Abstract
Myeloid-derived suppressor cells (MDSC) have recently emerged as one of the central regulators of the immune system. In recent years, interest in understanding MDSC biology and applying MDSC for therapeutic purpose has exploded exponentially. Despite recent progress in MDSC biology, the mechanisms underlying MDSC development from expansion and activation to polarization in different diseases remain poorly understood. More recent studies have demonstrated that two MDSC subsets, M (monocytic)-MDSC and G (granulocytic)-MDSC, are able to polarize from a classically activated phenotype (M1) to an alternatively activated one (M2), or vice versa, in tumor-bearing mice. This phenotypic polarization affects MDSC function and disease progression. In this article, we summarize and discuss polarization, mechanism and therapeutic potential of MDSC. An emphasis is placed on the emerging concept of reprogramming MDSC polarization as a therapeutic strategy.
The importance of myeloid-derived suppressor cells (MDSC) in regulating the immune system has been recognized for the past 10 years. Accumulating evidence indicates a role of MDSC in tumors, infections, trauma, transplantation and other diseases. MDSC represent a heterogeneous population of myeloid progenitor cells with immune suppressive function. They are recognized for their morphological, phenotypic, and functional heterogeneity (Supplementary Figure S1) (Gabrilovich and Nagaraj, 2009).
The development of MDSC under different circumstances is not well understood. In normal physiological situations, immature myeloid cells (IMC) differentiate from hematopoietic stem cells and, finally, mature into dendritic cells, macrophages, and neutrophils/granulocytes upon entering the periphery (Supplementary Figure S1A). In pathological situations, abundant growth factors associated with specific pathological conditions boost IMCs expansion and subsequently interfere with their normal differentiation in the bone marrow (Condamine and Gabrilovich, 2011). Furthermore, inflammatory mediators of pathology can aberrantly induce IMCs to activate and polarize into MDSC with different phenotypes (Supplementary Figure S1B).
More recently, interest in basic research and clinical applications of MDSC has dramatically increased. Despite rapid advances made in understanding MDSC biology, the molecular basis for MDSC development, from expansion to activation and particularly polarization, is largely unknown. Here we highlight and discuss MDSC polarization with a special emphasis on the likely mechanism and potential application.
Compelling evidence showed that tumor-associated MDSC exhibit predominantly M2-like phenotypes with pro-tumoral and immunosuppressive activities (Ochando and Chen, 2012). However, M1 and M2 phenotypes in MDSC co-existed in some mouse tumor models (Umemura et al., 2008). Phenotype and function of the polarized MDSC subpopulations can be differentiated by specific signature markers (Supplementary Figure S1). MDSC exert immune suppression through cross-communication with T cells and other immune cells (Ostrand-Rosenberg et al., 2012). M2 MDSC inactivate effector T cells (Teff) but activate regulatory T cells (Treg), whereas M1 counterparts have the opposing actions.
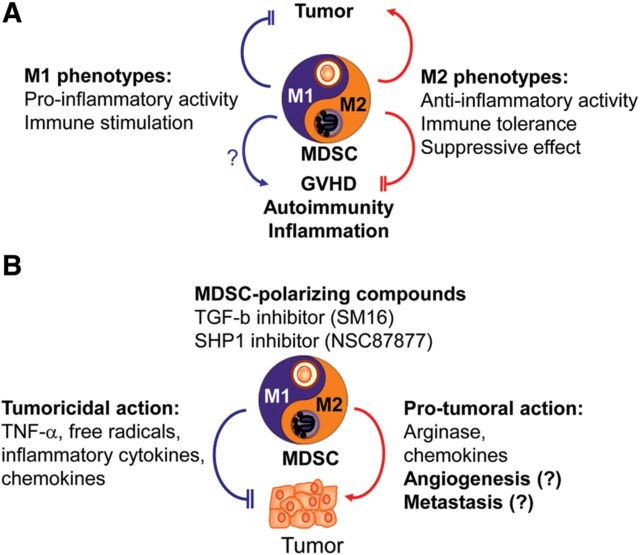
MDSC polarization from one phenotype to the other is accompanied by functional changes in different pathological situations. Overwhelming evidence demonstrates that tumor-related inflammation predominantly induces the accumulation of M2 MDSC. M2 MDSC accelerates tumor growth mainly by enhanced immunosuppression involving an increase in arginase and immunosuppressive cytokines (Figure 1) (Ma et al., 2011; Fridlender and Albelda, 2012). Moreover, M2 cells are relevant to angiogenesis (Ma et al., 2011; Fridlender and Albelda, 2012). Accordingly, pharmacological and genetic manipulation of reprogramming MDSC polarization to the M1 state was shown to suppress tumor growth in tumor-bearing hosts through the augmented free radicals, death ligand, and immunostimulating cytokines (Figure 1) (Ma et al., 2011; Fridlender and Albelda, 2012). Understanding MDSC polarization and related functional changes is prerequisite for their application.
