Summary
RNA interference (RNAi) is an ancient process by which noncoding RNAs regulate gene expression in a sequence-specific manner. The core components of RNAi are small regulatory RNAs, ~21 to 30 nucleotides in length, including small interfering RNAs (siRNAs) and microRNAs (miRNAs). The past two decades have seen considerable progress in our understanding of the molecular mechanisms underlying the biogenesis of siRNAs and miRNAs. Recent advances have also revealed the crucial regulatory roles played by small RNAs in such diverse processes as development, homeostasis, innate immunity, and oncogenesis. Accumulating evidence indicates that RNAi initially evolved as a host defense mechanism against viruses and transposons. The ability of the host small RNA biogenesis machinery to recognize viral double-stranded RNA replication intermediates and transposon transcripts is critical to this process, as is small RNA-guided targeting of RNAs via complementary base pairing. Collectively, these properties confer unparalleled specificity and precision to RNAi-mediated gene silencing as an effective antiviral mechanism.
Keywords: RNAi, siRNAs, miRNAs, antiviral immunity
Introduction
RNA interference (RNAi) is an evolutionarily conserved mechanism of gene regulation that functions in organisms as diverse as the unicellular fission yeast Saccharomyces pombe and humans (1-6). The central components of RNAi include two classes of short (~21–30 nucleotides) regulatory RNAs termed small interfering RNAs (siRNAs) and microRNAs (miRNAs) (5-8). RNAi can suppress gene expression at the transcriptional level by modulating chromatin structure, and under certain circumstances, small RNAs can also activate gene expression (9-11). However, the siRNAs and miRNAs most often act post-transcriptionally to destabilize and/or inhibit the translation of complementary target mRNAs in a sequence-specific manner. RNAi is a crucial component of the host defense system against exogenous and endogenous ‘parasitic’ nucleic acids in the form of viral genome replication intermediates and transposon transcripts, respectively. In this review, we summarize our progress in understanding the molecular mechanisms underlying the biogenesis of small RNAs and their roles in antiviral immunity. Readers are also directed to several excellent recent reviews that cover various aspects of these topics (12-14). We first outline the similarities and differences in the initiation (biogenesis) and effector steps (target gene repression) of RNAi in various organisms, with particular emphasis on the mechanism of RNAi in Drosophila. We then discuss how the siRNA pathway contributes to antiviral immunity in Drosophila and describe the diverse strategies employed by viruses to suppress RNAi. Finally, we describe examples of miRNAs that modulate host-virus interactions in mammals, emphasizing their role in combating human immunodeficiency virus (HIV) infection.
Biogenesis and function of miRNAs
miRNAs are endogenous small RNAs expressed in every cell type of higher organisms. miRNAs are encoded in the genome as independent intergenic transcription units, clusters of multiple genes within a single transcription unit, or within intronic regions of protein-coding genes, in which case they are spliced from the host gene transcript. Canonical miRNA biogenesis begins with the transcription, primarily by RNA polymerase II, of long stem-loop transcripts known as primary miRNA transcripts (pri-miRNAs) (Figs 1 and 2) (15). Pri-miRNA transcripts range in length from a few hundred to hundreds of thousands of nucleotides, and they undergo the same maturation steps as conventional pre-mRNA transcripts: namely, addition of a 5′ cap structure, removal of introns by the splicing machinery, and polyadenylation at the 3′ terminus. In animals, pri-miRNAs undergo initial processing in the nucleus by a microprocessor complex composed of the ribonuclease III (RNase III) Drosha and the double stranded (ds) RNA-binding protein (RBP) Pasha/DGCR8, which liberates ~60–70 nucleotide precursor miRNAs (pre-miRNAs) that carry a dinucleotide overhang at the 3′ terminus, a characteristic of RNase III products (16-19). The Exportin 5/Ran-GTP complex specifically recognizes this 3′ overhang and transports the pre-miRNAs to the cytoplasm (20-22), where they are processed by another RNase III complex consisting of the RNase III Dicer-1 and the dsRBP Loquacious (Loqs)-PB/TRBP/PACT. This complex produces ~22–24 nucleotide miRNA duplexes with dinucleotide overhangs at both 3′ ends (23-27). Both strands feature 5′ phosphate and 3′ hydroxyl groups, also characteristic of RNase III-mediated cleavage products. Interestingly, plants lack a Drosha ortholog, and in contrast to the two-step compartmentalized miRNA maturation process observed in animals, miRNA biogenesis in plants is carried out in the nucleus by a single Dicer-like 1 (DCL1) RNase III family protein. DCL1 and the RBP hyponastic leaves 1 (HYL1) process pri-miRNAs into miRNA duplexes (28-35). Efficient miRNA processing in plants also requires the zinc finger protein SERRATE, the RBP DAWDLE, and the cap-binding complex (CBC) (36-40). The CBC and Arsenite-resistance protein 2 (Ars2), an ortholog of SERRATE, have also been implicated in miRNA biogenesis in animals (41, 42). A number of additional RBPs, including La, KSRP, SMAD, and Lin28, as well as the terminal uridyl transferases TUT2/4/7 and the nuclease MCPIP1, have been shown to affect miRNA biogenesis (43-48). We expect that the list of proteins involved in miRNA biogenesis and function will continue to grow, as many candidate miRNA factors are currently awaiting functional characterization (49, 50).
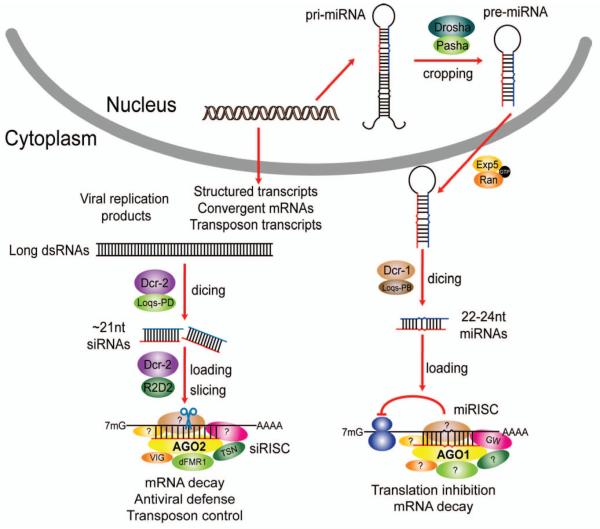
Fig. 1. Outline of the siRNA and miRNA pathways in Drosophila.
