 2).
Figure 2. Model for amino acid sensing pathways in DR benefits.
Modulation of activity of GCN2 or TOR pathways by dietary or pharmacological interventions can lead to benefits. Not all interventions necessarily lead to all benefits. Broken lines indicate indirect or hypothesized effects; solid lines indicate known or direct effects. In some cases, yeast/mammalian gene names are both indicated respectively. AMPK, AMP-activated protein kinase.
Aside from the essential role of GCN2 in surgical stress resistance by short-term tryptophan deficiency [29], there is little in the DR or longevity literature on the potential role of this amino-acid-deprivation sensing kinase. This is perhaps surprising in light of established models of longevity extension involving amino acid restriction in rodents or amino acid imbalance in flies, combined with data suggesting the ability of GCN2 activation to inhibit mTOR activity upon amino acid deprivation. It may be that redundancy in mammalian eIF2α kinases, as well as multiple additional routes of mTOR inhibition, may abrogate the specific genetic requirement for GCN2 in a number of settings. Nonetheless, activation of GCN2 via dietary or anti-metabolite means would seem a productive area of future research, including the role of translational control in downstream benefits (Figure 2).
In yeast, GCN4 stabilization has been implicated in both chronological and replicative longevity extension [102,103]. In the replicative longevity model, lifespan is extended by knocking out any of a number of 60S ribosomal subunits or inhibiting 60S subunit biogenesis with small molecules [102]. By reducing the 60S subunit, translational initiation becomes inefficient, mimicking the effects of GCN2 activation and eIF2α phosphorylation on downstream GCN4 stabilization. Thus transcriptional reprogramming by GCN4 may play a role in longevity extension, whether or not it is stabilized upon GCN2 activation or by some other process.
In addition to translational control, GCN2 and TOR control a number of other processes that could also contribute to the reported benefits of DR, including autophagy, energy metabolism, immune function and food intake. Autophagy is an adaptive process that provides biological material (amino acids and lipids) to sustain anabolic processes [104] under conditions of nutrient depletion, including DR. mTORC1 negatively regulates autophagy by suppressing the ULK1 complex via phosphorylation [43]. In yeast, GCN2 and GCN4 are required for the induction of autophagy upon amino acid depletion [105,106]. DR also results in changes in metabolic pathways regulated, at least in part, by mTORC1, including fatty acid synthesis, glycolysis and the pentose phosphate pathway via the transcription factors HIF1α (hypoxia-inducible factor 1α) and SREBP [89,107]. GCN2 can also have an effect on fat metabolism. In addition to activation of amino acid biosynthesis and transport, GCN2 is required for suppression of fatty acid synthesis in the liver upon leucine deprivation [88]. Although this particular function of GCN2 is independent of ATF4, the broader role of this transcription factor in GCN2-dependent effects remains to be explored.
Immune function, including autoimmunity, can have a major impact on lifespan and aging-related disease in multicellular organisms. Activation of GCN2 upon local tryptophan depletion by the tryptophan-catabolizing enzyme IDO is one strategy to reduce T-cell proliferation and induce tolerance towards such unintended targets as apoptotic cells in the spleen [26] or the allogeneic fetus. Activation of the amino acid starvation response by the small molecule halofuginone, a prolyl-tRNA synthase inhibitor [28], prevents differentiation of inflammatory Th17 cells [30]. mTOR inhibition by rapamycin can also induce tolerance by suppressing T-cell proliferation, but is paradoxically pro-inflammatory in the context of innate immune activation [108,109].
Finally, GCN2 and mTOR can both participate through different mechanisms in behavioural control of food intake. Central administration of leucine activates hypothalamic mTOR and reduces food intake [110]. Diets lacking one or more EAAs cause an aversion to food intake centred in a different brain region, the APC (anterior piriform cortex) [111]. GCN2 is activated rapidly in the APC upon dietary amino acid deprivation or stereotactic injection of amino acid alcohol derivatives that compete for tRNA synthetases, resulting in uncharged tRNA accumulation [3,23]. Nonetheless, mice lacking GCN2 still display aversion to incomplete diets over the period of days to weeks [24,29,88], suggesting a redundant mechanism of amino acid deprivation sensing. Future studies will be required to elucidate the role of food aversion, if any, to the benefits of amino acid restriction.
Pharmacological activators of the amino acid starvation response as DR mimetics?
