Abstract
The taxonomic position of three acidophilic actinobacteria, strains FGG38, FGG39 and FSCA67T, isolated from the fermentation litter layer of a spruce forest soil was established using a polyphasic approach. The strains were shown to have chemotaxonomic and morphological properties consistent with their classification in the genus Streptacidiphilus and formed a distinct phyletic line in the Streptacidiphilus 16S rRNA gene tree being most closely related to Streptacidiphilus albus DSM 41753T (99.4 % similarity). DNA:DNA relatedness data showed that isolate FSCA67T and the type strain of S. albus belonged to markedly distinct genomic species. The isolates had many phenotypic properties in common and were distinguished readily from their closest phylogenetic neighbours in the Streptacidiphilus gene tree using a broad range of these features. Based on the combined genotypic and phenotypic data the three isolates are considered to represent a new Streptacidiphilus species. The name Streptacidiphilus durhamensis sp. nov. is proposed for this taxon with isolate FSCA67T (=DSM 45796T = KACC 17154T = NCIMB 14829T) as the type strain.
Keywords: Actinobacteria, Polyphasic taxonomy, Streptacidiphilus durhamensis, Spruce forest soil
Introduction
Acidophilic sporoactinobacteria which share key chemotaxonomic and morphological properties with streptomycetes grow between pH 3.5 and 6.5 with optimal growth at pH 4.5–5.5. These organisms are numerous and widely distributed in acidic habitats, notably coniferous soils (Williams et al. 1971; Khan and Williams 1975; Goodfellow and Dawson 1978; Cho et al. 2008) where they have a role in the turnover of fungal biomass (Williams and Robinson 1981). They are a source of antifungal compounds (Williams and Khan 1974) and produce chitinases and diastases with pH optima below those of neutrophilic streptomycetes (Williams and Flowers 1978). In a comprehensive numerical taxonomic study, Lonsdale (1985) found that acidophilic streptomycetes fell into several distinct clusters (taxospecies), which were clearly distinguished using a combination of phenotypic tests. Subsequent studies underpinned the taxonomic heterogeneity of the group (Goodfellow and Simpson 1987; Seong et al. 1993).
Representatives of three of the taxospecies circumscribed by Lonsdale (1985) were assigned to the novel genus Streptacidiphilus as Streptacidiphilus albus, the type species, Streptacidiphilus carbonis and Streptacidiphilus neutrinimicus by Kim et al. (2003). In the meantime, five additional species have been recognised: Streptacidiphilus jiangxiensis (Huang et al. 2004), Streptacidiphilus oryzae (Wang et al. 2006), Streptacidiphilus anmyonensis, Streptacidiphilus melanogenes and Streptacidiphilus rugosus (Cho et al. 2008) bringing the total validly named species to eight.
All but one of the Streptacidiphilus species formed a monophyletic lineage (clade 53) in the extensive phylogenetic study of the family Streptomycetaceae undertaken by Labeda et al. (2012), whilst the exception, S. oryzae, was found in the related clade 54. The taxonomic integrity of clade 53 was well supported by all three of the tree-making algorithms used by Labeda and his colleagues and was closely related to the genus Kitasatospora, the species of which formed a large statistically unsupported clade. The species of both of these genera fell within the evolutionary radiation occupied by the genus Streptomyces and hence in the final analysis may only be considered taxonomically valid if the genus Streptomyces is found to be truly paraphyletic. However, to date, it is notable that all streptacidiphili form either white or gray aerial hyphae which differentiate into flexuous to straight chains of spores with smooth surfaces on oatmeal agar (International Streptomyces Project [ISP] medium 3; Shirling and Gottlieb 1966).
In a continuation of our bioprospecting studies on acidophilic and aciditolerant filamentous actinobacteria isolated from a spruce forest soil, several representative isolates were found to have colonial properties typical of streptacidiphili. The aim of the present study was to establish the taxonomic status of three of these isolates using a polyphasic approach. The resultant data showed that the strains belong to a new Streptacidiphilus species, Streptacidiphilus durhamensis sp. nov.