Exogenous dsRNAs (viral RNA replication intermediates or experimental reagents) or endogenous dsRNAs (structured transcripts, transposon transcripts, and convergently transcribed mRNAs that hybridize) are processed by the Dcr-2/Loqs-PD complex into siRNAs, which are then incorporated into AGO2-containing siRISCs in a Dcr-2/R2D2-dependent manner. There, siRNAs target mRNAs by complementary base pairing, which leads to degradation of the target mRNA. Primary miRNA transcripts are processed in the nucleus by the Drosha/Pasha complex into pre-miRNAs, which are subsequently exported to the cytoplasm by Exportin 5/Ran-GTP and further processed by the Dcr-1/Loqs-PB complex into miRNA duplexes. The miRNA strands are selectively incorporated into AGO1-containing miRISCs, where they target mRNAs via complementary base pairing between the miRNA seed sequence and miRNA-binding sites in target mRNAs, leading to translation inhibition and target mRNA destabilization.
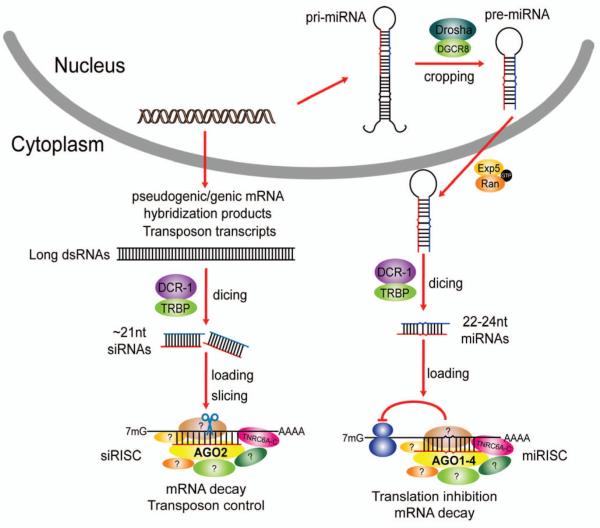
Fig. 2. Gene silencing pathways in mammals.
In oocytes and embryonic stem cells, dsRNAs are formed by hybridization between antisense pseudogenic transcripts and spliced genic mRNAs or between sense and antisense transposon transcripts. The dsRNAs are processed by Dicer into siRNAs, which are then incorporated into AGO2-containing siRISCs where they silence the expression of complementary target mRNAs or transposon transcripts via the slicer activity of AGO2. Guide strand perfectly matches the accessible sites in a target mRNA (178), forming an A-form helix required for the slicer activity of AGO2 (179-181). In all cell and tissue types, primary miRNA transcripts are processed in the nucleus by the Drosha/DGCR8 complex into pre-miRNAs, which are exported to the cytoplasm by Exportin 5/Ran-GTP and further processed into miRNA duplexes by the DCR-1/TRBP complex. The miRNA strand of the duplex is randomly partitioned among the four AGO proteins to form miRISCs. miRNAs guide the targeting of mRNAs via complementary base pairing between the miRNA seed sequence and miRNA-binding sites in target mRNAs, leading to translation inhibition, localization to cytoplasmic structures P bodies (182), and target mRNA destabilization (183).
To perform their gene regulatory functions, plant and animal miRNAs are incorporated into RNA-induced silencing complexes (RISCs) in a process referred to as loading. RISCs are multiprotein complexes composed of effector and accessory proteins together with the appropriate small RNAs. The key effector proteins of RISCs are the Argonaute (AGO) proteins, which mediate mRNA cleavage and/or translation inhibition. Typically, only one strand of the miRNA duplex, termed the mature miRNA strand, associates with the miRISC, and the remaining strand (known as the miRNA* or star strand) is discarded. There have been descriptions of both the mature miRNA strands and the miRNA star strands joining separate miRISC to carry out gene regulatory functions (51-53). In these cases, the two strands are most likely derived from distinct miRNA duplexes. In mammals, miRNAs randomly associate with one of four AGO proteins, all of which are competent to execute miRNA-mediated gene regulatory function. In contrast, plant miRNAs predominantly associate with AGO1. The Drosophila genome encodes two members of the AGO clade proteins, which bind different classes of small RNAs. AGO1 primarily binds miRNAs, whereas AGO2 tends to associate with siRNAs (54, 55). Exceptions to this bias include miR-277, which preferentially associates with AGO2 because of the high degree of complementary base pairing between the miRNA and miRNA* strand in the duplex (55, 56).
Once the mature miRNA, AGO, and accessory proteins are assembled into the miRISC, the target mRNAs are engaged via complementary base pairing between the seed region (positions 2–8) of the mature miRNA strand and the miRNA-binding sites (primarily in the 3′ UTR) of the target mRNAs. Gene expression is repressed by mRNA destabilization and/or translation inhibition, as described further below (57-62). The endoribonuclease or ‘slicer’ activity of AGO proteins is dependent on substantial miRNA–mRNA complementarity. In animals, miRNA–target mRNA base pairing is typically imperfect and inhibition of translation is the primary mode of miRNA-mediated target gene repression. This subsequently leads to mRNA destabilization by removal of the 5′ cap and 3′ polyA tail (63, 64). In contrast, plant miRNAs often display perfect complementarity with their target mRNAs. Thus, it has been proposed that plant miRNAs repress target gene expression predominantly through mRNA destabilization. Interestingly, a recent study reported widespread translation inhibition in plants (65). It remains to be determined whether translation inhibition precedes or takes place in parallel with mRNA destabilization. Consistent with the different mechanisms of miRNA-mediated gene silencing in plants and animals, plant AGO1 mainly binds miRNAs and possesses robust slicer activity (66); in contrast, Drosophila AGO1 has poor slicer activity (55). In mammals, the four AGO proteins all inhibit translation, but the catalytic core residues required for slicer activity are preserved only in AGO2 (67). Plants and animals also differ in the 3′ modifications of miRNAs. During miRNA biogenesis in plants, both duplex strands are 2′-O-methylated at the 3′ end by the methyltransferase Hua Enhancer 1 (HEN1) (68-70). The biological relevance of this 3′ modification had been unclear but was recently found to be required to maintain the stability of miRNAs. In Drosophila, the 3′ terminal 2′-O-methyl group is missing from miRNAs associated with AGO1, but is present on those bound to AGO2. Consequently, AGO1-bound miRNAs are subject to 3′ deadenylation and subsequent degradation upon perfect complementary base pairing with target mRNAs (71), whereas the AGO2-bound miRNAs remain stable. Similar observations have been made for mammalian miRNAs (71). These findings confirmed that the HEN1-mediated 3′ modification is critical for maintaining the stability of plant miRNAs, which predominantly show perfect base pairing with their target mRNAs.
