Abstract
Fibroblast growth factors (FGFs) mediate a broad range of functions in both the developing and adult organism. The accumulated wealth of structural information on the FGF signalling pathway has begun to unveil the underlying molecular mechanisms that modulate this system to generate a myriad of distinct biological outputs in development, tissue homeostasis and metabolism. At the ligand and receptor level, these mechanisms include alternative splicing of the ligand (FGF8 subfamily) and the receptor (FGFR1–FGFR3), ligand homodimerization (FGF9 subfamily), site-specific proteolytic cleavage of the ligand (FGF23), and interaction of the ligand and the receptor with heparan sulphate cofactor and Klotho co-receptor.
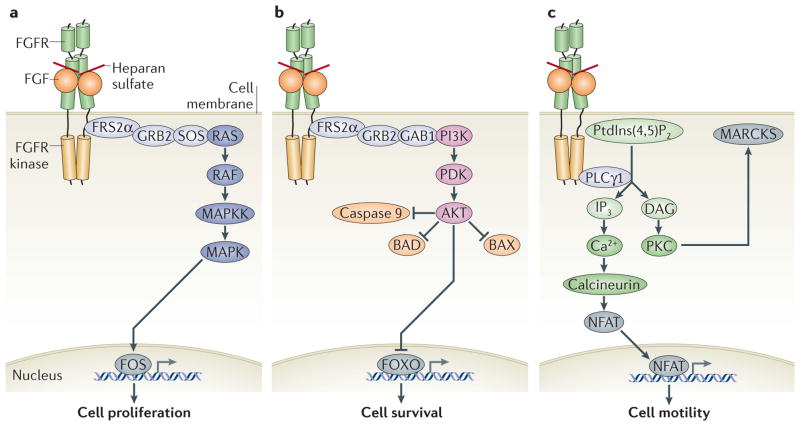
Fibroblast growth factors (FGFs) are secreted protein ligands that act in a paracrine or endocrine fashion to carry out their pleiotropic functions in development, tissue homeostasis and metabolism. The mammalian FGF family comprises 18 ligands, which are grouped into five paracrine-acting subfamilies and one endocrine-acting subfamily on the basis of sequence homology and phylogenetic and structural analysis1,2 (FIG. 1a). FGF ligands signal through cell surface FGF receptor (FGFR) Tyr kinases, which are encoded by four distinct genes in mammals (FGFR1–FGFR4)3 (FIG. 1b). Ligand binding induces FGFR dimerization4 (FIG. 1c,d), which brings the intracellular receptor kinase domains in the correct proximity and orientation to each other such that transphosphorylation, and hence activation, of the kinases can occur5,6. The activated receptor kinases, in turn, phosphorylate and activate their intracellular substrates, chief among which are FGFR substrate 2α (FRS2α) and phospholipase Cγ1 (PLCγ1)7,8 (FIG. 2). Activated FRS2α initiates downstream signalling through the RAS–MAPK pathway or the PI3K–AKT pathway, whereas the activation of PLCγ1 leads to release of calcium ions from intracellular stores and activation of protein kinase C (PKC)7–11 (FIG. 2). Although the biological outcome of activation of these pathways by FGFs varies depending on the cellular context, the RAS–MAPK pathway mostly generates a mitogenic cell response, the PI3K–AKT pathway promotes cell survival and the PLCγ1 pathway is thought to have a role in mediating cell motility7–11 (FIG. 2).
Figure 1. The FGF signalling system.
a | The 18 mammalian fibroblast growth factor (FGF) ligands are listed, grouped by subfamily and mode of action. The ligand which each subfamily is named after is boxed in orange. The crystal structures of FGF2, a prototypical paracrine FGF (Protein Databank identifier (PDB ID): 1FQ9)4, and FGF19, an endocrine FGF (PDB ID: 2P23)14 are shown. The conserved globular core domain consists of 12 β-strands in FGF2 and 11 β-strands in FGF19. Residues from the loop connecting β-strand 1 and β-strand 2 and from the region between β-strand 10 and β-strand 12 comprise the heparan sulphate-binding site (shown in blue). The conformation of the FGF19 heparan sulphate-binding site differs from the conserved conformation of the heparan sulphate-binding site in paracrine FGFs, which are represented here by FGF2. b | The domain architecture of a prototypical FGF receptor and Klotho co-receptor are shown. Alternative splicing in the D3 domain of FGFR1, FGFR2 and FGFR3 generates FGFRb and FGFRc isoforms with distinct ligand-binding specificity37,39,66,72. FGFRs interact with cell surface heparan sulphate via their D2 domain. Heparan sulphate is the obligatory cofactor for paracrine FGF signalling. A schematic of an heparan sulphate proteoglycan and the crystal structure of an heparan sulphate octasaccharide (PDB ID: 1FQ9)4 are shown. c | The crystal structure of the 2:2:2 ternary complex of FGF2, FGFR1c and heparan sulphate (PDB ID: 1FQ9)4 as well as a schematic of the paracrine FGF signal transduction unit based on this structure are shown. In the ternary complex, each FGF ligand interacts extensively with one receptor through the primary ligand-binding site comprising D2, D3 and the D2 D3 linker of the receptor. Each ligand also binds to the adjoining receptor in the complex through a secondary ligand-binding site on the D2 domain, which is adjacent to the site mediating the receptor receptor interaction. Heparan sulphate promotes the formation of the 2:2 paracrine FGF FGFR complex through concomitant interaction with both ligand and receptor.d | A schematic of two working models for the endocrine FGF FGFR Klotho co-receptor signal transduction unit is shown. A recent study on the ternary complex formation between FGF21, FGFR1c and β-Klotho supports the 1:2:1 model112 rather than the 2:2:2 model. AB, acid box; TMD, transmembrane domain. The FGF19 structure in parta is reproduced, with permission, from REF. 14 © (2007) American Society for Microbiology.
