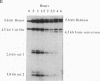
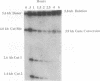
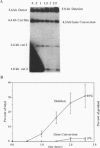
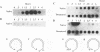Abstract
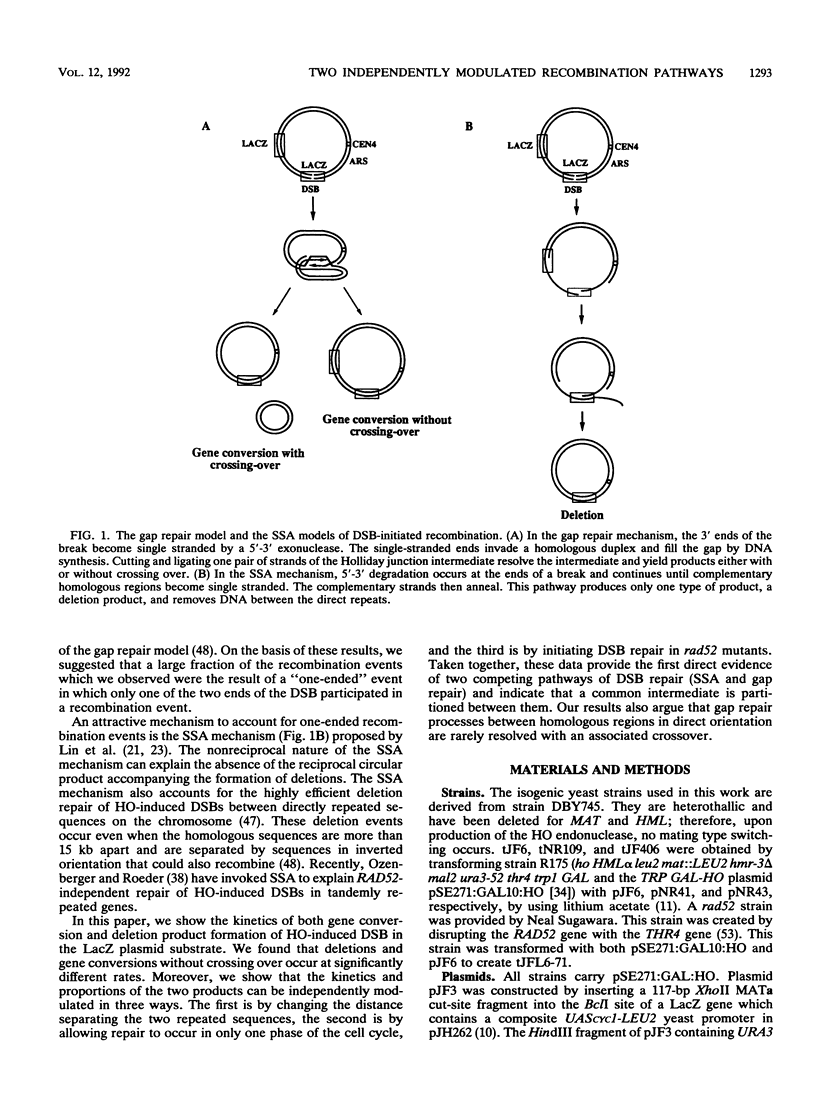
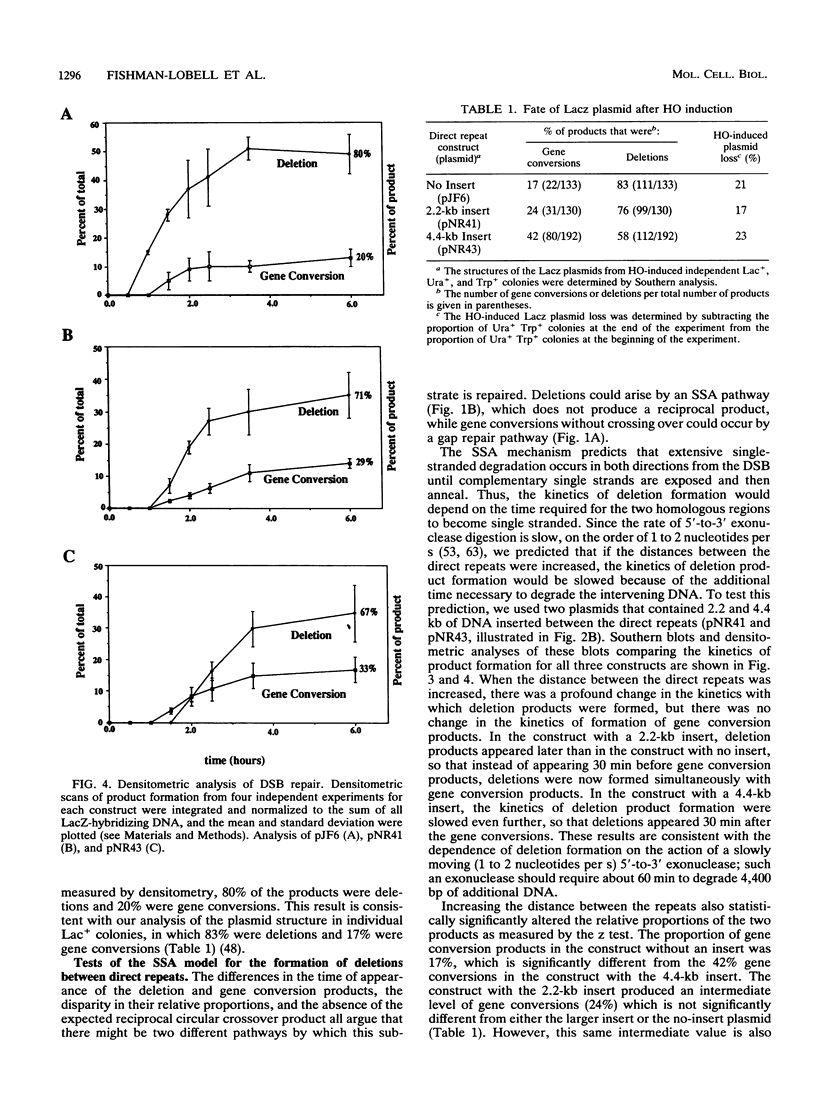
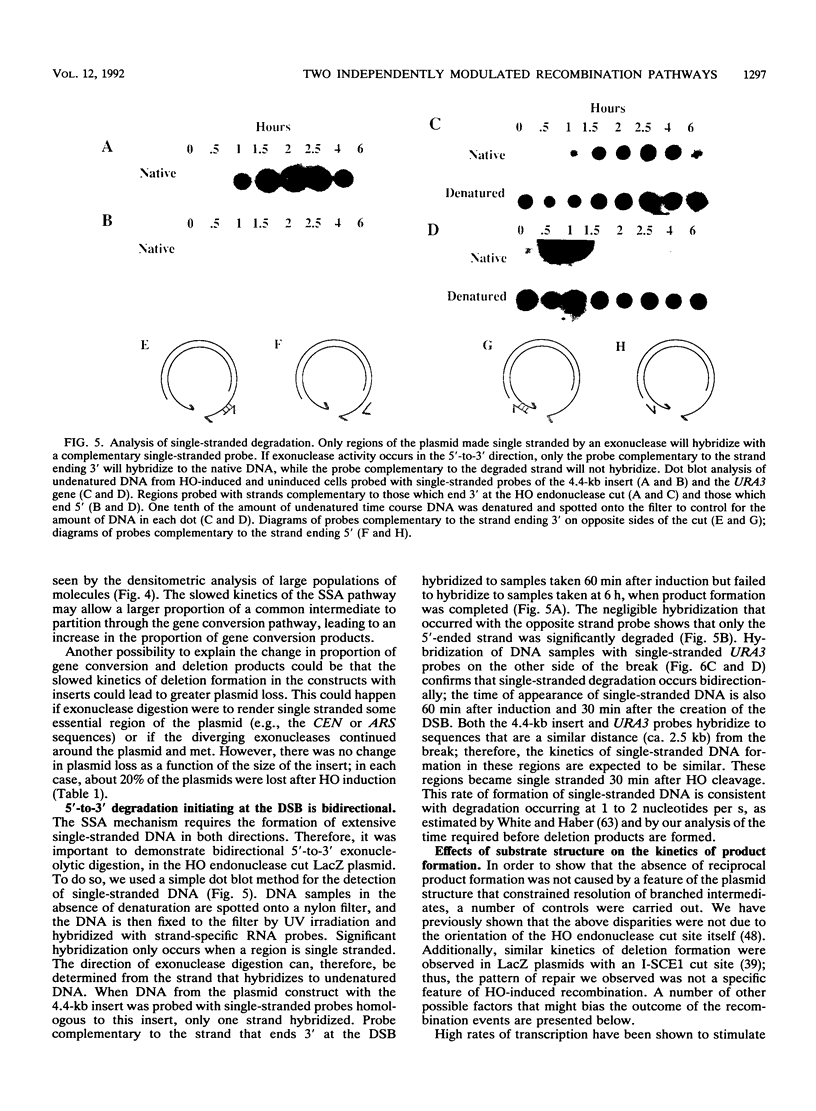
HO endonuclease-induced double-strand breaks in Saccharomyces cerevisiae can undergo recombination by two distinct and competing pathways. In a plasmid containing a direct repeat, in which one repeat is interrupted by an HO endonuclease cut site, gap repair yields gene conversions while single-strand annealing produces deletions. Consistent with predictions of the single-strand annealing mechanism, deletion formation is not accompanied by the formation of a reciprocal recombination product. Deletions are delayed 60 min when the distance separating the repeats is increased by 4.4 kb. Moreover, the rate of deletion formation corresponds to the time at which complementary regions become single stranded. Gap repair processes are independent of distance but are reduced in rad52 mutants and in G1-arrested cells.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Double-strand gap repair results in homologous recombination in mouse L cells. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1762–1766. doi: 10.1073/pnas.83.6.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B., White C. I., Haber J. E. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desautels L., Brouillette S., Wallenburg J., Belmaaza A., Gusew N., Trudel P., Chartrand P. Characterization of nonconservative homologous junctions in mammalian cells. Mol Cell Biol. 1990 Dec;10(12):6613–6618. doi: 10.1128/mcb.10.12.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M. S. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985 Jun;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Jasin M., Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988 Nov;2(11):1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Jessberger R., Berg P. Repair of deletions and double-strand gaps by homologous recombination in a mammalian in vitro system. Mol Cell Biol. 1991 Jan;11(1):445–457. doi: 10.1128/mcb.11.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Hicks J. B. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell. 1981 Aug;25(2):517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N. Resolution of recombination intermediates generated during yeast mating type switching. 1984 Aug 30-Sep 5Nature. 310(5980):744–748. doi: 10.1038/310744a0. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990 Jan;10(1):103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990 Jan;10(1):113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L., Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Thomas K. R., Capecchi M. R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988 Nov 24;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Marshall C. J. Tumor suppressor genes. Cell. 1991 Jan 25;64(2):313–326. doi: 10.1016/0092-8674(91)90641-b. [DOI] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991 Jun;11(6):3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989 Nov;9(11):4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Involvement of single-stranded tails in homologous recombination of DNA injected into Xenopus laevis oocyte nuclei. Mol Cell Biol. 1991 Jun;11(6):3268–3277. doi: 10.1128/mcb.11.6.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls R. D., Fischel-Ghodsian N., Higgs D. R. Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell. 1987 May 8;49(3):369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991 Mar;11(3):1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh D., Hughes S. H., Shafer B. K., Strathern J. N. Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989 Dec;220(1):33–42. [PubMed] [Google Scholar]