miRNAs can also be generated through several non-canonical pathways; for example, the biogenesis of a subset of miRNAs occurs in a microprocessor-independent and Dicer-dependent manner. In Caenorhabditis elegans, Drosophila, and vertebrates, an miRNA subset called mirtrons are derived from short group II introns. These introns, in the form of lariat structures that are excised from the canonical splicing reaction, are linearized by debranching enzymes, refolded into stem-loop pre-miRNA configurations, exported into the cytoplasm by Exportin 5/Ran-GTP, and further processed into mature miRNAs by Dicer proteins (72-74). In a variation of this process, some atypically longer mirtron precursors are first trimmed from the 5′ end by a currently unidentified nuclease(s) or from the 3′ end by exosomes before being processed into mirtrons by Dicer proteins (75-77). The precursor of tRNA-Ile/mir-1983, as well as certain small nucleolar RNAs (snoRNAs), can also adopt pre-miRNA–like hairpin conformations and are similarly exported into the cytoplasm and processed into miRNAs (76, 78). These small RNAs appear to be bona fide miRNAs. For example, snoRNA-derived RNAs (sdRNAs) can associate with AGO proteins and are capable of repressing expression of the complementary target mRNAs (78). Furthermore, some tRNA-derived RNAs (tdRNAs) that are processed by Dicer, as well as certain 3′ trailer fragments released by tRNAse Z cleavage during tRNA biogenesis, can associate with AGO proteins, thus resembling functional miRNAs (79). Moreover, miRNAs encoded by murine γ-herpesvirus 68 (MHV68) reside immediately downstream of a tRNA. The 5′ and 3′ ends of these miRNAs are defined by tRNAse Z cleavage and RNA polymerase III transcriptional termination, respectively (80-82). Finally, the biogenesis of a small number of miRNAs requires AGO, but not Dicer proteins. For example, microprocessor-mediated processing of the primary transcripts of miR-451, a conserved miRNA in vertebrates, releases pre-miRNAs that have unusually short stem-loop structures and are thus suboptimal substrates for Dicer proteins. Instead, the second step of miRNA processing is performed by the slicer activity of AGO2 followed by 3′ end maturation by a currently unidentified nuclease(s) (83-85).
Biogenesis and function of siRNAs
siRNAs are typically derived from dsRNA or hairpin RNA precursors (Figs 1 and 2). These precursor RNAs may be produced from exogenous sources such as viral RNA replication intermediates or from endogenous sources such as retrotransposon transcripts, which form dsRNAs, at least in part, by hybridization of sense and antisense transcripts in trans. In Drosophila, some transcripts can adopt extensively structured dsRNA configurations. Furthermore, the overlapping regions of convergently transcribed RNAs can hybridize to form dsRNAs (86-89). In mouse oocytes, dsRNAs can be generated by pairing between pseudogene transcripts and genic transcripts or between sense and antisense transposon transcripts (90, 91). Endogenous small hairpin RNAs have also been discovered in mouse embryonic stem cells (76). Regardless of their origin, dsRNAs and hairpin RNAs are processed by Dicer proteins to generate siRNAs. In worms and mammals, a single Dicer protein, Dicer-1, is responsible for the biogenesis of both siRNAs and miRNAs, whereas in Drosophila, Dicer-2 is dedicated to siRNA biogenesis. Dicer proteins are assisted by the RBPs Loqs-PD/TRBP, which associate with Dicer and play a critical role in defining the substrate specificity of Dicer proteins and in maintaining the precision of Dicer-mediated small RNA biogenesis (92-94). For example, R2D2, the binding partner for Drosophila Dcr-2, acts in concert with inorganic phosphate to inhibit Dcr-2–mediated processing of pre-miRNAs, thus restricting Dcr-2 activity to siRNA biogenesis (95). In addition, Drosophila Loqs-PB and mammalian TRBP contribute to the precise processing of a subset of miRNAs by Dicer proteins (96, 97).
As is observed for miRNAs, siRNA duplexes are incorporated into AGO-containing siRISCs. The molecular mechanism of siRISC formation is best characterized in Drosophila, where the Dcr-2/R2D2 heterodimer plays a critical role in gauging the thermodynamic stability of both ends of the siRNA duplex, which results in a polarized binding pattern. R2D2 binds the 5′ end with the greatest double-stranded character; that is, the more thermodynamically stable end, whereas Dcr-2 favors the end with less stable base pairing, thereby orienting the heterodimer on the siRNA duplex to form the RISC-loading complex (98, 99). Given the involvement of Dcr-2 in the processing of long dsRNAs into siRNAs, which typically starts from the ends of the dsRNA and proceeds progressively, it is conceivable that the processed siRNA duplexes are first released from Dcr-2 and then rejoin the Dcr-2/R2D2 heterodimer in the RISC-loading complex in an orientation dictated by the relative thermodynamic stability of the duplex ends. A number of other proteins then associate with the RISC-loading complex, including AGO2 and the Hsc70/Hsp90 chaperone machinery, to form the pre-RISC (100). Assembly of the pre-RISC, or the handover of the siRNA duplex from Dcr-2/R2D2 to AGO2, is an ATP-dependent process. It has been proposed that the chaperone machinery induces an ATP-dependent conformational change in AGO2 to accommodate the bulky siRNA duplex (100). Subsequently, the passenger strand (complementary to the guide strand) of the duplex is cleaved by the slicer activity of AGO2, a process referred to as passenger strand nicking (101-104). Finally, the siRNA duplex is unwound in an ATP-independent manner, resulting in the formation of a mature RISC containing only the siRNA guide strand. Duplex unwinding and removal of the sliced passenger strand are facilitated by the Mg2+-dependent endoribonuclease complex C3PO, consisting of Traslin and Trax (105-107). Target mRNAs are recruited to the mature AGO2-containing siRISC via perfect base pairing with the siRNA guide strand. AGO2 slices the target mRNA at the position complementary to bases 10 and 11 of the siRNA guide strand (67, 108, 109). Drosophila AGO2 can also repress target gene expression by translation inhibition (110). As was described for miRNAs in plants, AGO2-bound small RNAs in Drosophila (primarily siRNAs and a subset of miRNAs) are 2′-O-methylated at their 3′ ends by the methyltransferase DmHen1/Pimet (111, 112). A key difference between the two species is that in Drosophila, the 3′ end is modified after removal of the passenger strand, ensuring that only the guide strand is modified. In contrast, both strands of the duplex are modified in plants. Among the four mammalian AGO proteins, only AGO2 possesses slicer activity and siRNA-mediated gene silencing in mammals is therefore mediated primarily by AGO2.