Figure 2. FGF signalling pathways.
Binding of fibroblast growth factor (FGF) to the FGF receptor (FGFR) induces FGFR dimerization, which juxtaposes the intracellular Tyr kinase domains of the receptors so that kinase activation by transphos- phorylation can occur5,6. Activated FGFR kinase in turn activates its intracellular substrates by phosphorylation, setting in motion distinct but potentially interactive signalling pathways that generate diverse cellular responses9,11. Major substrates of FGFR kinase are FGFR substrate 2α (FRS2α), which is constitutively associated with the receptor kinase, and phospholipase Cγ1 (PLCγ1)7,8. Activated FRS2α binds the adaptor protein growth factor receptor-bound 2 (GRB2)113. GRB2 then recruits either the guanine nucleotide exchange factor son of sevenless (SOS) (a) or the adaptor protein GRB2-associated binding protein 1 (GAB1) (b) to the signalling complex. Recruited SOS activates RAS GTPase, which initiates activation of the MAPK cascade113 (a), whereas recruited GAB1 leads to PI3K-mediated activation of AKT kinase (also known as protein kinase B)114 (b). Activated MAPK translocates from the cytoplasm to the nucleus, where it phosphorylates and hence activates immediate early gene transcription factors, such as FOS to induce transcription of specific genes (a). The outcome of this pathway is primarily cell proliferation but can also lead to cell differentiation, cell migration or another cellular response9,11. Activated AKT kinase, on the other hand, inactivates pro-apoptotic effectors such as the BCL-2 antagonist of cell death (BAD) and forkhead box class O (FOXO) transcription factors, thereby promoting cell survival115,116 (b). Recruitment and phosphorylation of PLCγ1 by FGFR kinase initiates a distinct signalling pathway that is thought to have roles in cell migration and cell differentiation and that can influence the RAS–MAPK and PI3K–AKT pathways (c). Activated PLCγ1 catalyses the hydrolysis of the membrane phospholipid phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)(P2) into diacylglycerol (DAG) and inositol-1,4,5,-trisphosphate (IP3)8. DAG activates protein kinase C (PKC), which in turn activates its substrates byphosphorylation, including the myristoylated Ala-rich C kinase substrate (MARCKS), a regulator of cell motility117. IP3 stimulates the release of calcium ions from intracellular stores, and this triggers the activation of calcium-dependent proteins such as the phosphatase calcineurin. Activated calcineurin induces nuclear translocation of the transcription factor nuclear factor of activated T cells (NFAT), which stimulates the expression of proteins essential for cell motility118. BAX, BCL-2-associated X protein; MAPKK, MAPK kinase; PDK, phosphoinositide-dependent protein kinase.
A common characteristic of FGFs is that they interact with heparan sulphate glycosaminoglycan chains of heparan sulphate proteoglycans12, which are abundantly present both at the cell surface and in the pericellular and extracellular matrix13 (FIG. 1b). Paracrine FGFs exhibit a much higher affinity for heparan sulphate than endocrine FGFs12, and therefore, paracrine FGFs become immobilized in the pericellular and extracellular matrix near the site of their secretion and can only act on cells within the same organ. By contrast, owing to their poor affinity for heparan sulphate12,14, endocrine FGFs can freely diffuse away from the cells secreting them and enter the blood circulation to reach their target cells in distant organs. Whereas paracrine FGFs require heparan sulphate as a cofactor to activate FGFR15,16 (FIG. 1c), endocrine FGFs rely on Klotho co-receptors to do so17–23 (FIG. 1b,d) (see below).
Paracrine FGFs have essential roles in virtually every step during embryonic development, including the induction and patterning of germ cell layers, the formation of body axes, the induction of organogenesis and morphogenesis and the patterning of tissues1,9,10,24. These ligands also fulfil crucial functions in a number of home-ostatic processes in the adult organism, most notably in the repair and remodelling of tissues25–28. Endocrine FGFs regulate multiple metabolic processes, including phosphate, glucose and lipid metabolism29–32. The broad range of FGF functions is reflected in the diversity of disorders caused by dysregulated FGF signalling, including developmental syndromes, metabolic disorders and cancer1,11,33,34.
The structural information gained about the FGF signalling system in recent years has begun to illuminate the complexity of its molecular control, which is in place to fine-tune this system to enable it to produce specific signalling outputs in diverse biological contexts. This Review discusses, through the lens of structural biology, the molecular mechanisms by which the FGF signalling system is regulated extracellularly at the level of the ligand, the receptor, the heparan sulphate cofactor and the Klotho co-receptor. The specific mechanisms covered include autoinhibition of the ligand by homo-dimerization, ligand inactivation by site-specific proteolytic cleavage, modulation of the receptor-binding affinity of the ligand by alternative splicing and differences in ligand-induced receptor dimerization between different ligand–receptor pairs. We also discuss the mechanism by which Klotho co-receptors convert FGFRs into specific receptors for endocrine FGFs.
