Abstract
The major DNA-binding protein, or infected-cell protein 8 (ICP8), encoded by herpes simplex virus can localize to the cell nucleus independently of other viral proteins. To define the nuclear localization signals within ICP8, we performed several forms of mutagenesis on the cloned ICP8 gene. Deletion analysis of the ICP8 gene showed that several portions of ICP8 are involved in its nuclear localization. To determine whether these regions were independent localization signals, we introduced various portions of the ICP8 gene into a series of cassette plasmids which allowed expression of fusion proteins containing pyruvate kinase, normally a cytoplasmic protein, fused to various portions of ICP8. These results showed that the carboxyl-terminal 28 residues are the only portion of ICP8 capable of targeting protein kinase into the nucleus. However, inclusion of certain additional regions of ICP8 into the fusion protein led to an inhibition of nuclear localization. Therefore, the carboxyl-terminal 28 residues of ICP8 can act independently as a nuclear localization signal, but certain conformational constraints or folding or assembly requirements in the remainder of the protein can affect the nuclear localization of the protein. Our results demonstrate that sequences distant from a nuclear localization signal can affect its ability to function. A set of fusion vectors has been isolated which should be of general use for making 5' or 3' fusions in any reading frame to rapidly map localization signals.
Full text
PDF


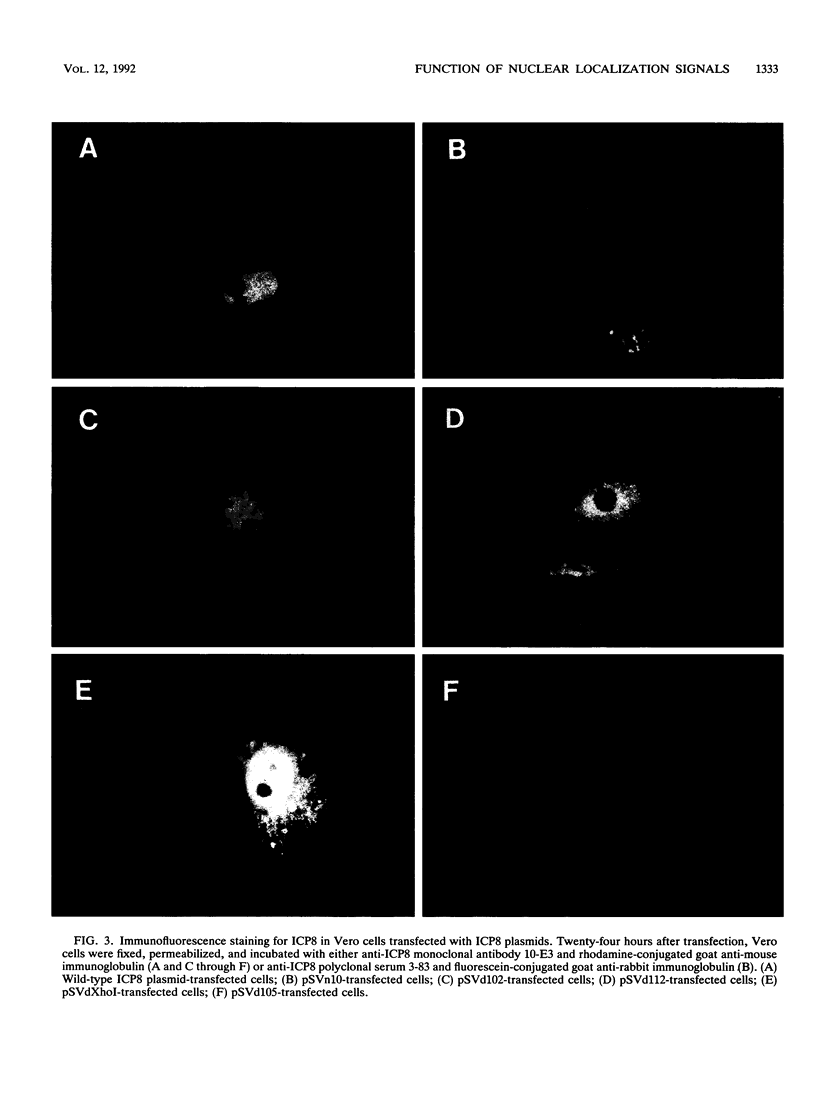






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayliss G. J., Marsden H. S., Hay J. Herpes simplex virus proteins: DNA-binding proteins in infected cells and in the virus structure. Virology. 1975 Nov;68(1):124–134. doi: 10.1016/0042-6822(75)90154-3. [DOI] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D. S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990 Mar 23;60(6):999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., De Robertis E. M. The nuclear migration signal of Xenopus laevis nucleoplasmin. EMBO J. 1987 Sep;6(9):2617–2625. doi: 10.1002/j.1460-2075.1987.tb02552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J., Dimmock N. J., Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985 Mar;40(3):667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Robbins J., Dilworth S. M., Roberts B., Richardson W. D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol. 1988 Sep;107(3):841–849. doi: 10.1083/jcb.107.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Bouchey J., Curtin K., Knipe D. M. Genetic identification of a portion of the herpes simplex virus ICP8 protein required for DNA-binding. Virology. 1988 Apr;163(2):319–329. doi: 10.1016/0042-6822(88)90272-3. [DOI] [PubMed] [Google Scholar]
- Gao M., Knipe D. M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989 Dec;63(12):5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Knipe D. M. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991 May;65(5):2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharakhanian E., Takahashi J., Kasamatsu H. The carboxyl 35 amino acids of SV40 Vp3 are essential for its nuclear accumulation. Virology. 1987 Apr;157(2):440–448. doi: 10.1016/0042-6822(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Godowski P. J., Knipe D. M. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci U S A. 1986 Jan;83(2):256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan D., Palese P., Krystal M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol. 1988 Aug;62(8):3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiochon-Mantel A., Loosfelt H., Lescop P., Sar S., Atger M., Perrot-Applanat M., Milgrom E. Mechanisms of nuclear localization of the progesterone receptor: evidence for interaction between monomers. Cell. 1989 Jun 30;57(7):1147–1154. doi: 10.1016/0092-8674(89)90052-4. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Knipe D. M., Senechek D., Rice S. A., Smith J. L. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987 Feb;61(2):276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R. E., Butel J. S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984 Jul;37(3):801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Lee C. K., Knipe D. M. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J Virol. 