In addition to primary siRNAs, worms and plants can generate secondary siRNAs through the action of RNA-dependent RNA polymerases (RdRPs) and/or Dicer proteins. In worms, secondary siRNAs are synthesized de novo in a non-primed, Dicer-independent manner. These secondary siRNAs feature a 5′ triphosphate group (113, 114). In contrast, secondary siRNA biogenesis in plants occurs via RdRP-dependent synthesis of long dsRNAs that are subsequently processed by Dicer. The resulting secondary siRNAs resemble primary siRNAs in that they carry monophosphate and hydroxyl groups at their 5′ and 3′ termini, respectively. Although Drosophila and mammals are not known to express RdRPs, a recent study has identified a complex capable of producing dsRNAs. The complex was composed of the catalytic subunit of human telomerase reverse transcriptase and the RNA component of the mitochondrial RNA-processing endoribonuclease, and the resulting dsRNAs were subsequently processed into siRNAs by Dicer (115). Thus, it seems likely that secondary siRNAs may be produced in other organisms, although this remains to be confirmed.
Crosstalk between the siRNA and miRNA biogenesis and effector pathways
As is evident from the preceding sections, there are several points of overlap and crosstalk between the siRNA and miRNA biogenesis and/or effector pathways, and the extent of crosstalk varies considerably in different organisms. At first glance, the siRNA and miRNA pathways of Drosophila appear to be relatively compartmentalized, and their biogenesis and effector machineries are distinct. For example, Dcr-1 and AGO1 operate in the miRNA pathway and Dcr-2 and AGO2 operate in the siRNA pathway. Moreover, multiple Loqs RBP isoforms are produced by alternative splicing and polyadenylation of mRNAs. Loqs-PB preferentially associates with Dcr-1 and participates in miRNA biogenesis, whereas Loqs-PD specifically binds Dcr-2 and plays a critical role in siRNA biogenesis (24, 25, 92, 93). In addition, newly formed miRNAs and siRNAs partition between AGO1- and AGO2-containing RISCs according to the 5′ modification and the sequence complementarity at the center of the duplex, thus dictating the distinct modes of gene silencing (51-53, 55, 56). Despite their seemingly disparate characteristics, recent studies have revealed several layers of crosstalk between the miRNA and siRNA pathways. Some RNAs and proteins are present in both siRISCs and miRISCs, which results in competition between the pathways when the concentrations of key components are limiting. For example, depletion of AGO2 can enhance miRNA-mediated gene silencing (49), presumably because miRISC formation is increased when previously limiting factors become available to complex with AGO1. Furthermore, both exogenous siRNA and endogenous siRNA pathways (referred to as siRNAs and endo-siRNA pathways, respectively) may be operating simultaneously. The biogenesis and function of siRNAs and endo-siRNAs are mediated through the same sets of proteins, which results in three-way competition for components between the siRNA, endo-siRNA, and miRNA pathways (116). Finally, our recent genome-wide RNAi screening effort in cultured Drosophila cells has identified a number of proteins that are implicated in the siRNA, endo-siRNA, and miRNA pathways, further supporting the notion of crosstalk between all three pathways of RNAi (49).
In mammals, the lone Dicer protein Dcr-1 is responsible for the biogenesis of both siRNAs and miRNAs, but there is little additional evidence of crosstalk between the two pathways. Indeed, endogenous siRNAs have been identified only in oocytes and embryonic stem cells thus far (76, 90, 91). A similar lack of crosstalk between the siRNA and miRNA pathways is apparent in C. elegans. Of the >300 genes identified in two extensive parallel genome-wide RNAi screens of this organism, only two genes affected the biogenesis of both siRNAs and miRNAs, one of which was Dcr-1 (50, 117). In contrast, multiple layers of crosstalk between the siRNA and endo-siRNA pathways have been uncovered in C. elegans. For example, proteomic analysis of the Dcr-1 complex identified four proteins, including ERI-1, mutations in which are associated with enhanced RNAi in response to exogenous dsRNAs. ERI proteins are also required for the biogenesis of endogenous siRNAs (116, 118), suggesting that ERI proteins may be examples of the ‘limiting factors’ in the siRNA and endo-siRNA pathways in worms that could influence the relative activities of the two pathways.
The role of the siRNA pathway in antiviral immunity
RNAi regulates the expression of diverse target mRNAs and thus plays a key role in numerous biological processes including cancer, development, homeostasis, and antiviral defense. Although RNAi was discovered in C. elegans as a gene silencing process that unifies miRNA-mediated regulation of developmental gene expression and dsRNA-mediated gene silencing, it seems likely that the process evolved as a sequence-specific defense mechanism against viruses and transposons (119). In fact, some of the first siRNAs discovered were virus-derived small RNAs from tobacco cells infected by a pathogenic RNA virus, potato virus X (4).
Viruses are obligate intracellular parasites that rely on the host organism for successful replication and dissemination. Upon infection of the host cell, viruses subvert the host protein and nucleic acid synthesis machinery to facilitate viral genome replication and subsequent particle packaging and release. Organisms have evolved various defense mechanisms to combat viral infection and RNAi has emerged as an integral and critical component of antiviral defense. siRNAs and miRNAs have been shown to modulate host-virus interactions in a variety of organisms ranging from plants to mammals.