Figure 1.
Novel vista on the use of reprogramming MDSC polarization as a novel therapeutic approach to treat cancers and other disorders. (A) ‘Yin-Yang’ balance in M1/M2 polarization of MDSC may relate to disease and health, and can be harnessed for the treatment of various diseases. Polarization of MDSC toward M1 phenotype can inhibit tumor growth, whereas undesirable immune responses, e.g. autoimmunity, transplant rejection and graft-versus-host diseases (GVHD) can be suppressed through the use of M2-polarized MDSC as a cell-based tolerogenic therapy. M2-polarized MDSC are characterized by their suppressive effect, anti-inflammatory activity and immune tolerance. In contrast, M1 cells possess pro-inflammatory and immunostimulatory activities. (B) MDSC-polarizing compounds can convert pro-tumoral cells M2 MDSC into tumoricidal M1 cells. In this case, TGF-β inhibitor, SM16, acts to inhibit its kinase activity and, in turn, skews G2 G-MDSC to G1 G-MDSC. SHP1 inhibitor, NSC87877, can shift M2 M-MDSC to M1 M-MDSC. Both compounds suppress tumor growth. Overall, control of M1/M2 MDSC reprogramming has emerged as a novel approach to treat cancers and other diseases.
Growth factors and inflammatory mediators have been shown to dictate functional polarization of MDSC. IFN-γ, IL-4, and IL-13 are well documented to be present in the tumor microenvironment. IFN-γ induces iNOS expression, while IL-4 or IL-13 increases arginase expression in MDSC as well as macrophages (Ma et al., 2011; Ochando and Chen, 2012). Furthermore, activation from toll-like receptor (TLR), IFN-γR, IL-4R and IL-13R could modulate MDSC function (Ma et al., 2011; Ochando and Chen, 2012). IFN-γR engagement induces the activation of JAK1/2 and STAT-1 and the expression of downstream genes like iNOS in different cell types. Similarly, IL-4R engagement activates JAK1/3 and STAT-6, whereas IL-13R engagement activates JAK1, TYK2 and STAT-3/6. TLR pathways are involved in MDSC development during infection. Collectively, most studies suggest TLR and IFN-γR are important for M1 function, while the IL-4R and IL-13R pathways are important for M2 function.
Murine PIR-A and PIR-B (Takai, 2005), and human PIR homologues (Andre et al., 2001), belong to the immunoglobulin super family (Supplementary Table S1). Herein we use PIR-A and PIR-B as a prototype to illustrate the mechanism controlling M-MDSC polarization. Following the ligand ligation, PIR-A, in complex with FcR, initiates a positive signaling cascade including activation of the Src family, phosphorylation of immune receptor tyrosine-based activatory motif (ITAM), and cytokine expression in myeloid cells (Takai, 2005). In contrast, PIR-B engagement induces activation of the Src family, ITIM phosphorylation, and the recruitment of SHP-1 as well as SHP-2 and SHIP-1, finally resulting in a negative signal transduction (Takai, 2005). PIR-A transduces a positive signal in MDSC, resulting in elevated expressions of M1 signature genes (Supplementary Figure S2A) (Ma et al., 2011). On the contrary, PIR-B inhibited the signals mediated by PIR-A as well as TLR, IFN-γR, IL-4R, and IL-13R (Supplementary Figure S2A) (Ma et al., 2011). As expected, M-MDSC exhibit an M2 phenotype in tumor-bearing mice (Ma et al., 2011). However, in tumor-bearing PIR-B knockout mice or wild-type mice in the presence of SHP inhibitor, which can block PIR-B-mediated downstream signaling, M-MDSC exhibited an M1 functional phenotype.