Regulation of FGF ligand activity
FGFs share a core homology domain of about 120 amino acids, which folds into a globular β-trefoil. Paracrine FGFs have a regular β-trefoil domain that is composed of 12 β-strands (β1–β12)2, whereas endocrine FGFs have an atypical β-trefoil due to the lack of the β11 strand14. The conserved core domain is flanked by highly divergent amino-terminal and carboxy-terminal sequences, which have a key role in conferring distinct functional properties on FGFs35–39. FGF activity is regulated by multiple mechanisms, including heparan sulphate binding, N-terminal alternative splicing, homodimerization and site-specific proteolytic cleavage of the FGF ligand. Among these, the interaction of FGFs with heparan sulphate is the most important in determining the biological activity of the ligand. It not only defines the mode of action of FGFs, that is, whether an FGF acts in a paracrine or endocrine fashion, but also contributes to the distinct biological activities of paracrine ligands within the same subfamily.
Interaction of FGF ligand with heparan sulphate
The interaction between heparan sulphate and paracrine FGFs serves, among other roles, to sequester ligand near the site of action, provide a reservoir for ligand storage, enhance ligand stability and limit the radius of ligand signalling. In this subsection, we discuss the emerging concept that differences in heparan sulphate-binding affinity among paracrine FGFs, including ligands of the same subfamily, underlie the formation of different, ligand-specific gradients in the extracellular matrix, which produce distinct biological responses.
Heparan sulphate is a linear glycan composed of repeating disaccharide units of N-substituted glucosamine and glucuronic acid13,40. N-sulphated domains of 5–10 disaccharide units in length that harbour various modifications alternate along the glycan chain with N-acetylated regions that are mostly unmodified13,40. Modifications at N-sulphated domains include C5 epimerization of glucuronic acid to iduronic acid, 2-O-sulphation of glucuronic or iduronic acid and 3-O- and 6-O-sulphation of N-sulphated glucosamine (REFS 13,40). These modified sulphated domains are the principal binding sites for FGFs40. Notably, the modification of heparan sulphate by sulphation and epimerization is regulated in a tissue-specific fashion, and hence different tissues can produce heparan sulphate chains with a distinct ‘fine structure’13,40, which influences FGF activity.
Heparan sulphate mediates the formation of paracrine FGF gradients in the extracellular matrix36,41–43, which determine cellular processes such as cell migration, cell proliferation and specification of cell fate during tissue morphogenesis and patterning in embryonic development. Recent studies on FGF7 subfamily ligands, which play essential parts in the branching morphogenesis of a number of glandular organs, have provided new insight into how these FGF gradients are formed and how the biological response to an FGF ligand is dictated by the shape of this gradient43–45. FGF10 has greater binding affinity for heparan sulphate than its subfamily member FGF7 (REF. 43) (FIG. 3a). As a result, the diffusion of FGF10 in the extracellular matrix is more restricted than that of FGF7, and hence FGF10 forms a short, steep gradient close to its source, whereas FGF7 forms a long, shallow gradient spreading away from its source43 (FIG. 3b). In explants of developing submandibular gland tissue, FGF10 causes elongation of epithelial gland buds, whereas FGF7 induces bud branching43 (FIG. 3c,d). Reducing the heparan sulphate-binding affinity of FGF10 by replacing Arg187 in its heparan sulphate-binding site with Val, which is the corresponding residue in FGF7, converts FGF10 functionally into an FGF7-like morphogen43 (FIG. 3a–c). By contrast, reducing the receptor-binding affinity of FGF10 affects the degree, but not the nature, of the morphogenetic activity of FGF10 (REF. 43). Thus, the differences in heparan sulphate-binding affinity between FGF7 and FGF10, which translate into differently shaped extracellular ligand gradients, alone account for the distinct morphogenetic activities of these ligands. Similarly, the aberrant activity of the FGF9 mutant carrying a N143T mutation42, a spontaneous dominant mutation causing Elbow knee synostosis (Eks) in mice46, is due to altered heparan sulphate-binding affinity and hence altered gradient formation of the mutant FGF9 compared with wild-type ligand42.
Figure 3. Heparan sulphate-binding affinity determines the morphogenetic activity of FGF7 subfamily ligands.
a | Structural model of heparan sulphate binding to fibroblast growth factor 7 (FGF7) and FGF10. FGF7 (Protein Databank identifier (PDB ID): 1QQK)119 and FGF10 (PDB ID: 1NUN)39 were superimposed onto FGF2 in the FGF2–FGFR1c–heparan sulphate complex (PDB ID: 1FQ9)4. The heparan sulphate octasaccharide from this complex and the α-carbon traces of the heparan sulphate-binding regions of FGF7 (orange) and FGF10 (teal) are shown. Note that Arg187 of FGF10, a crucial residue for heparan sulphate-binding affinity, makes hydrogen bonds (dashed black lines) with the heparan sulphate octasaccharide, whereas Val174, the corresponding residue of FGF7, cannot interact with the cofactor.b | Diffusion profiles of FGF10, FGF7 and an FGF10 mutant with reduced heparan sulphate-binding affinity (FGF10R187V) in the extracellular matrix. Heparan sulphate beads loaded with fluorescently labelled FGF ligand were embedded in a laminin-rich hydrogel (also known as Matrigel), and ligand diffusion was monitored by measuring fluorescence intensity along a line passing through the centre of the bead. The dashed circles indicate the radii of ligand diffusion; FGF10 forms a short, steep gradient, FGF7 forms a long, shallow gradient and the FGF10 mutant (FGF10R187V) diffuses similarly to FGF7. c | Epithelial growth response of developing submandibular gland tissue to FGF10, FGF7 and FGF10R187V mutant. Explants of prebranched submandibular gland epithelial buds were embedded in a Matrigel matrix and exposed to FGF ligand. FGF10 causes elongation of the epithelial gland buds, whereas FGF7 and FGF10R187V induce bud branching. d | Model for the regulation of branching morphogenesis by paracrine FGFs with different heparan sulphate-binding affinity. An FGF ligand with high binding affinity for heparan sulphate has a restricted diffusion range in the extracellular matrix (red sector), and hence can only signal to cells (red circles) closest to the FGF source (black asterisk). This leads to elongation of an epithelial gland bud towards the FGF source. Conversely, an FGF with low heparan sulphate-binding affinity can diffuse over a greater distance in the extracellular matrix, reaching cells further from the FGF source. The activation of cells both close to and distant from the FGF source results in branching of an epithelial gland bud. Image in part a is modified and the images in parts b, c, and d are reproduced, with permission, from REF. 43 © (2009) AAAS.