Materials and methods
Organisms, maintenance and biomass preparation
Acidophilic and aciditolerant filamentous sporoactinobacteria were sought from the L, F, H, A1 and C horizons of the spruce forest soil at Hamsterley Forest, County Durham; details of the site and the dilution plate procedure used are given elsewhere (Golinska et al. 2013). Aliquots (100 μl) of serial dilutions were spread over the surfaces of oven dried plates of starch-casein medium (Kűster and Williams 1964) with agar (SCA) and gellan gum (GG) as gelling agents, the media were supplemented with cycloheximide and nystatin (each at 50 μg ml−1) and adjusted to pH 4.5 with 1 N HCl. The inoculated plates were incubated at 28 °C for 4 weeks, at which point Streptacidiphilus-like colonies were detected from suspensions prepared from each of the environmental samples. The 56 Streptacidiphilus-like isolates were subcultured from starch-casein media prepared using each of the gelling agents and found to grow well on starch-casein agar at pH 5.5 but poorly on this medium at pH 4.5 and 6.0 with no growth at pH 7.0. All of the isolates were maintained on acidified modified Bennett’s agar slopes (Jones 1949) at room temperature and as suspensions of mycelial fragments and spores in glycerol (20 %, w/v) at −80 °C. Three strains from the F-horizon, isolates FGG38, FGG39 and FSCA67T, were chosen for detailed taxonomic analyses.
Biomass for the molecular systematic study and most of the chemotaxonomic studies was prepared by cultivating the three isolates in shake flasks of yeast extract-malt extract broth (pH 5.5) at 150 revolutions per minute at 28 °C for 3 weeks. Cells were harvested by centrifugation and washed twice in distilled water; biomass for the chemotaxonomic analyses was freeze dried and that for the molecular systematic work stored at −20 °C. Biomass for the fatty acid analysis conducted on isolate FSCA67T was harvested from modified Bennett’s broth (Jones 1949), adjusted to pH 5.5, after incubation at 28 °C for 7 days.
Phylogenetic analyses
Extraction of genomic DNA, PCR-mediated amplification of the 16S rRNA genes of the three isolates and direct sequencing of the purified PCR products were carried out as described by Golinska et al. (2013). The search for the closest phylogenetic neighbours based on 16S rRNA gene similarly was performed using the EzTaxon server (http://eztaxon-e.ezbiocloud.net/, Kim et al. 2012). Phylogenetic analyses were carried out using MEGA 5 (Tamura et al. 2011) and PHYML (Guindon and Gascuel 2003) software packages following multiple alignment using Clustal W. Evolutionary distances were calculated and clustering determined using the maximum-likelihood, maximum-parsimony and neighbour-joining methods to generate phylogenetic trees. The topologies of the evolutionary trees were evaluated by a bootstrap analysis (Felsenstein 1985) derived from 1,000 resamplings of the neighbour-joining dataset using MEGA 5 software. The tree was rooted using the 16S rRNA gene sequence of Streptomyces albus subsp. albus DSM 40313T (GenBank accession number AJ621602).
DNA:DNA relatedness
DNA:DNA relatedness values (∆Tm) between isolate FSCA67T and S. albus DSM 41753T were determined in duplicate using a fluorimetric method (Gonzalez and Saiz-Jimenez 2005), the optimal temperature for renaturation (Tm) was calculated using the equation Tor − 0.51 (% GC) + 47. The melting temperature (Tm) at which 50 % of the initial double-stranded DNA denatured into single-stranded DNA for isolate FSCA67T and the isolate FSCA67T: S. albus DSM 41753T hybrid DNA preparations were compared and the differences (∆Tm) calculated.
Chemotaxonomy
Isolates FGG38, FGG39 and FCSA67T were examined for chemical markers known to be of value in the systematics of genera classified in the family Streptomycetaceae (Kämpfer 2012a). Standard chromatographic methods were used to determine the isomers of diaminopimelic acid (Staneck and Roberts 1974), isoprenoid quinones (Collins 1985), polar lipids (Minnikin et al. 1984) and whole-organism sugars (Hasegawa et al. 1983), using appropriate controls. Cellular fatty acids of isolate FSCA67T were extracted, methylated and determined by gas chromatography (Hewlett Packard instrument 6890) and analysed using the standard Sherlock Microbial Identification (MIDI) system, version 5 (Sasser 1990). The G+C vol% of the DNA of this isolate was determined following the procedure described by Gonzalez and Saiz-Jimenez (2002).