With the exception of retroviruses, all RNA viruses generate dsRNA intermediates during replication in the cytoplasm of host cells. These dsRNAs are perfect substrates for Dicer complexes and are thus readily destroyed. This process is well documented in Drosophila. Flies carrying mutations in Dcr-2 and R2D2 succumb more rapidly than wildtype flies to flock house virus (FHV) and cricket paralysis virus (CrPV) infections. The hypersensitivity of these mutants correlated with the accelerated accumulation of viral genomic RNAs and coat proteins (120, 121). Consistent with a role for RNAi in control of infection, 21-nucleotide viral siRNAs (vsiRNAs) were detected in cultured FHV-infected Drosophila cells (122). The zinc finger protein Ars2 is required for optimal Dcr-2–mediated siRNA biogenesis and is also critical for viral defense in Drosophila. Flies or fly cells depleted of Ars2 accumulate viral genomic RNAs and proteins and are more susceptible to infection by a number of RNA viruses, including Drosophila C virus (DCV), FHV, vesicular stomatitis virus, and Sindbis virus (41). Similarly, cultured Drosophila cells depleted of Cbp80 and Cbp20, components of the Dcr-2–interacting CBC important in siRNA biogenesis, support more robust viral replication than control cells (41). Finally, Drosophila cells infected with positive (+) strand viruses contain approximately equal numbers of (+) and negative-strand (−) vsiRNAs, despite the fact that (+) strands are >50-fold more abundant than (−) strands during the genome replication cycle (123-125). This is in good agreement with the notion that vsiRNAs are derived from the viral dsRNA replication intermediates, rather than from the single-stranded viral genomic RNAs. In the case of FHV, the vsiRNAs do not map uniformly across the viral genome but instead are derived mainly from specific hot spots, one of which is the 5′-terminal 400-bp region of the (+) strand viral RNA1. This finding suggests that the predominant source of FHV vsiRNAs is the dsRNA region of the viral replication intermediates between the nascent 5′-terminal region of the progeny (+) RNA1 and the 3′-terminal region of the (−) RNA1 template formed during initiation of the progeny (+) RNA synthesis (126). Collectively, these data illustrate the critical contribution of Dcr-2–mediated destruction of viral dsRNA replication intermediates to antiviral immunity in Drosophila.
Host organisms also mobilize the effector step of RNAi in antiviral defense. vsiRNAs processed by Dicer proteins associate with AGO-containing RISCs to target viral genomic RNAs with exquisite specificity. In FHV-infected Drosophila cells, for example, a significant proportion of AGO2-associated siRNAs was shown to be derived from the viral genome (86). In addition, flies carrying homozygous mutations in AGO2 are more susceptible to lethal infection by DCV and CrPV than are wildtype animals, despite the presence of wildtype Dcr-2 (127). This hypersensitivity also correlated with higher viral RNA levels and viral titers (127). In addition, flies carrying mutations in genes encoding Rm62 or VIG, components of siRISCs, are hypersensitive to infection by Drosophila X virus (DXV), a bisegmented dsRNA virus (128). These data clearly indicate that the RNAi effector step is required for effective antiviral immunity in Drosophila. Of note, no viral RNA cleavage products of AGO2-containing siRISCs have yet been identified. This may be due to the diverse sequences of vsiRNAs. Thus, the abundance of cleavage products from individual vsiRNA-programmed RISCs may be too low to detect. Alternatively, only a very small fraction of vsiRNAs may be loaded into AGO2. The latter possibility is supported by observations with Drosophila S2 cells persistently infected by FHV, in which even the most abundant vsiRNAs failed to repress the expression of a perfectly complementary sensor mRNA transcript. Furthermore, the bulk of these vsiRNAs failed to load into either AGO1 or AGO2 (125). In another study, cultured Drosophila cells were acutely infected with a mutant FHV that lacked B2, a suppressor of RNAi. In these cells, only a minor fraction of vsiRNAs was loaded into AGO2 siRISCs, and these vsiRNAs were 2′-O-methylated at their 3′ ends (126). Thus, Dicer-mediated destruction of viral RNA replication intermediates appeared to be the major antiviral mechanism in S2 cells persistently infected with FHV, whereas the RNAi effector step contributes to antiviral defense during acute infection. It will be interesting to determine whether the differential requirement for Dicer and AGO proteins in Drosophila antiviral immunity results from fundamental differences in the host cellular responses to acute and persistent viral infection.
Several unique features of RNAi illustrate the remarkable specificity of RNA-based antiviral defense. First, at the initiation phase of RNAi, Dicer recognition of the unique double-stranded structure of the viral RNA replication intermediates allows the host RNAi machinery to control viral RNA replication in a sequence-independent manner. Second, at the effector phase of RNAi, vsiRNAs are mobilized to target viral RNAs with perfectly complementary sequences. This ensures that viral genomes are targeted for destruction with remarkable precision even when the viral genome is extensively mutated, which is a common strategy to evade the host antigen-specific response to viral proteins. Third, RNAi-based antiviral defense mechanisms in Drosophila show characteristics reminiscent of the recall response in adaptive immunity. Thus, exposure to viral dsRNAs or a sublethal dose of virus can confer protection against a subsequent lethal dose of the same virus, essentially ‘vaccinating’ the flies (129).
Both the host and virus are under intense selective pressure to evolve more effective defense and counter-defense mechanisms, respectively, as supported by the observation that genes encoding RNAi-related factors are among the fastest evolving genes in Drosophila (130). On the other hand, viruses have evolved counter-defense strategies to evade or suppress the host RNAi machinery (Fig. 3). For example, one evasion strategy is for the virus to create a physical barrier within the host cell. Replication of the FHV genome in cultured Drosophila cells takes place in viral RdRP-containing membranous vesicles called spherules, which form on the outer mitochondrial membrane (131-133). Such compartmentalization serves to shield the viral RNA replication intermediates from the cellular RNAi machinery. In addition, many viruses encode viral suppressors of RNAi (VSR) in their genome, indicating that RNAi exerts active selective pressure on viruses. VSR-mediated binding and sequestration of components of the host RNAi machinery appears to be an evasion strategy employed by many viruses. The 1A protein encoded by DCV specifically binds to long dsRNAs, but not siRNAs, and consistent with this, DCV-1A inhibits Dcr-2–mediated processing of dsRNAs into siRNAs, but leaves siRNA-mediated gene silencing intact (127). In contrast, the FHV-B2 protein sequesters viral dsRNAs and suppresses Dcr-2–mediated destruction of viral RNA replication intermediates. FHV-B2 adopts a four-helix bundle conformation that allows it to bind to one side of an A-form RNA duplex independently of the RNA sequence or length (134-136). In addition, FHV-B2 binding and sequestration of vsiRNAs blocks their association with AGO2, thereby suppressing siRISC-mediated cleavage of viral genomic RNAs (136). The Cymbidium ringspot virus P19, on the other hand, adopts a head-to-tail homodimeric conformation that permits exclusive binding of 21-bp duplex siRNAs and prevents their loading into siRISCs (137, 138). VSRs operate not only by interfering with vsiRNA biogenesis and siRISC assembly but also by suppressing the catalytic activity of siRISCs. CrPV-1A physically associates with AGO2 and inhibits its slicer activity but does not affect siRNA loading or assembly of the mature siRISC (139). Many other counter-defense mechanisms are mediated through VSRs, ranging from targeted degradation of AGO proteins to degradation of vsiRNAs (140-142). Collectively, these observations highlight the diverse strategies employed by viruses to suppress the antiviral RNAi pathway in Drosophila. FHV-B2 also functions as a potent VSR in non-host plant cells, underscoring the functional conservation of VSRs in countering the host RNAi-based defense system (122).