The cells that secrete FGF ligand also produce hep-aran sulphate, the ‘fine structure’ of which is crucial for mediating the formation of an FGF gradient in the extracellular matrix. In addition, the ‘fine structure’ of the heparan sulphate produced by the cells that are responding to FGF is crucial for ligand signalling. This is suggested by the findings that abrogation of glycosaminoglycan biosynthesis selectively in mesenchymal cells of the developing eye, which secrete FGF10, leads to excessive diffusion of FGF10 through the extracellular matrix and, as a consequence, a lack of FGF10-induced budding of the prospective lacrimal gland epithelium45. Moreover, the formation of the extracellular FGF10 gradient that results in induction of lacrimal gland budding requires heparan sulphate produced by the mesenchyme around the developing gland to be modified by N-sulphation but not 2-O- or 6-O-sulphation45. By contrast, FGF10 signalling to the epithelium of the developing gland requires heparan sulphate produced by the epithelium to be modified by 2-O-and 6-O-sulphation44. Thus, distinct modifications of heparan sulphate in the mesenchyme and epithelium may provide another layer of control for directional FGF signalling from the mesenchyme to the epithelium and vice versa (see below). In addition, distinct hepa-ran sulphate modifications in different FGF-secreting cells, or in the same cell at different stages of embryonic development, may give rise to the formation of distinct gradients of specific FGF ligands, resulting in diverse biological responses.
Dimerization of FGF ligand
Unique among FGFs, ligands of the FGF9 subfamily undergo reversible homodimerization, which controls their biological activity36,38,42. The crystal structures of the homodimers of FGF9 and FGF20 have illuminated the role of homodimerization as an autoinhibitory mechanism. In the structures, both the β-trefoil core and the N-terminal and C-terminal regions flanking the core participate in the dimer formation, with extensive interactions between the N-terminal and C-terminal regions of each monomer being the driving force36,38 (FIG. 4a). Notably, about half of the residues involved in ligand homodimerization are also predicted to mediate receptor binding36,38 (FIG. 4b). In other words, the ligand homodimer is incompatible with receptor binding, and hence the dimer must dissociate into monomers in order to enable ligand–receptor interactions. These structural findings suggest that homodimerization autoinhibits the activity of FGF9 subfamily ligands by downregulating the effective concentration of ligand monomer that can bind and activate the receptor. Consistent with this mechanism, mutations that disrupt contacts in the dimer interface of FGF9 or FGF20 cause a shift in the ligand monomer to dimer equilibrium in solution towards the monomeric ligand form36. The mutated FGF9 and FGF20 ligands exhibit increased receptor binding and, as a consequence, increased biological activity compared with wild-type ligands36. Notably, the extent of increased receptor binding caused by a dimer interface mutation correlates with the degree of ligand monomerization that is caused36.
Figure 4. Autoinhibition of FGF9 by homodimerization.
a | The structure of the crystallographic fibroblast growth factor 9 (FGF9) homodimer (Protein Databank identifier (PDB ID): 1IHK)38. The dimer interface can be divided into two regions (black circles), one containing residues of the β-trefoil core domain and the other containing the amino-terminal and carboxy-terminal regions flanking the core domain. Extensive hydrophobic and hydrogen-bonding contacts between the N-terminal and C-terminal regions of the two monomers in the dimer drive homodimerization. Note that Asn143, which is spontaneously mutated in mice with Elbow knee synostosis (Eks), is located in the core region of the dimer interface. b | The molecular surface of an FGF9 monomer (dark yellow; PDB ID: 1IHK) 38 is shown with residues that exclusively mediate ligand dimerization depicted in light blue, and residues that mediate dimerization and are also predicted to bind FGF receptors shown in dark blue. Note that the bifunctional residues account for about half of the residues that form the ligand dimer interface (red boundary).c | Joint synostosis in developing limb tissue caused by the N143T mutation leading to Eks. Forelimbs of a newborn mouse with Eks and a newborn wild-type mouse are shown. The elbow joints (marked by arrows) are also shown at greater magnification. In the forelimb of the mouse with Eks, humerus (h) and radius (r) bones are fused together at the prospective elbow joint, and humerus and ulna (u) bones are barely separated by cartilaginous joint tissue. d | Hyperdiffusion of the FGF9 mutant that causes Eks in developing limb tissue in mice. Beads loaded with FGF9 or the FGF9N143T mutant were implanted into forelimb buds of an Fgf9−/− embryo around the initiation stage of joint synostosis, and ligand diffusion was detected immunochemically. As FGF9N143T exhibits reduced binding affinity for heparan sulphate, it diffuses further than wild-type FGF9. s, scapula. The image in partc is reproduced, with permission, from REF. 46 © (2002) Springer, and the image in partd is reproduced, with permission, from REF. 42 © (2009) Macmillan Publishers Limited. All Rights Reserved.