1985 Jun;54(3):731–738. doi: 10.1128/jvi.54.3.731-738.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg N., Gilbert W. Primary structure of chicken muscle pyruvate kinase mRNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3661–3665. doi: 10.1073/pnas.80.12.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R. H., Ferguson B. Q., Rosenberg M. Pentapeptide nuclear localization signal in adenovirus E1a. Mol Cell Biol. 1987 Jul;7(7):2451–2456. doi: 10.1128/mcb.7.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Nam H. G., Hereford L. M., Fried H. M. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin N., Delsert C., Klessig D. F. Nuclear localization of the adenovirus DNA-binding protein: requirement for two signals and complementation during viral infection. Mol Cell Biol. 1989 Oct;9(10):4372–4380. doi: 10.1128/mcb.9.10.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S. T., Nayak D. P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol Cell Biol. 1990 Aug;10(8):4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D. D., Forbes D. J. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell. 1988 Mar 11;52(5):641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Orberg P. K., Schaffer P. A. Expression of herpes simplex virus type 1 major DNA-binding protein, ICP8, in transformed cell lines: complementation of deletion mutants and inhibition of wild-type virus. J Virol. 1987 Apr;61(4):1136–1146. doi: 10.1128/jvi.61.4.1136-1146.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Yamamoto K. R. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987 Nov;6(11):3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. L., Littler E., Purifoy D. J. Nonstructural proteins of herpes simplex virus. II. Major virus-specific DNa-binding protein. J Virol. 1981 Sep;39(3):894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purifoy D. J., Powell K. L. DNA-binding proteins induced by herpes simplex virus type 2 in HEp-2 cells. J Virol. 1976 Aug;19(2):717–731. doi: 10.1128/jvi.19.2.717-731.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan M. P., Chen L. B., Knipe D. M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984 Apr;36(4):857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- Quinlan M. P., Knipe D. M. Nuclear localization of herpesvirus proteins: potential role for the cellular framework. Mol Cell Biol. 1983 Mar;3(3):315–324. doi: 10.1128/mcb.3.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Mills A. D., Dilworth S. M., Laskey R. A., Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988 Mar 11;52(5):655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Richardson W. D., Roberts B. L., Smith A. E. Nuclear location signals in polyoma virus large-T. Cell. 1986 Jan 17;44(1):77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- Roberts B. L., Richardson W. D., Smith A. E. The effect of protein context on nuclear location signal function. Cell. 1987 Jul 31;50(3):465–475. doi: 10.1016/0092-8674(87)90500-9. [DOI] [PubMed] [Google Scholar]
- Rose D. S., Shriver K., Latchman D. S., LaThangue N. B. A filamentous distribution for the herpes simplex virus type 2-encoded major DNA-binding protein. J Gen Virol. 1986 Jul;67(Pt 7):1315–1325. doi: 10.1099/0022-1317-67-7-1315. [DOI] [PubMed] [Google Scholar]
- Shaulsky G., Goldfinger N., Ben-Ze'ev A., Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990 Dec;10(12):6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P. A., Keegan L. P., Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Kalderon D., Roberts B. L., Colledge W. H., Edge M., Gillett P., Markham A., Paucha E., Richardson W. D. The nuclear location signal. Proc R Soc Lond B Biol Sci. 1985 Oct 22;226(1242):43–58. doi: 10.1098/rspb.1985.0078. [DOI] [PubMed] [Google Scholar]
- Spang A. E., Godowski P. J., Knipe D. M. Characterization of herpes simplex virus 2 temperature-sensitive mutants whose lesions map in or near the coding sequences for the major DNA-binding protein. J Virol. 1983 Jan;45(1):332–342. doi: 10.1128/jvi.45.1.332-342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Wychowski C., Benichou D., Girard M. A domain of SV40 capsid polypeptide VP1 that specifies migration into the cell nucleus. EMBO J. 1986 Oct;5(10):2569–2576. doi: 10.1002/j.1460-2075.1986.tb04536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]