Fridlender et al. (2009) showed that tumor-associated inflammation polarized G-MDSC from the G1 to G2 phenotype. Inhibition of TGF-βR by the kinase inhibitor, SM16, reprogrammed G-MDSC from G2 to G1 (Fridlender et al., 2009). Accordingly, anti-TGF-β antibody drove a shift from the G2 to G1 phenotype in MDSC (Umemura et al., 2008). Currently, exactly how the TGF-β/TGF-βR pathway regulates G-MDSC polarization through fine-tuning of the signaling elicited by LPS/IFN-γ, IL-4/IL-13 and PIR remains mostly unknown. A scheme delineating the molecular mechanism for G1/G2 polarization is proposed in Supplementary Figure S2B.
Given the role of PIR-B and TGF-βR in the polarization of M-MDSC and G-MDSC, respectively, ligands, receptors and downstream molecules of PIR-B and TGF-βR are potential targets for manipulating MDSC polarization. More information on the signaling pathways underlying MDSC polarization is required prior to further clinical application.
MDSC are one of the pivotal players in immunosuppression induced in pathologies such as malignancies, infections, transplants, and autoimmune disorders. They act as an ‘information hub’ to interact with immune cells via multiple mechanisms (Ostrand-Rosenberg et al., 2012). Besides tumoricidal activity, some anti-cancer agents kill or block signal regulation pathways on MDSC to restore immunity against cancers. Hence, reprogramming MDSC as a therapeutic strategy represents a paradigm shift.
Mounting data clearly point to a critical role for MDSC in the regulation of immune responses in various diseases, e.g. tumors, infections, allografts, autoimmune disorders, etc. MDSC polarization links their phenotypic and functional changes to disease development. Therefore, understanding the molecular basis of MDSC polarization may facilitate their clinical application and drug development. Indeed, reprogramming M2 MDSC to M1 state by manipulating PIR-B and TGF-βR pathways can cause remarkable tumor suppression. In conclusion, reprogramming disease-promoting MDSC into therapeutic one against the diseases is evolving into a new therapeutic approach.
[Supplementary material is available at Journal of Molecular Cell Biology online. We thank the authors whose publications we cited for their contributions. W.C.Y. is a visiting scholar of Agricultural Biotechnology Research Center, Academia Sinica.]
Supplementary Material
References
- Andre P., Biassoni R., Colonna M., et al. New nomenclature for MHC receptors. Nat. Immunol. 2001;2:661. doi: 10.1038/90589. [DOI] [PubMed] [Google Scholar]
- Condamine T., Gabrilovich D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender Z.G., Albelda S.M. Tumor associated neutrophils: friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- Fridlender Z.G., Sun J., Kim S., et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: ‘N1’ versus ‘N2’ TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Pan P.Y., Eisenstein S., et al. Paired immunoglobin-like receptor-B regulates the suppressive function and fate of myeloid-derived suppressor cells. Immunity. 2011;34:385–395. doi: 10.1016/j.immuni.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochando J.C., Chen S.H. Myeloid-derived suppressor cells in transplantation and cancer. Immunol. Res. 2012;54:275–285. doi: 10.1007/s12026-012-8335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S., Sinha P., Beury D.W., et al. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin. Cancer Biol. 2012;22:275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology. 2005;115:433–440. doi: 10.1111/j.1365-2567.2005.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura N., Saio M., Suwa T., et al. Tumor-infiltrating myeloid-derived suppressor cells are pleiotropic-inflamed monocytes/macrophages that bear M1- and M2-type characteristics. J. Leukoc. Biol. 2008;83:1136–1144. doi: 10.1189/jlb.0907611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.