In addition to downregulating the concentration of monomeric ligand that is available to interact with receptors, homodimerization autoinhibits ligand activity by restricting the signalling radius of the ligand. The mutated FGF9 and FGF20 ligands, which have a reduced ability to homodimerize, also exhibit reduced binding affinity for heparan sulphate compared with wild-type ligands36. As a result, the mutated ligands diffuse over a greater distance in the extracellular matrix than their wild-type counterparts and hence have a greater signalling radius than them36. A recent study demonstrated the biological importance of homodimerization in the regulation of FGF9 activity42. This study identified an N143T mutation in FGF9 as the spontaneous mutation that causes a primarily skeletal phenotype in mice with Eks, a hallmark of which is the synostosis of elbow and knee joints42,46 (FIG. 4c). Replacement of Asn143, which lies in the FGF9 dimer interface, with Thr weakens FGF9 homodimerization42. As a consequence, the N143T mutant exhibits reduced binding affinity for heparan sulphate, and hence diffuses over a greater distance in developing tissue than wild-type FGF9 (REF. 42) (FIG. 4d). This hyperdiffusion leads to ectopic FGF9 signalling42, which underlies the Eks phenotype. These findings demonstrate that FGF ligand homodimerization enhances the binding affinity of ligand for heparan sulphate, thereby restricting ligand diffusion in the extracellular matrix and, as a consequence, limiting the signalling radius of the ligand. Taken together, ligand homodimerization autoinhibits the activity of FGF9 subfamily ligands at two levels: it inhibits ligand–receptor binding by occluding receptor-binding sites on the ligand, and it reduces the ability of the ligand to diffuse in the extracellular matrix by promoting ligand–heparan sulphate binding.
Alternative splicing of FGF ligand
Unlike other FGFs, the biological activity of ligands of the FGF8 subfamily is regulated by alternative splicing. The genes encoding FGF8 and FGF17, but not the FGF18 gene, are alternatively spliced to generate ligand isoforms with N termini of varying length and sequence47–50. In humans, there are four isoforms of FGF8 (FGF8a, FGF8b, FGF8e and FGF8f)48 and two isoforms of FGF17 (FGF17a and FGF17b) (FIG. 5a). The importance of N-terminal splicing in regulating the biological activity of these ligands has been demonstrated by studies on the roles of FGF8a and FGF8b in midbrain and hindbrain patterning. Both splice isoforms are expressed by the isthmic organizer, which is a signalling centre within the anterior neural plate that directs the patterning of midbrain and anterior hindbrain (the cerebellum)47,51. When ectopically expressed in the neural plate of chick embryos, FGF8a induces an expansion of midbrain tissue into the fore-brain region, whereas FGF8b induces cerebellum formation in regions of prospective midbrain and caudal forebrain51 (FIG. 5b). Likewise, in mouse embryos, ectopic expression of FGF8a in the midbrain causes an overgrowth of midbrain tissue, whereas FGF8b transforms the midbrain into cerebellum52,53. FGF8a and FGF8b also differ in their ability to induce mesoderm formation54,55, and mutations at the spliced N-terminal region of FGF8 found in patients with idiopathic hypogonadotropic hypogonadism impair the biological activity of the affected isoforms56. These findings illustrate the biological significance of N-terminal alternative splicing.
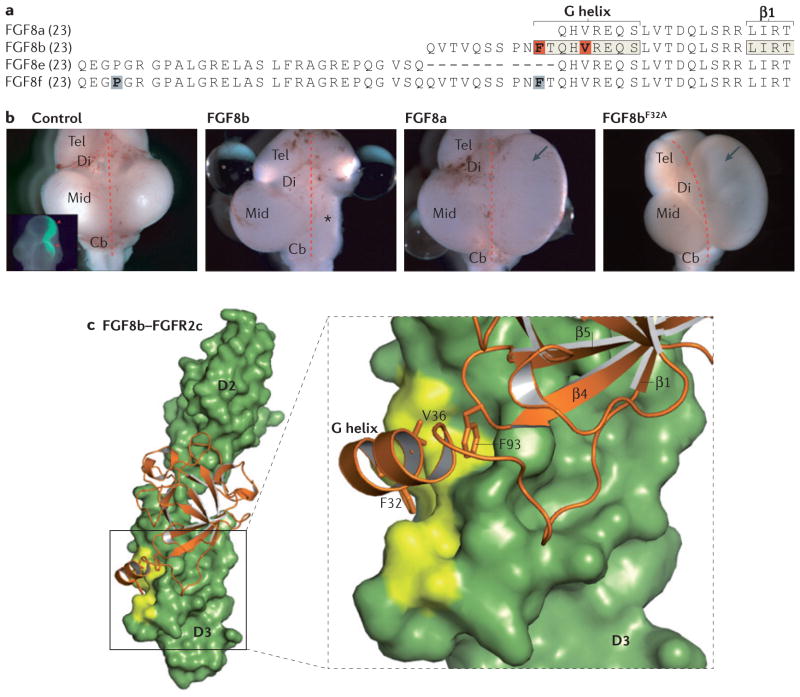
Figure 5. N-terminal alternative splicing regulates the biological activity of FGF8.