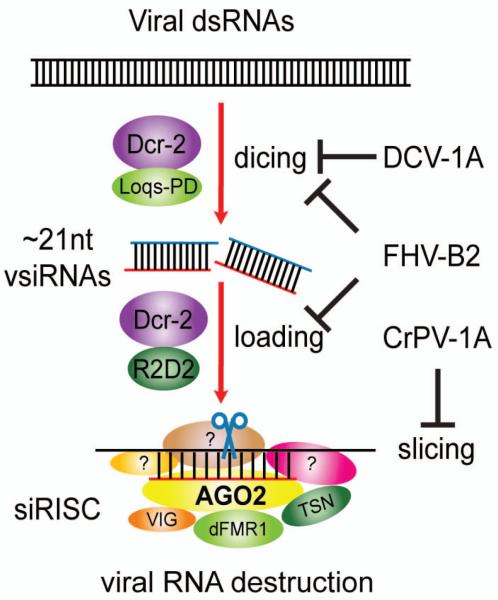
Fig. 3. Viral suppressors of RNAi impinge on various steps in RNAi.
DCV-1A selectively associates with long dsRNAs and inhibits Dcr-2–mediated processing of viral dsRNAs into vsiRNAs. FHV-B2 binds both siRNAs and long dsRNAs, thereby inhibiting Dcr-2–mediated viral dsRNA destruction and the loading of vsiRNAs into AGO2. CrPV-1A associates with AGO2 and inhibits its slicer activity, thereby impairing the effector step of RNAi.
In plants, worms, and flies, RNAi can operate at sites remote from primary site of gene silencing, a process known as systemic RNAi. For instance, silencing signals in plants can travel between cells through plasmodesmata and over longer distances through the phloem (143). In C. elegans, dsRNA uptake and transport between cells is mediated by dedicated RNA transporter proteins such as SID-1 (144-146). In flies, receptor-mediated endocytosis is essential for systemic RNAi. This process mediates cellular uptake of long RNAs but not short RNAs of the lengths typically observed in siRNAs, suggesting that systemic antiviral RNAi in flies depends on long dsRNAs (129, 147). The ability of long viral dsRNAs and short vsiRNAs to mediate systemic silencing in plants, worms, and flies underscores the remarkable versatility of RNAi-dependent antiviral mechanisms in combating invading viruses at both the primary infection site and remote sites throughout the organism.
In contrast to the well-documented role of the siRNA pathway in antiviral immunity in plants and invertebrates, the role played by siRNAs in mammalian antiviral defense is less clear. This is due in part to the dominant role played by the mammalian interferon (IFN) response to viral infection. Cells of the innate immune system detect viral infection predominantly through cell surface or intracellular pattern recognition receptors such as Toll-like receptors (TLRs), nucleotide-oligomerization domain receptors (NLRs), and the RNA helicase family of retinoic acid-inducible gene I-like receptors (RLRs). Two RLRs, retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), play particularly important roles in RNA virus defense. RIG-I detects viral RNAs that carry 5′ triphosphates and adopt short dsRNA conformations with base-paired 5′ ends, as opposed to the cap structure in host cellular mRNAs (148, 149). MDA5 is thought to bind long dsRNAs such as viral RNA replication intermediates (150). Interestingly, Drosophila Dcr-2 shows greater sequence homology at the helicase domain to RIG-I and MDA5 than to other RNA helicases (151). The dominant role played by IFN in mammalian antiviral immunity is illustrated by the fact that endogenous siRNAs derived from long dsRNA precursors have been identified only in oocytes and embryonic stem cells, and notably, the IFN pathway is less active in these cells (76, 90, 91). In other cell types, long dsRNAs are readily detected by the innate receptors, which trigger the IFN response. Therefore, it seems likely that the RNAi machinery in mammals is primarily dedicated to miRNA biogenesis and function, and not to antiviral defense. In support of this notion, a recent study identifying HIV-encoded small RNAs in virus-infected cells is the only convincing evidence for the existence of vsiRNAs in mammalian cells (152), and furthermore, VSRs encoded by viruses that specifically infect mammals have not yet been identified. Thus, although the siRNA pathway first evolved as the primary antiviral defense mechanism in lower eukaryotes, this role has been assumed by the more effective IFN response in mammals.
The role of miRNAs in modulating host-virus interactions
The interaction between host and virus is crucial for successful viral replication. Host factors may have beneficial or detrimental effects on virion production and infectivity, and viral regulation of the expression of host factors is a key component in ensuring its survival. Although viruses encode proteins that inactivate or suppress most steps of the host antiviral immune response, they have also evolved miRNA-based mechanisms as a complementary strategy to evade host responses. In invertebrates, viral miRNAs have not yet been identified in infected cells, making the potential role of this pathway unclear. Moreover, there are no reports of invertebrate miRNAs that can effectively target viral RNAs. In contrast, there is a large body of literature documenting the role of host and viral miRNAs as modulators of host-virus interactions in mammals. Many viruses encode miRNAs and exploit the host processing machinery for their biogenesis. Viral infections also influence miRNA production by the host cell. Host and viral miRNAs use a number of mechanisms to regulate the viral life cycle and to evade the host immune system, respectively. For example, viral infection may downregulate expression of host miRNAs that participate in antiviral pathways or that are barriers to establishing latency or immune evasion. In turn, host cells may respond to viral infection by upregulating antiviral miRNAs or miRNAs that interfere with viral latency and replication. Some examples of these mechanisms will be discussed below.