DR mimetics can be loosely defined as interventions that mimic some beneficial aspect of DR, for example lifespan extension, maintenance of metabolic fitness upon challenge with a high-fat diet or increased stress resistance. Metformin and rapamycin both extend lifespan in rodents, and are thus considered DR mimetics [98,112] (Figure 2). Halofuginone is a prolyl-tRNA synthetase inhibitor that activates the amino acid starvation response by mimicking proline deprivation. Like short-term EAA deprivation, halofuginone can increase resistance to renal ischaemia/reperfusion injury in a GCN2-dependent manner [29]. This serves as proof-of-principle that compounds that activate GCN2 and stimulate the amino acid starvation response can have benefits similar to dietary protein/amino acid restriction. Nonetheless, this anti-metabolite and others in its class are toxic due to on-target effects of tRNA synthetase inhibition. More desirable would be a DR mimetic that would activate the GCN2 kinase directly without inhibiting tRNA charging, or selectively inhibit amino acid sensing through the mTOR pathway. Our understanding of the mechanisms underlying amino acid sensing will probably improve the chances of identifying such a compound with potential beneficial uses in humans.
ACKNOWLEDGEMENTS
We thank Brendan Manning, Chris Hine and members of the Mitchell laboratory for critical feedback on the text before submission.
FUNDING
Work of the authors is funded, in part, by the National Institutes of Health National Institute on Aging [grant number AG036712], National Institute of Diabetes and Digestive and Kidney Diseases [grant number DK090629], Interdisciplinary Training in Genes and the Environment [grant number T32 ES016645], the American Federation for Aging Research, the Ellison Medical Foundation and the Glenn Foundation for Medical Research.
Abbreviations used
- APC
anterior piriform cortex
- ATF4
activating transcription factor 4
- BCAA
branched-chain amino acid
- DR
dietary restriction
- EAA
essential amino acid
- 4E-BP
eIF4E-Binding protein
- eIF
eukaryotic initiation factor
- GAAC
general amino acid control
- GAP
GTPase-activating protein
- GCN
general amino acid control non-derepressible
- HisRS
histidyl-tRNA synthetase
- IDO
indoleamine 2,3-dioxygenase
- IRES
internal ribosome entry site
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- NRF2
NF-E2-related factor 2
- ORF
open reading frame
- µORF
microORF
- PKR
double-stranded-RNA-dependent protein kinase
- Rheb
Ras homologue enriched in brain
- siRNA
small interfering RNA
- S6K
S6 kinase
- SREBP
sterol-regulatory-element-binding protein
- TOP
terminal oligopyrimidine
- TOR
target of rapamycin
- TSC
tuberous sclerosis complex
- UTR
untranslated region
- V-ATPase
vacuolar ATPase
REFERENCES
- 1.McCay CM, Crowel MF, Maynard LA. The effect of retarded growth upon the length of the life span and upon the ultimate body size. J. Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 2.Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci. SAGE KE. 2003;2003 doi: 10.1126/sageke.2003.8.re2. re2. [DOI] [PubMed] [Google Scholar]
- 3.Ross MH. Length of life and nutrition in the rat. J. Nutr. 1961;75:197–210. doi: 10.1093/jn/75.2.197. [DOI] [PubMed] [Google Scholar]
- 4.Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila . PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila . Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J. Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 7.Segall PE, Timiras PS. Patho-physiologic findings after chronic tryptophan deficiency in rats: a model for delayed growth and aging. Mech. Ageing Dev. 1976;5:109–124. doi: 10.1016/0047-6374(76)90012-9. [DOI] [PubMed] [Google Scholar]
- 8.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 10.Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 11.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol. Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 12.Cashel M, Gentry DM, Hernandez VJ, Vinella D. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology Press; 1996. [Google Scholar]
- 13.Goldman E, Jakubowski H. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol. Microbiol. 1990;4:2035–2040. doi: 10.1111/j.1365-2958.1990.tb00563.x. [DOI] [PubMed] [Google Scholar]
- 14.Krohn M, Wagner R. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J. Biol. Chem. 1996;271:23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- 15.Diallinas G, Thireos G. Genetic and biochemical evidence for yeast GCN2 protein kinase polymerization. Gene. 1994;143:21–27. doi: 10.1016/0378-1119(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 16.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 17.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2a protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acid. Mol. Cell. Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narasimhan J, Staschke KA, Wek RC. Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 2004;279:22820–22832. doi: 10.1074/jbc.M402228200. [DOI] [PubMed] [Google Scholar]