a | Sequence alignment of the amino-terminal regions of the mature human fibroblast growth factor 8 (FGF8) splice isoforms. Residues that make up the secondary structure elements known for FGF8b (the N-terminal G helix and the β1 strand) are indicated. Phe32 and Val36 of the FGF8b G helix (shaded orange) are key residues that interact with the D3 domain of FGF receptor (FGFR) c isoforms. Mutations at Pro26 and Phe40 of FGF8f (shaded grey) have been associated with idiopathic hypogonadotropic hypogonadism in humans56. b | FGF8 splice isoforms possess distinct abilities to transform midbrain (Mid) into cerebellum (Cb) in chick embryos. Shown are dorsal views of developing chick brains transfected on the right side (represented by green fluorescence in the inset in the first image) with plasmids encoding FGF8a, FGF8b or FGF8bF32A or empty vector (control). The asterisk to the right of the midline of the brain (dashed red line) marks transformation of midbrain into cerebellum, and the arrows point to the expansion of midbrain tissue. FGF8b, but not FGF8a, is able to transform midbrain into cerebellum, and mutation of Phe32 to Ala in FGF8b abrogates this isoform-specific patterning ability.c | Crystal structure of the FGF8b–FGFR2c complex (Protein Databank identifier: 2FDB)37. A view of the whole structure and a close-up view of the ligand receptor D3 domain interface are shown. Phe32 and Val36 from the G helix and Phe93 from the β4–β5 loop of FGF8b engage a hydrophobic groove in the D3 domain of FGFR2c (coloured yellow). Note that Phe32 is the sole isoform-specific residue of FGF8b that interacts with the receptor groove. Residues that are specific to the FGFRc splice isoforms of FGFR1, FGFR2 and FGFR3 mainly form this hydrophobic groove, thus the groove is unique to the FGFRc isoforms. Note that Klotho co-receptors also engage this receptor groove. Di, diencephalon; Tel, telencephalon. The image in part b is reproduced, with permission, from REF. 37 © (2006) Cold Spring Harbor Laboratory Press.
The crystal structure of FGF8b in complex with FGFR2c has unveiled the molecular mechanism by which N-terminal splicing regulates the biological activity of FGF8 (REF. 37) (FIG. 5c). In the structure, hydrophobic residues from the N-terminal G helix and the β4–β5 loop of FGF8b bind to a hydrophobic groove in the D3 domain of FGFR2c37. Notably, among the FGF8b residues that interact with the receptor groove, a single residue, Phe32, is from the isoform-specific N-terminal sequence37. As FGF8a lacks this sequence, the number of hydrophobic contacts is reduced in an FGF8a–receptor complex as compared to an FGF8b–receptor complex, and hence FGF8a binds more weakly to receptors than FGF8b. Indeed, replacement of Phe32 with Ala in FGF8b reduces its receptor-binding affinity to a similar level to that of FGF8a37, and converts FGF8b functionally into an FGF8a-like ligand37 (FIG. 5b). Thus, N-terminal alternative splicing regulates ligand activity by modulating the binding affinity of ligand for receptor, and differences in receptor-binding affinity underlie the distinct biological activities of the FGF8 splice isoforms. It is likely that differences in receptor-binding affinity among the members of other ligand subfamilies also contribute to the distinct biological activities of those ligands.
Proteolytic cleavage of FGF ligand
Specific proteolytic processing of FGF ligands also regulates their activity. FGF23, which functions as a hormone that controls phosphate and vitamin D metabolism30,31, is inactivated by proteolytic cleavage at the 176Arg-Xaa-Xaa-Arg179 motif (where Xaa represents any amino acid) that is located at the boundary between its β-trefoil core domain and its 72 amino acid long C-terminal tail57,58. The Arg-Xaa-Xaa-Arg motif is recognized by proprotein convertases that cleave specifically at basic amino acid residues and belong to the family of subtilisin-like Ser endoproteases59. These convertases cleave secretory proteins in subcellular organelles such as the Golgi complex, at the cell surface or in the extracellular matrix, and in most cases they convert a protein precursor into an active protein59. The cleavage occurs C-terminally to the second Arg residue of the Arg-Xaa-Xaa-Arg motif59, and can be inhibited by O-glycosylation of Ser or Thr residues within or adjacent to the cleavage site60. Although the endoprotease that cleaves FGF23 is unidentified, recent studies have provided insight into the mechanism by which proteolytic cleavage inactivates FGF23 (REF. 35). It was shown that FGF23 binds to preformed binary complexes of FGFR and α-Klotho co-receptor35. The ligand interacts with a de novo binding site that is generated at the composite receptor– co-receptor interface in these binary complexes35. The region on FGF23 that binds to this de novo site was mapped to the 72 amino acid long C-terminal tail, which follows the β-trefoil core domain35. Thus, the N-terminal fragment of proteolytic cleavage (Tyr25 to Arg179, which includes the β-trefoil core domain) is metabolically inactive57 because it lacks the binding site for the FGFR–α-Klotho complex. The C-terminal proteolytic fragment (Ser180 to Ile251), however, can compete with full-length FGF23 for binding to the FGFR–α-Klotho complex to antagonize the metabolic activity of FGF23, because this fragment contains the binding site for the FGFR–α-Klotho complex35.