Cultural and morphological properties
The isolates were examined for cultural and morphological features following growth on standard media at 28 °C for 3 weeks. Cultural properties were determined using acidified tryptone-yeast extract, yeast extract-malt extract, oatmeal, inorganic salts-starch, glucose–asparagine, peptone-yeast extract-iron and tyrosine agars (ISP media 1–7, respectively; Shirling and Gottlieb 1966). The arrangements of hyphae and spore chains were detected on acidified oatmeal agar (ISP medium 3) after 14 days at 28 °C, using the coverslip technique of Kawato and Shinobu (1959). Spore arrangement and spore surface ornamentation of isolate FSCA67T were observed by examining a gold-coated dehydrated preparation from the oatmeal agar plate with a scanning electron microscope (Cambridge Stereoscan 240) and the procedure described by O’Donnell et al. (1993).
Phenotypic tests
An extensive range of phenotypic tests were carried out on all of the isolates using media and methods described by Williams et al. (1983), albeit with acidified media. The ability of the isolates to grow at various temperatures (10, 30, 35 and 40 °C), pH values (4, 5, 6 and 7) and NaCl concentrations (1, 3, 5, 7 and 10 %, w/v) was determined using acidified modified Bennett’s agar (Jones 1949).
Results
16S rRNA gene sequencing and DNA:DNA relatedness studies
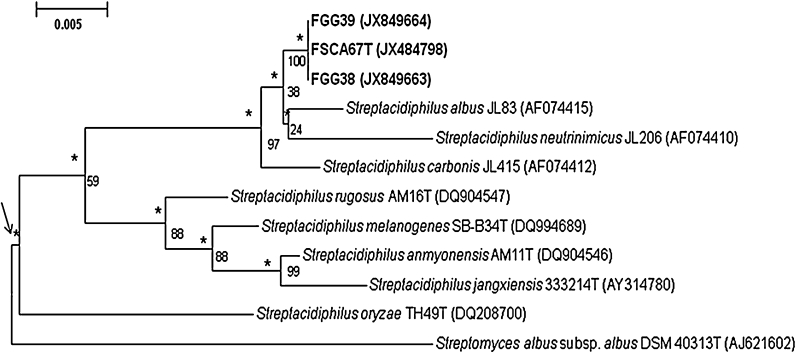
Near complete 16S rRNA gene sequences of isolates (1,423–1,463 nucleotides [nt]) FSCA67T, FGG38 and FGG39 were determined (GenBank accession numbers: JX484798, JX849663 and JX849664, respectively). The strains were found to have identical sequences and to form a branch in the Streptacidiphilus 16S rRNA gene tree that was shown to be supported by all of the tree-making algorithms and by a 100 % bootstrap value (Fig. 1). They were found to form a subclade in the Streptacidiphilus 16S rRNA tree together with the type strains of S. albus, S. carbonis and S. neutrinimicus, a taxon that was shown to be supported by a 97 % bootstrap value and all of the tree-making algorithms. The isolates were most closely related to S. albus JL83T and found to share a 16S rRNA similarity with the latter of 99.4 %, a value found to correspond to 8 nt differences. The corresponding 16S rRNA gene similarities with the type strains of S. carbonis and S. neutrinimicus were shown to be 99.3 and 98.8 %, respectively, values equivalent to 10 and 17 nt differences. The 16S rRNA sequence similarities with the type strains of the remaining Streptacidiphilus species was found to be within the range 96.2–97.3 %. Notably, the 16S rRNA tree also supported the previously observed separation of S. oryzae from the other Streptacidiphilus species (Labeda et al. 2012).
Fig. 1.
Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1,376–1,511 nucleotides) showing relationships between the isolates and the type strains of Streptacidiphilus species. Asterisks indicate branches that were also found using the maximum-likelihood and maximum-parsimony tree-making algorithms. Numbers at the nodes are percentage bootstrap values based on 1,000 re-sampled datasets, only values above 50 % are given. T type strain. Bar 0.005 substitutions per nucleotide position. The root position of the tree was determined using Streptomyces albus subsp. albus DSM 40313T as outgroup
The ∆Tm between isolate FSCA67T g DNA and isolate FSCA67T: S. albus DSM 41753T hybrid DNA was determined to be 9.5 °C (±2.83), a result that corresponds to a DNA similarity value of 45 % (Gonzalez and Saiz-Jimenez 2005), a value well below the 70 % cut-off point recommended for assigning strains to the same genomic species (Wayne et al. 1987).
Chemotaxonomic, cultural, morphological and phenotypic properties
The isolates were found to produce an extensively branched substrate mycelium which carried abundant gray aerial hyphae that were shown to differentiate into straight to flexuous spore chains on oatmeal agar. Isolate FSCA67T was found to produce long chains of smooth spores (Fig. 2). The isolates were shown to grow well on ISP media 2, 3 and 5; poorly on ISP 1; and not at all on ISP 4 (Table 1). In general, they were shown to form a variety of substrate mycelial pigments and, when formed, a white to medium gray aerial spore mass.
Fig. 2.
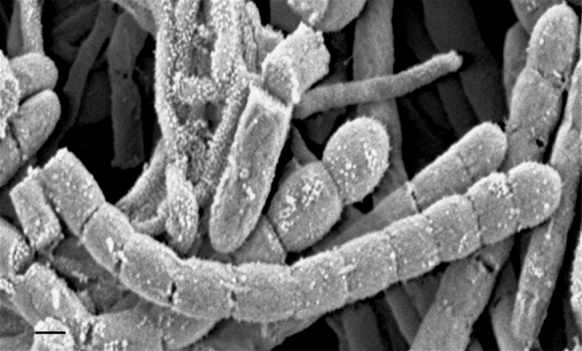
Scanning electron micrograph of isolate FSCA67T showing straight chains of smooth-surfaced, cylindrical spores on oatmeal agar after growth for 3 weeks at 28 °C. Bar 0.5 μm
Table 1.
Growth and cultural characteristics of isolates on ISP media after incubation for 3 weeks at 28 °C
| Isolates | FGG38 | FGG39 | FSCA67T | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Medium | Growth | Colour of aerial spore mass | Colour of substrate mycelium | Growth | Colour of aerial spore mass | Colour of substrate mycelium | Growth | Colour of aerial spore mass | Colour of substrate mycelium |
| Yeast extract-malt extract agar (ISP 2) | ++ | Light gray | Yellowish brown | +++ | Light gray | Yellowish brown | ++ | Light gray | Yellowish brown |
| Oatmeal agar (ISP 3) | +++ | Medium gray | Yellowish gray | ++ | Medium gray | Light yellowish brown | +++ | Medium gray | Light grayish yellowish brown |
| Glucose-asparagine agar (ISP 5) | ++ | Light gray | Moderate yellowish brown | +++ | Light gray | Moderate yellowish brown | ++ | Light gray | Moderate yellowish brown |
| Peptone-yeast extract-iron agar (ISP 6) | + | − | − | − | − | − | − | − | − |
| Tyrosine agar (ISP 7) | + | Light gray | Moderate yellowish brown | +++ | Light gray | Moderate yellowish brown | + | Light gray | Moderate yellowish brown |
The isolates did not grow on medium 4 and grew poorly on ISP medium 1 forming a light yellow substrate mycelium and a white aerial spore mass
+++ abundant, ++ moderate, + poor, − no growth
Whole-organism hydrolysates of the isolates were shown to be rich in ll-diaminopimelic acid (ll-A2pm) and to contain major amounts of galactose and rhamnose. The major polar lipids were established to be diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol and phosphatidylinositol mannosides, as exemplified in Fig. 3. The predominant isoprenologues of isolates FGG39, FGG38 and FSCA67T were determined to be hexa- and octahydrogenated menaquinones with nine isoprene units, components found in ratios of 1:1.3, 1:1.4 and 1.8:1, respectively. The cellular fatty acid profile of isolate FSCA67T was shown to consist of major proportions of (>10 %) of iso-C15:0 (15.8 %), anteiso-C15:0 (12.1 %) and iso-C16:0 (30.3 %), lower proportions (>2.8 %) of iso-C14:0 (6.2 %), iso-C16:1 H (3.4 %), C16:0 (9.1 %), anteiso-C17:1 ω9c (3.6 %), iso-C17:0 (2.8 %), anteiso-C17:0 (4.8 %) and C17:0 cyclo (3.4 %), summed features C16:1 ω7c/C16:1 ω6c (3.6 %) and iso-C17:1 ω9c/10 methyl C16:0 (2.8 %) plus trace amounts of other components (<0.5 %). The genomic G+C content of isolate FSCA67T was 71.0 mol%.