- Ray A., Siddiqi I., Kolodkin A. L., Stahl F. W. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J Mol Biol. 1988 May 20;201(2):247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- Resnick M. A. Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics. 1969 Jul;62(3):519–531. doi: 10.1093/genetics/62.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rudin N., Haber J. E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988 Sep;8(9):3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985 Sep 19;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Subramani S., Rubnitz J. Recombination events after transient infection and stable integration of DNA into mouse cells. Mol Cell Biol. 1985 Apr;5(4):659–666. doi: 10.1128/mcb.5.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992 Feb;12(2):563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Surosky R. T., Tye B. K. Construction of telocentric chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2106–2110. doi: 10.1073/pnas.82.7.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Vnencak-Jones C. L., Phillips J. A., 3rd Hot spots for growth hormone gene deletions in homologous regions outside of Alu repeats. Science. 1990 Dec 21;250(4988):1745–1748. doi: 10.1126/science.1980158. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiffenbach B., Haber J. E. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen P. H., Li X. M., Tsai S. P., Johnson C., Mohandas T., Shapiro L. J. Frequent deletions of the human X chromosome distal short arm result from recombination between low copy repetitive elements. Cell. 1990 May 18;61(4):603–610. doi: 10.1016/0092-8674(90)90472-q. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell. 1985 Apr;40(4):887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]