These findings suggest a dual mechanism by which proteolytic cleavage at the Arg-Xaa-Xaa-Arg motif inactivates FGF23; the cleavage removes the binding site for the FGFR–α-Klotho complex from FGF23 and concomitantly generates an endogenous inhibitor of FGF23. Inhibition of this proteolytic cleavage by missense mutations within the Arg-Xaa-Xaa-Arg motif in FGF23 leads to accumulation of full-length, bioactive FGF23, causing renal phosphate wasting disease in humans57,58,61. Conversely, enhanced FGF23 cleavage due to impaired O-glycosylation of FGF23 leads to a deficit in full-length FGF23, which manifests as hyperphosphatemia and soft tissue calcification in humans62,63. Therefore, specific proteolytic processing of FGF23 is essential for regulating the activity of this hormone. It is currently not known whether FGF19 and FGF21, the other two members of the endocrine FGF subfamily, undergo proteolytic processing in order to be inactivated.
Interestingly, ligands of the FGF7 subfamily contain an Arg-Xaa-Xaa-Arg cleavage motif in the N-terminal region preceding the β-trefoil core domain, and cultured mammalian cells secrete both full-length FGF7 ligand and the C-terminal fragment that results from proteolytic cleavage at the Arg-Xaa-Xaa-Arg motif64. However, whether specific proteolytic cleavage of FGF7 subfamily ligands occurs in vivo and is crucial for regulating the activity of these ligands is currently unknown. Proteolytic processing may also have a role in limiting the activity of FGF8 subfamily ligands. This is suggested by the findings that high temperature requirement A1 (HTRA1), a Staphylococcus aureus V8-like Ser-endoprotease, cleaves FGF8, and that loss of htra1 gene function in zebrafish embryos leads to an increase in fgf8 gene expression and defects in dorsoventral patterning reminiscent of those caused by ectopic FGF8 signalling65.
Regulation of FGFR function
A prototypical FGFR consists of three extracellular immunoglobulin-like domains (D1–D3), a single-pass transmembrane domain and an intracellular Tyr kinase domain2 (FIG. 1b). The ectodomain region encompassing D2, D3 and the D2–D3 linker is necessary and sufficient for ligand binding2 (FIG. 1b,c). Specificity of ligand binding by FGFR1, FGFR2 and FGFR3 is primarily determined by alternative splicing in the D3 domain of these receptors, which generates FGFRb and FGFRc isoforms37,39,66–72. This splicing event is fundamental to the establishment of directional FGF signalling between epithelial and mesenchymal tissues. FGF–FGFR binding induces FGFR dimerization4, which juxtaposes the intracellular kinase domains of the receptors so that one kinase can phosphorylate and hence activate the other5,6. Subtle differences in receptor dimerization are induced by distinct ligands, and these may translate into differences in receptor activation that yield distinct biological responses.
Alternative splicing in the D3 domain of FGFR
Ligand-binding specificity of FGFR1, FGFR2 and FGFR3 is regulated by alternative splicing in the D3 domain of these receptors37,39,66–72. The N-terminal portion of the D3 domain is encoded by exon 7 (also known as exon IIIa), whereas the C-terminal portion is encoded by one of two mutually exclusive exons, exon 8 (also known as exon IIIb) or exon 9 (also known as exon IIIc)66,68,69. This alternative splicing is largely tissue-specific, with the expression of the FGFRb splice isoform generally being restricted to epithelial cell lineages and the expression of the FGFRc splice isoform by and large limited to mesenchymal cell lineages69,70,73. These receptor splice isoforms exhibit a distinct ligand-binding specificity profile67,69–72. The epithelial FGFRb isoforms specifically bind FGF ligands that are secreted from mesenchymal tissue and, conversely, the mesenchymal FGFRc isoforms preferentially interact with FGF ligands that are secreted from epithelial tissue74. This reciprocal expression of receptor splice isoforms and cognate ligands creates specific paracrine FGF signalling loops between the epithelium and the mesenchyme, which are crucial for orchestrating developmental processes and also for regulating tissue homeostasis in the adult75–81 (FIG. 6a). In the developing lung, for example, bidirectional FGF signalling between the epithelial layer and the surrounding mesenchyme underlies the coordination of mesenchymal proliferation and epithelial branching that governs normal lung organogenesis. Specifically, FGF9 secreted by the epithelium stimulates proliferation of the mesenchyme76, whereas FGF10 secreted by the mesenchyme acts on the epithelium to induce budding and branching78,79. Notably, the reciprocal expression of receptor splice isoforms and cognate ligands not only establishes directional paracrine FGF signalling but safeguards against illegitimate autocrine FGF signalling.