Fig. 3.
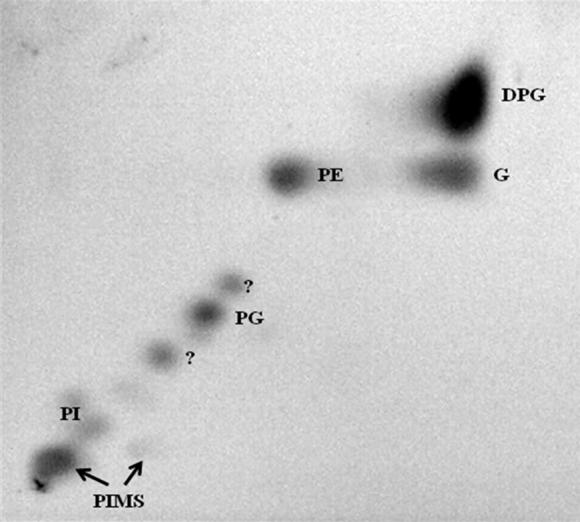
Two dimensional thin-layer chromatography of polar lipids of isolate FSCA67T stained with molybdenum blue (Sigma). Chloroform:methanol:water (32.5:12.5:2.0 v/v) were used in the first direction and chloroform:acetic acid:methanol:water (40:7.5:6:2 v/v) in the second direction. DPG diphosphatidylglycerol, PE phosphatidylethanolamine, PG phosphatidylglycerol, PI phosphatidylinositol, PIMS phosphatidylinositol mannosides and G glycolipid
The phenotypic properties of the isolates were compared with those of the type strains of S. albus, S. carbonis and S. neutrinimicus which had previously been studied using the same media and methods. It can be seen from Table 2 that the isolates can be distinguished from the type strains of their closest phylogenetic neighbours using a range of phenotypic features. Thus, the isolates, unlike S. albus DSM 41753T, their closest phylogenetic relative, were shown to metabolize Tween 40 and to grow on d-melezitose and d-xylose as sole carbon sources and at 10 and 30 °C. In contrast, the S. albus strain, unlike the isolates, degrades xanthine, uses sodium pyruvate and sodium propionate as sole sources of carbon and grows at pH 4.0. The isolates shared many other phenotypic features, as cited in the species description.
Table 2.