Virus-encoded miRNAs
The first report of virus-encoded miRNAs was made in the human γ-herpesvirus Epstein-Barr virus (EBV) (153). Subsequently, viral miRNAs have been found in a large number of DNA viruses, including other herpesviruses, polyomaviruses, and human adenoviruses. For the most part, viral miRNA biogenesis is analogous to that of host miRNAs, and proceeds through RNA polymerase II-mediated transcription of pri-miRNAs and Drosha- and Dicer-mediated production of mature miRNAs (154). The few exceptions to this mechanism include RNA polymerase III-mediated transcription of viral miRNA precursor transcripts and tRNase Z-mediated processing of mature miRNAs (82). In contrast to the numerous reports of miRNAs expressed by DNA viruses, only a few RNA virus-derived miRNAs have been documented (152, 155, 156), and the biological significance of these findings is under debate (80, 157, 158). Viral miRNA biogenesis invariably results in cleavage of the primary transcripts encoding them and sacrifices large segments of genomic RNA, which seems a remarkably inefficient way to produce ~22–24-nucleotide miRNAs. Furthermore, most RNA viruses replicate their genomes in the cytoplasm, separated from the primarily nuclear microprocessor complexes. Interestingly, a recent study reported the production of virus-derived pri-miRNAs by a microprocessor-like activity in the cytoplasm (159). Further studies will be necessary to determine whether miRNAs can be derived from an RNA virus and, if so, to unravel the molecular mechanisms that underlie their biogenesis.
Virus-encoded miRNAs can modulate the expression of both viral and host mRNAs. One class of host target mRNAs encodes proteins that increase the sensitivity of virus detection by the host innate or adaptive immune systems or encode pro-apoptotic proteins that contribute to the elimination of virally infected host cells. In contrast, the target viral mRNAs primarily encode proteins that promote entrance into the lytic replication cycle, and thus tilt the balance away from latency.
Host mRNA targets
MICB mRNA is a target of miR-UL112, an miRNA expressed by human cytomegalovirus (hCMV), a member of the β-herpesvirus subfamily (160). MICB encodes a stress-induced ligand of the natural killer (NK) cell receptor NKG2D, which plays a crucial role in activating NK cell-mediated lysis of virus-infected cells (161). MICB is also regulated by miRNAs from a number of other herpesviruses, including the Kaposi’s sarcoma-associated herpesvirus (KSHV) miR-K7 and the EBV miR-BART2 (162). Another EBV miRNA, miR-BART5, decreases the expression of the mRNA encoding the pro-apoptotic protein PUMA (p53 upregulated modulator of apoptosis) (163), which renders the infected cells less sensitive to pro-apoptotic agents. Accordingly, inhibition of miR-BART5 by an anti–miR-BART5 oligonucleotide promotes apoptosis of the host cells. This effect is reversed by concomitant siRNA-mediated repression of PUMA (163), establishing PUMA as a bona fide target of miR-BART5. Bcl-2 interacting mediator of cell death (Bim), another pro-apoptotic protein, is repressed by multiple EBV-encoded miRNAs (164). Finally, the pro-apoptotic protein Bcl2-associated factor 1 (BCLAF1) is also a target of multiple miRNAs, including miR-K5, miR-K9, and miR-K10, encoded by the γ-herpesvirus KSHV (165). Collectively, these studies highlight the role of viral miRNAs in reducing the production of proteins involved in the host immune response and in suppressing apoptosis of virally infected host cells.
Viral mRNA targets
The α-herpesviruses herpes simplex viruses 1 and 2 (HSV-1 and HSV-2) maintain lifelong latency in their animal hosts. The HSV-1 latency-associated noncoding transcript (LAT) is a precursor of several miRNAs in infected cells. Among these is miR-H2, which is transcribed in an antisense orientation relative to ICP0, a viral immediate-early transcriptional activator that is important for productive HSV-1 replication and reactivation from latency. Such perfect base-pairing between miR-H2 and ICP0 mRNA suggests that miR-H2 represses ICP0 most likely by slicer-mediated mRNA cleavage and destabilization (166, 167). In addition, the seed region of the HSV-1 miR-H6 undergoes base pairing with ICP4 mRNA and inhibits its expression. ICP4 is a second HSV-1 transcription factor required for expression of most HSV-1 genes during productive infection (167). Thus, virus-encoded miRNAs contribute to the establishment and maintenance of viral latency.
Host-encoded miRNAs
A number of cellular miRNAs also modulate host-virus interactions. The most notable example is the liver-specific miR-122, which plays a key role in hepatitis C virus (HCV) genome replication through interactions between its seed region and partially complementary sites at the 5′ UTR of the HCV RNA genome (168, 169). In addition, miR-122 enhances the translation of HCV mRNAs through the same binding sites (170). These observations reveal that functional interactions between miR-122 and HCV genomic RNA contribute to the tissue tropism of HCV. Viruses have also developed effective strategies to compromise the action of antiviral cellular miRNAs and to promote viral replication. Herpesvirus saimiri (HVS), a simian virus closely related to KSHV, expresses high levels of small noncoding RNAs called HSURs (H. saimiri U-rich RNAs). HSUR1 and HSUR2 interact with cellular miR-27 through partially complementary sites. HSUR expression significantly reduces miR-27 levels and concomitantly upregulates expression of cellular miR-27 target genes (171). HSURs are thought to serve as natural miRNA sponges by sequestering miRNAs from their normal cellular mRNA targets. In another example, a recent report showed that m169, a spliced and highly abundant transcript from murine cytomegalovirus (MCMV), binds through its 3′ UTR to cellular miR-27a and miR-27b and facilitates their degradation (172). Remarkably, mutant viruses in which the m169–miR-27a/b interaction was disrupted showed significantly attenuated replication in vivo (172).
Viral evolution
To determine whether miRNAs might also play a role in immunity to the human immunodeficiency virus HIV-1, Nathans et al. (173) examined miRNA expression in infected human T lymphocytes. A subset of miRNAs was upregulated in these cells, including miR-29a, which specifically targets the HIV-1 3′ UTR region. Inhibition of miR-29a enhanced HIV-1 viral production and infectivity, and conversely, expression of a miR-29a mimic suppressed viral replication. In addition, the interaction between miR-29a and HIV-1 mRNAs enhanced viral mRNA association with RISCs and cytoplasmic structures called P bodies. RNAi-mediated disruption of P bodies released the suppression of translation and enhanced HIV-1 production and infectivity. P bodies could play a role in modulating viral replication by several mechanisms. For example, as supported by the results of Nathans et al., viral mRNA translation could be suppressed by miRNAs and the mRNAs shuttled to P bodies. In addition, viral mRNAs located in P bodies could be released by host or environmental cues to activate viral replication (Fig. 4). This possibility is supported by the observation that cellular proteins such as APOBEC3G are localized to P bodies and assembled into RNP complexes for packaging into viral particles (174, 175).