- 19.Bushman JL, Asuru AI, Matts RL, Hinnebusch AG. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci., U.S.A. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, Gietzen DW. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 24.Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J. Biol. Chem. 2004;279:36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 25.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, Mellor AL. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J. Biol. Chem. 2009;284:32742–32749. doi: 10.1074/jbc.M109.047910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller TL, Zocco D, Sundrud MS, Hendrick M, Edenius M, Yum J, Kim YJ, Lee HK, Cortese JF, Wirth DF, et al. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 2012;8:311–317. doi: 10.1038/nchembio.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng W, Robertson L, Gallinetti J, Mejia P, Vose S, Charlip A, Chu T, Mitchell JR. Surgical stress resistance induced by single amino acid deprivation requires gcn2 in mice. Sci. STKE. 2012;4 doi: 10.1126/scitranslmed.3002629. 118ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, Waldner H, et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J. Biol. Chem. 2007;282:16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 32.Malmberg SE, Adams CM. Insulin signaling and the general amino acid control response. Two distinct pathways to amino acid synthesis and uptake. J. Biol. Chem. 2008;283:19229–19234. doi: 10.1074/jbc.M801331200. [DOI] [PubMed] [Google Scholar]
- 33.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 35.Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 36.Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2α kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol. Genomics. 2009;38:328–341. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Williams BR. The B56α regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol. Cell. Biol. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Laplante M, Sabatini DM. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol. Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 45.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 46.Fox HL, Kimball SR, Jefferson LS, Lynch CJ. Amino acids stimulate phosphorylation of p70S6k and organization of rat adipocytes into multicellular clusters. Am. J. Physiol. 1998;274:C206–C213. doi: 10.1152/ajpcell.1998.274.1.C206. [DOI] [PubMed] [Google Scholar]
- 47.Xu G, Kwon G, Marshall CA, Lin TA, Lawrence JC, Jr, McDaniel ML. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic β-cells. A possible role in protein translation and mitogenic signaling. J. Biol. Chem. 1998;273:28178–28184. doi: 10.1074/jbc.273.43.28178. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Campbell LE, Miller CM, Proud CG. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J. Biol. Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 50.Krause U, Bertrand L, Maisin L, Rosa M, Hue L. Signalling pathways and combinatory effects of insulin and amino acids in isolated rat hepatocytes. Eur. J. Biochem. 2002;269:3742–3750. doi: 10.1046/j.1432-1033.2002.03069.x. [DOI] [PubMed] [Google Scholar]
- 51.Peyrollier K, Hajduch E, Blair AS, Hyde R, Hundal HS. L-Leucine availability regulates phosphatidylinositol 3-kinase, p70 S6 kinase and glycogen synthase kinase-3 activity in L6 muscle cells: evidence for the involvement of the mammalian target of rapamycin (mTOR) pathway in the L-leucine-induced up-regulation of system A amino acid transport. Biochem. J. 2000;350:361–368. [PMC free article] [PubMed] [Google Scholar]
- 52.Nakajo T, Yamatsuji T, Ban H, Shigemitsu K, Haisa M, Motoki T, Noma K, Nobuhisa T, Matsuoka J, Gunduz M, et al. Glutamine is a key regulator for amino acid-controlled cell growth through the mTOR signaling pathway in rat intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2005;326:174–180. doi: 10.1016/j.bbrc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Ban H, Shigemitsu K, Yamatsuji T, Haisa M, Nakajo T, Takaoka M, Nobuhisa T, Gunduz M, Tanaka N, Naomoto Y. Arginine and leucine regulate p70 S6 kinase and 4E-BP1 in intestinal epithelial cells. Int. J. Mol. Med. 2004;13:537–543. [PubMed] [Google Scholar]
- 54.Deldicque L, Sanchez Canedo C, Horman S, De Potter I, Bertrand L, Hue L, Francaux M. Antagonistic effects of leucine and glutamine on the mTOR pathway in myogenic C2C12 cells. Amino Acids. 2008;35:147–155. doi: 10.1007/s00726-007-0607-z. [DOI] [PubMed] [Google Scholar]
- 55.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 57.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 58.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 59.Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–2829. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 60.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaw RJ. mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem. Sci. 2008;33:565–568. doi: 10.1016/j.tibs.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki T, Inoki K. Spatial regulation of the mTORC1 system in amino acids sensing pathway. Biochim. Biophys. Acta. 2011;43:671–679. doi: 10.1093/abbs/gmr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+ -ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 70.Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 73.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 74.Richardson CJ, Broenstrup M, Fingar DC, Julich K, Ballif BA, Gygi S, Blenis J. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr. Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 75.Li W, Thakor N, Xu EY, Huang Y, Chen C, Yu R, Holcik M, Kong AN. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010;38:778–788. doi: 10.1093/nar/gkp1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of nrf2 protein in activation of antioxidant response by oxidants. Mol. Pharmacol. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 77.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thakor N, Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2α-independent manner during stress. Nucleic Acids Res. 2012;40:541–552. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Layman DK. The role of leucine in weight loss diets and glucose homeostasis. J. Nutr. 2003;133:261S–267S. doi: 10.1093/jn/133.1.261S. [DOI] [PubMed] [Google Scholar]
- 80.Harper AE, Benevenga NJ, Wohlhueter RM. Effects of ingestion of disproportionate amounts of amino acids. Physiol. Rev. 1970;50:428–558. doi: 10.1152/physrev.1970.50.3.428. [DOI] [PubMed] [Google Scholar]
- 81.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 82.Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J. Biol. Chem. 2003;278:20457–20460. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- 83.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 2010;285:16893–16911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iiboshi Y, Papst PJ, Hunger SP, Terada N. L-asparaginase inhibits the rapamycin-targeted signaling pathway. Biochem. Biophys. Res. Commun. 1999;260:534–539. doi: 10.1006/bbrc.1999.0920. [DOI] [PubMed] [Google Scholar]
- 86.Iiboshi Y, Papst PJ, Kawasome H, Hosoi H, Abraham RT, Houghton PJ, Terada N. Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J. Biol. Chem. 1999;274:1092–1099. doi: 10.1074/jbc.274.2.1092. [DOI] [PubMed] [Google Scholar]
- 87.Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, et al. Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes. 2011;60:746–756. doi: 10.2337/db10-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo F, Cavener DR. The GCN2 eIF2α kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell. Metabolism. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, Manning BD. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell. Metabolism. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watanabe R, Tambe Y, Inoue H, Isono T, Haneda M, Isobe K, Kobayashi T, Hino O, Okabe H, Chano T. GADD34 inhibits mammalian target of rapamycin signaling via tuberous sclerosis complex and controls cell survival under bioenergetic stress. Int. J. Mol. Med. 2007;19:475–483. [PubMed] [Google Scholar]
- 91.Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, Munakata Y, Satake C, Katagiri H, Tashiro F, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell. Metabolism. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 92.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res. Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 94.Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell. Mol. Life Sci. 2007;64:752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pamplona R, Barja G. Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochim. Biophys. Acta. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 96.Ooka H, Segall PE, Timiras PS. Histology and survival in age-delayed low-tryptophan-fed rats. Mech. Ageing Dev. 1988;43:79–98. doi: 10.1016/0047-6374(88)90099-1. [DOI] [PubMed] [Google Scholar]
- 97.Perrone CE, Mattocks DA, Hristopoulos G, Plummer JD, Krajcik RA, Orentreich N. Methionine restriction effects on 11-HSD1 activity and lipogenic/lipolytic balance in F344 rat adipose tissue. J. Lipid Res. 2008;49:12–23. doi: 10.1194/jlr.M700194-JLR200. [DOI] [PubMed] [Google Scholar]
- 98.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 101.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila . Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae . Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ecker N, Mor A, Journo D, Abeliovich H. Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy. 2010;6:879–890. doi: 10.4161/auto.6.7.12753. [DOI] [PubMed] [Google Scholar]
- 106.Talloczy Z, Jiang W, Virgin HW, III, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 109.Fielhaber JA, Carroll SF, Dydensborg AB, Shourian M, Triantafillopoulos A, Harel S, Hussain SN, Bouchard M, Qureshi ST, Kristof AS. Inhibition of mammalian target of rapamycin augments lipopolysaccharide-induced lung injury and apoptosis. J. Immunol. 2012;188:4535–4542. doi: 10.4049/jimmunol.1003655. [DOI] [PubMed] [Google Scholar]
- 110.Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 111.Gietzen DW, Hao S, Anthony TG. Mechanisms of food intake repression in indispensable amino acid deficiency. Annu. Rev. Nutr. 2007;27:63–78. doi: 10.1146/annurev.nutr.27.061406.093726. [DOI] [PubMed] [Google Scholar]
- 112.Anisimov VN, Berstein LM, Egormin PA, Piskunova TS, Popovich IG, Zabezhinski MA, Kovalenko IG, Poroshina TE, Semenchenko AV, Provinciali M, et al. Effect of metformin on life span and on the development of spontaneous mammary tumors in HER-2/neu transgenic mice. Exp. Gerontol. 2005;40:685–693. doi: 10.1016/j.exger.2005.07.007. [DOI] [PubMed] [Google Scholar]