Phenotypic properties that distinguish the isolates from the type strains of their nearest taxonomic neighbours
| Characteristic | Isolates FGG38, FGG39 and FSCA67T | S. albus DSM 41753T | S. carbonis DSM 41754T | S. neutrinimicus DSM 41755T |
|---|---|---|---|---|
| Growth on acidified oatmeal agar | ||||
| Aerial spore mass | Medium grey | White | White | White/greyish white |
| Substrate mycelium | V (see Table 1) | Cream | Cream | Cream |
| Degradation of | ||||
| Tween 40 | + | − | − | − |
| Tween 60 | + | + | + | − |
| Xanthine | − | + | + | − |
| Growth on sole carbon sources | ||||
| At 1 %, w/v | ||||
| d-glucosamine | + | + | − | + |
| Meso-inositol | − | − | + | − |
| d-melezitose | + | − | − | − |
| l-rhamnose | + | + | + | − |
| d-xylose | + | − | + | + |
| At 0.1 %, w/v | ||||
| Sodium pyruvate | − | + | + | + |
| Sodium succinate | − | + | + | + |
| Growth on l-isoleucine (0.1 %, w/v) as sole nitrogen source | + | + | + | − |
| Growth at | ||||
| 10 °C | + | − | − | − |
| 30 °C | + | − | + | − |
| pH 4.0 | − | + | + | + |
| pH 6.0 | + | + | + | − |
| G+C content of DNA (mol %) | 71 | 70–72 | 70–72 | 70–72 |
Discussion
The three acidophilic isolates from the F-horizon of the spruce forest soil at Hamsterley Forest were found to share a range of cultural, chemotaxonomic and morphological properties typical of members of the genus Streptacidiphilus (Kim et al. 2003; Huang et al. 2004; Wang et al. 2006; Cho et al. 2008). They were shown to form extensively branched substrate mycelia, white to medium gray aerial hyphae that were found to differentiate into straight to flexuous chains of spores on oatmeal agar; and to have whole-organism hydrolysates rich in ll-A2pm, galactose and rhamnose, MK-9 (H6, H8) as the predominant isoprenologues; and diphosphatidylglycerol, phophatidylethanolamine (diagnostic lipid), phosphatidylinositol and phosphatidylinositol mannosides as major polar lipids (phospholipid pattern 2 after Lechevalier et al. 1977). Isolate FSCA67T was shown to form long chains of smooth spores, contains fatty acids rich in saturated, iso- and anteiso- components (fatty acid type 2c; Kroppenstedt 1985) and a DNA G+C ratio of 71 mol%. The isolates were found to have identical 16S rRNA gene sequences and to form a subclade in the Streptacidiphilus 16S rRNA gene tree together with the type strains of S. albus (their nearest taxonomic neighbour), S. carbonis and S. neutrinimicus. DNA:DNA relatedness data showed that isolate FSCA67T and S. albus DSM 41753T belonged to distinct genomic species using the 70 % cut-off point recommended by Wayne et al. (1987). The isolates had identical phenotypic properties, some of which clearly distinguished them from the type strains of their closest phylogenetic neighbours. In light of all of these genotypic and phenotypic data, it is proposed that the isolates represent a new Streptacidiphilus species for which the name S. durhamensis is proposed with isolate FSCA67T as the type strain. The species description is given below.
Filamentous spore-forming acidophilic actinobacteria remain a neglected group despite being common in acidic habitats (Williams et al. 1971; Khan and Williams 1975; Goodfellow and Dawson 1978; Goodfellow and Simpson 1987) and a potential source of novel acid-stable antibiotics and enzymes (Williams and Khan 1974; Williams and Flowers 1978; Williams and Robinson 1981). The present study provides further evidence that the genus Streptacidiphilus is underspeciated and common in acidic habitats, notably in coniferous litter (Lonsdale 1985; Goodfellow and Simpson 1987; Golinska, unpublished observations). Indeed, like isolates FGG38, FGG39 and FSCA67T, two out of the eight validly named Streptacidiphilus species were isolated from a single location, the spruce forest soil at Hamsterley Forest, County Durham. Additional comparative taxonomic surveys of streptacidiphili isolated from diverse acidic habitats can be expected to throw additional light on the evolutionary radiation of these organisms and thereby help underpin the continued recognition of the genus or indicate whether it should be seen as a genus incertae sedis (Kämpfer 2012b).
Description of Streptacidiphilus durhamensis sp. nov.
Streptacidiphilus durhamensis. N.L. masc. adj. durhamensis, belonging to Durham, a country in the North East of England, the source of the isolates.