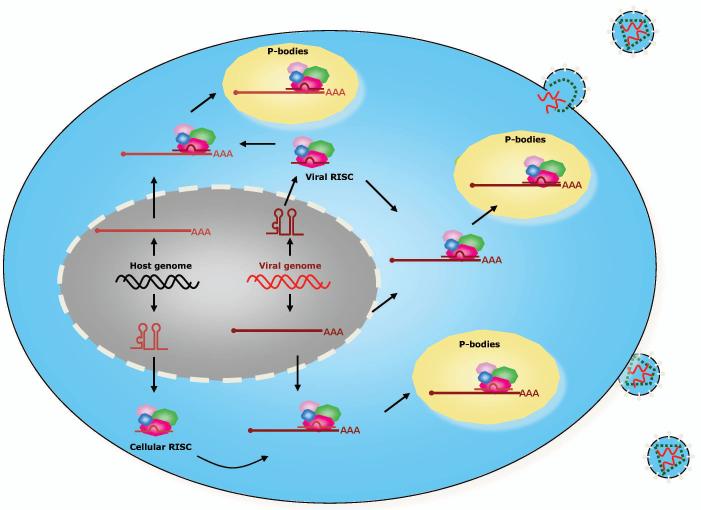
Fig 4. Plausible mechanisms for modulation of host-virus interactions by miRNAs.
Viruses encode miRNA genes and use the host RNAi machinery to assemble viral RISCs that can target host and viral mRNAs. Viral infections also affect the expression of cellular miRNAs and the concentrations of specific miRISCs. Cellular RISCs can target viral mRNAs as part of the host antiviral response. RISCs suppress translation and localize mRNA-RISC RNPs to P bodies. Depending on the prevailing cellular conditions, the suppressed mRNAs may be destroyed or re-enter the translation process. Another mechanism (not shown) could use cellular RISCs to suppress cellular mRNAs integral to the host antiviral response, which would allow the virus to evade the immune response.
Since miR-29 is a conserved miRNA that emerged many millions of years before HIV-1, the findings of Nathans et al. (13) raise the possibility that HIV-1 has evolved to exploit miR-29a to modulate its own life cycle. For example, miRNA-mediated suppression of HIV-1 mRNAs could act as an essential checkpoint in the cycle of viral latency to activation. Such mechanisms would allow viruses to evade the immune system or create viral reservoirs shielded from chemotherapy. This hypothesis can be tested by analyzing miRNA target-site sequence conservation in various HIV-1 subtypes or viral sequences isolated from patients and correlating them with viral infectivity. Interestingly, HIV-1 group O viruses contain non-conserved nucleotides in the region predicted to interact with the 5′ end of the miR-29a seed region, suggesting that this HIV-1 group would not be suppressed by miR-29a. Notably, the main HIV-1 group responsible for the global AIDS epidemic, group M, is about 100-fold more infectious than HIV-1 group O strains, which are endemic only in western and central Africa (176). Similarly, HIV-1 strains with deletions in nef and in the U3 region of the LTR, which contains a miR-29a target-site deletion, failed to cause disease in humans 10 to 14 years after infection (177). Taken together, these findings raise the possibility that the loss of infectivity of HIV-1 group O and nef-deleted HIV-1 may be linked to the reduced ability of viral mRNAs to be targeted by miR-29a for transport to P bodies.
Viruses have thus evolved to exploit the ancient RNAi antiviral defense mechanism by subverting the host RISC machinery to regulate various stages of their own life cycles. A deeper understanding of RNA-based host-virus interactions will not only provide insights into the fundamental mechanisms by which viruses evade the immune response but could also aid in the development of new broad-spectrum therapeutics and vaccines that exploit RNA-based immunity.
Acknowledgements
We are grateful to members of our laboratories for helpful discussions. This work was supported in part by grants from the National Institutes of Health to T.M.R.
Footnotes
The authors have no conflicts of interest to declare.
References Cited
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 7.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 9.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 10.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 11.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 21.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 24.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 31.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 35.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 38.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabin LR, et al. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber JJ, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang C, et al. Sjogren’s syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J Biol Chem. 2012 doi: 10.1074/jbc.M112.401323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heo I, et al. Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 47.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Czech B, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 60.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 61.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- 63.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 66.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 79.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 81.Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang JS, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 89.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 92.Zhou R, et al. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cenik ES, et al. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer Partner Proteins Tune the Length of Mature miRNAs in Flies and Mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee HY, Doudna JA. TRBP alters human precursor microRNA processing in vitro. RNA. 2012;18:2012–2019. doi: 10.1261/rna.035501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 99.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 100.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 101.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 103.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 104.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ye X, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian Y, et al. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol. 2011;18:658–664. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 109.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 110.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 111.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 113.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 114.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 115.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 117.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 118.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 119.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 120.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 122.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 123.Ho T, Pallett D, Rusholme R, Dalmay T, Wang H. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J Virol Methods. 2006;136:217–223. doi: 10.1016/j.jviromet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 124.Yoo BC, et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flynt A, Liu N, Martin R, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci U S A. 2009;106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 129.Saleh MC, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 131.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol. 2001;75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Venter PA, Schneemann A. Recent insights into the biology and biomedical applications of Flock House virus. Cell Mol Life Sci. 2008;65:2675–2687. doi: 10.1007/s00018-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fenner BJ, Goh W, Kwang J. Dissection of double-stranded RNA binding protein B2 from betanodavirus. J Virol. 2007;81:5449–5459. doi: 10.1128/JVI.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu R, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 137.Vargason JM, Szittya G, Burgyan J, Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 138.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nayak A, et al. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pazhouhandeh M, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci U S A. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 144.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 145.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 146.Hunter CP, et al. Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 2006;71:95–100. doi: 10.1101/sqb.2006.71.060. [DOI] [PubMed] [Google Scholar]
- 147.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 152.Schopman NC, et al. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 2012;40:414–427. doi: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 154.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Omoto S, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ouellet DL, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shapiro JS, Langlois RA, Pham AM, Tenoever BR. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 162.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 163.Choy EY, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 169.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marcinowski L, et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathogens. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wichroski MJ, Ichiyama K, Rana TM. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J Biol Chem. 2005;280:8387–8396. doi: 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- 176.Arien KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J Virol. 2005;79:8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Deacon NJ, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 178.Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 179.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 180.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 181.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li Z, Rana TM. Molecular mechanisms of RNA-triggered gene silencing machineries. Accounts Chem Res. 2012;45:1122–1131. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]