Aerobic, Gram-positive, non-acid alcohol fast, acidophilic actinobacteria which form an extensively branched substrate mycelium that carries aerial hyphae which differentiate into long straight to flexuous chains of smooth, cylindrical spores (0.7 × 1.0 μm). Grows from 10 to 30 °C, optimally ~26 °C, from pH 4.5–6.0, optimally ~pH 5.5, and in the presence of 1 %, but not 3, 5, 7 and 10 %, w/v NaCl. Aesculin and arbutin are hydrolysed, but not allantoin or urea. Does not reduce nitrate. Degrades starch, but not adenine, casein, chitin, elastin, guanine, l-tyrosine or xylan. d-arabitol, l-arabinose, d-cellobiose, d-fructose, d-galactose, d-glucosamine, d-glucose, d-glycerol, d-lactose, d-mannitol, d-raffinose, d-salicin, d-sucrose and d-trehalose are used as sole carbon sources for energy and growth, but not l-arabitol, dextran, meso-erythritol, d-glucosamine or xylitol (all at 1 %, w/v). Does not use acetate, adipate, benzoate, butyrate, citrate, fumarate, hippurate, oxalate or propionate (sodium salts) or para-hydroxybenzoic acid as sole carbon sources (all at 0.1 %, w/v). l-alanine, l-isoleucine, l-phenylalanine and l-valine are used as sole nitrogen sources, but not l-arginine, l-cysteine or l-histidine (all at 0.1 %, w/v). Aminoethanol, l-cyteine, l-hydroxyproline, l-isoleucine, l-phenylalanine, l-serine and l-valine are used as sole carbon and nitrogen sources, but not acetamide, l-histidine or l-methionine (all at 0.1 %, w/v). Additional phenotypic properties are cited in Tables 1 and 2. The major fatty acids of the type strain are anteiso-C15:0, iso-C15:0 and iso-C16:0. The remaining chemotaxonomic markers are typical of the genus Streptacidiphilus. The G+C content of the DNA of the type strain is 71.0 mol%.
The species currently contains the type strain FSCA67T (=DSM 45796T = KACC 17155T = NCIMB 14829T) and strains FGG38 and FGG39, all of which were isolated from the F-horizon of a Picea sitchensis forest at Hamsterley Forest, County Durham, England.
Acknowledgments
Patrycja Golinska was supported by a research fellowship from Nicolaus Copernicus University (Torun, Poland) “Enhancing Educational Potential of Nicolaus Copernicus University in the Disciplines of Mathematical and Natural Sciences” (project no. POKL.04.01.01-00-081/10). The authors are indebted to Dr. Byung-Yong Kim for carrying out the fatty acid analysis on the type strain of S. durhamensis.
References
- Cho S-H, Han J-H, Ko H-Y, Kim SB. Streptacidiphilus anmyonensis sp. nov., Streptacidiphilus rugosus sp. nov. and Streptacidiphilus melanogenes sp. nov., acidophilic actinobacteria isolated from Pinus soils. Int J Syst Evol Microbiol. 2008;58:1566–1570. doi: 10.1099/ijs.0.65480-0. [DOI] [PubMed] [Google Scholar]
- Collins MD. Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE, editors. Chemical methods in bacterial systematics. London: Academic Press; 1985. pp. 267–287. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Golinska P, Wang D, Goodfellow M. Nocardia aciditolerans sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek. 2013;103:1079–1088. doi: 10.1007/s10482-013-9887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Saiz-Jimenez C. A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol. 2002;4:770–773. doi: 10.1046/j.1462-2920.2002.00362.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Saiz-Jimenez C. A simple fluorimetric method for the estimation of DNA–DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles. 2005;9:75–79. doi: 10.1007/s00792-004-0417-0. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Dawson D. Qualitative and quantitative studies of bacteria colonizing Picea sitchensis litter. Soil Biol Biochem. 1978;10:303–307. doi: 10.1016/0038-0717(78)90027-5. [DOI] [Google Scholar]
- Goodfellow M, Simpson KE. Ecology of streptomycetes. Front Appl Microbiol. 1987;2:97–125. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- Huang Y, Cui Q, Wang L, Rodriguez C, Quintana E, Goodfellow M, Liu Z. Streptacidiphilus jiangxiensis sp. nov., a novel actinomycete isolated from acidic rhizosphere soil in China. Antonie Van Leeuwenhoek. 2004;86:159–165. doi: 10.1023/B:ANTO.0000036124.18820.ae. [DOI] [PubMed] [Google Scholar]
- Jones KL. Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J Bacteriol. 1949;57:141–145. doi: 10.1128/jb.57.2.141-145.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer P. Family I. Streptomycetaceae Waksman and Herrici 1943, 339AL emend. Rainey, Ward-Rainey and Stackebrandt 1997, 486 emend. Kim, Lonsdale, Seong and Goodfellow 2003b, 113 emend Zhi, Li and Stackebrandt 2009, 600. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, 2nd Edition, vol 5. The Actinobacteria, Part B. 2. New York: Springer; 2012. pp. 1446–1454. [Google Scholar]
- Kämpfer P. Genus incertae sedis. II. Streptacidiphilus Kim, Lonsdale, Seong and Goodfellow 2003a, 1219VP (Effective publication: Kim, Lonsdale, Seong and Goodfellow 2003b, 115) In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, 2nd Edition, vol 5. The Actinobacteria, Part B. New York: Springer; 2012. pp. 1777–1781. [Google Scholar]
- Kawato M, Shinobu R. On Streptomyces herbaricolor sp. nov., supplement: as simple technique for microscopical observation. Mem Osaka Univ Lib Arts Educ B Nat Sci. 1959;8:114–119. [Google Scholar]
- Khan MR, Williams ST. Studies on the ecology of actinomycetes in soil. VIII. Distribution and characteristics of acidophilic actinomycetes. Soil Biol Biochem. 1975;7:345–348. doi: 10.1016/0038-0717(75)90047-4. [DOI] [Google Scholar]
- Kim SB, Lonsdale J, Seong C-N, Goodfellow M. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici 1943)AL emend. Rainey et al. 1977. Antonie Van Leeuwenhoek. 2003;83:107–116. doi: 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e, a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kroppenstedt RM. Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE, editors. Chemical methods in bacterial systematics. London: Academic Press; 1985. pp. 173–199. [Google Scholar]
- Kűster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vancanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Tase A, Takahashi M, Sakane T, Suzuki K-I, Hatano K, Yamaguchi A. Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek. 2012;101:73–104. doi: 10.1007/s10482-011-9656-0. [DOI] [PubMed] [Google Scholar]
- Lechevalier MP, De Biévre C, Lechevalier HA. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–260. doi: 10.1016/0305-1978(77)90021-7. [DOI] [Google Scholar]
- Lonsdale JT. Aspects of the biology of acidophilic actinomycetes. PhD thesis. Newcastle upon Tyne: University of Newcastle; 1985. [Google Scholar]
- Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- O’Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E. Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek. 1993;64:325–340. doi: 10.1007/BF00873091. [DOI] [PubMed] [Google Scholar]
- Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, MIDI Inc. Newark
- Seong CN, Goodfellow M, Ward AC, Hah YC. Numerical classification of acidophilic actinomycetes isolated from acid soil in Korea. Korean J Microbiol. 1993;31:355–363. [Google Scholar]
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega 5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huang Y, Liu Z, Goodfellow M, Rodriguez C. Streptacidiphilus oryzae sp. nov., an actinomycete isolated from rice field soil in Thailand. Int J Syst Evol Microbiol. 2006;56:1257–1261. doi: 10.1099/ijs.0.64165-0. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, et al. International committee on systematic bacteriology. Report on the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–465. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- Williams ST, Flowers TH. The influence of pH on starch hydrolysis by neutrophilic and acidophilic actinomycetes. Microbios. 1978;20:99–106. [PubMed] [Google Scholar]
- Williams ST, Khan MR. Antibiotics—a soil microbiologist’s viewpoint. Post Hig I Med Dosw. 1974;28:395–408. [PubMed] [Google Scholar]
- Williams ST, Robinson CS. The role of streptomycetes in decomposition of chitin in acidic soils. J Gen Microbiol. 1981;127:55–63. [Google Scholar]
- Williams ST, Davies FL, Mayfield CI, Khan MR. Studies on the ecology of actinomycetes in soil. II. The pH requirements of streptomycetes from two acid soils. Soil Biol Biochem. 1971;3:187–195. doi: 10.1016/0038-0717(71)90014-9. [DOI] [Google Scholar]
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]