Figure 6. Alternative splicing confers ligand-binding specificity on FGFR.
a | Fibroblast growth factor (FGF) ligands secreted from mesenchymal tissue exhibit binding specificity for epithelial FGF receptor (FGFR) b isoforms, whereas ligands secreted from epithelial tissue specifically bind to mesenchymal FGFRc isoforms. These paracrine FGF signalling loops are essential for controlling developmental processes75,81. b | D3 domain sequence alignment of FGFR2b and FGFR2c. The location of the β-strands in the D3 domain of FGFR2c are indicated by brackets, and the dashed line across the alignment marks the junction between the common amino-terminal portion and the spliced carboxy-terminal portion of the D3 domain. Isoform-specific residues of the receptor βC′–βE loop that are crucial in determining ligand-binding specificity are shaded orange. Boxes indicate the positions of mutations in FGFR2c that cause skeletal disorders in humans. c | The ligand–receptor D3 domain interface in the FGF10–FGFR2b structure (Protein Databank identifier (PDB ID): 1NUN)39 is shown, along with a view of the whole structure. The ligand is coloured orange, the receptor D3 domain is coloured green (common N-terminal portion) and teal (alternatively spliced C-terminal portion). A network of hydrogen bonds (dashed lines) is formed between FGF10 and the isoform-specific βC′–βE loop of FGFR2b. Within this network, the hydrogen bonds between Asp76 of FGF10 (a residue unique to FGF10 and other FGF7 subfamily ligands) and Ser315 of FGFR2b (an isoform-specific residue of the βC′–βE loop) determine binding specificity. The hydrogen-bonding network is buttressed by hydrophobic contacts between Phe146 of FGF10 and Ile317 of FGFR2b.d | The ligand–receptor D3 domain interface in the FGF2–FGFR2c structure (PDB ID: 1EV2)82 is shown. Ligand and receptor are coloured as in partc. Among the hydrogen bonds (dashed lines) that are formed at the ligand–receptor βC′–βE loop interface, unique bonds between Gln65 of FGF2 and Asp321 of FGFR2c dictate binding specificity. Mutation of Asp321 to Ala, which causes Pfeiffer syndrome in humans, overrides the binding specificity of FGFR2c by eliminating steric clashes and electrostatic repulsion between Asp76 of FGF10 and Asp321 of FGFR2c. This enables FGF10 to bind to the mutant receptor and signal illegitimately through FGFR2c. Hydrophobic contacts between Tyr82, Val97 and Phe102 of FGF2 and Val317 of FGFR2c strengthen the hydrogen bonding interactions at the ligand receptor βC′–βE loop interface.
The crystal structures of ligand-bound FGFR2b and FGFR2c splice isoforms have provided molecular insight into how alternative splicing in the D3 domain controls ligand-binding specificity37,39,82 (FIG. 6b–d). In the structure of FGF10 in complex with its cognate receptor, FGFR2b, FGF10 residues bind to a cleft in the D3 domain of FGFR2b39. One side of this cleft is formed by the FGFR2b splice isoform-specific βC′–βE loop. At the ligand–D3 domain cleft interface, unique hydrogen bonding contacts between Asp76 of FGF10 (a residue specific to FGF10 and other FGF7 subfamily ligands) and Ser315 of FGFR2b (a residue specific for FGFR2b) are crucial in determining the binding specificity of FGFR2b towards FGF10 (REF. 39) (FIG. 6b,c). The crucial role of the Asp76–Ser315 interaction for the ligand-binding specificity of FGFR2b is underscored by the finding that replacement of Ser315 in FGFR2b with Ala, the corresponding residue of FGFR2c, greatly reduces the ability of FGFR2b to bind FGF7 (REF. 83). FGF2, a member of the FGF1 subfamily, specifically binds to FGFR2c. The crystal structure of the FGF2–FGFR2c complex shows that, similar to the mode of FGF10 binding to the D3 domain of FGFR2b, FGF2 residues interact with the D3 domain cleft of FGFR2c, which is formed in part by the receptor isoform-specific βC′–βE loop82 (FIG. 6d). As is the case for FGF10–FGFR2b binding, unique hydrogen bonding contacts between FGF2 and the βC’–βE loop of FGFR2c determine binding specificity. These are the hydrogen bonds formed between Gln65 of FGF2, a residue unique to FGF2, and Asp321 of FGFR2c, an isoform-specific residue of the βC′–βE loop of the receptor (FIG. 6d). Together, the structural data show that the alternative splicing in the D3 domain of FGFR2 confers ligand-binding specificity by changing the sequence of key ligand-binding residues or ligand-binding pockets.
Receptor mutations that disrupt ligand-binding specificity set by alternative splicing cause developmental skeletal disorders. The D321A mutation in FGFR2c, for example, causes Pfeiffer syndrome84,85, an autosomal dominant disorder characterized by premature synostosis of cranial sutures. The mutation greatly reduces the binding affinity of FGFR2c for FGF2 and, at the same time, enables FGFR2c to interact with the non-cognate ligand FGF1086. The loss in binding affinity of the mutant receptor for FGF2 can be explained by the loss of highly specific hydrogen bonding contacts between Asp321 in FGFR2c and Gln65 in FGF2 (REF. 82) (FIG. 6d). The concomitant gain in the ability of the mutant receptor to bind FGF10 is likely due to the loss of steric clashes and electrostatic repulsion between Asp321 of FGFR2c and Asp76 of FGF10, which preclude binding of FGF10 to the wild-type receptor. By disrupting ligand-binding specificity, the D321A mutation enables autocrine FGF signalling in the mesenchyme, and such illegitimate signalling in the developing cranial suture leads to craniosynostosis as seen in Pfeiffer syndrome. The effects of the mutation of key ligand-binding residues in the receptor, similarly to the D321A mutation in FGFR2c, underscore the central role of alternative splicing in regulating the specificity of FGF signalling.
Ligand-induced dimerization of FGFR
Paracrine FGFs require heparan sulphate as a cofactor to robustly bind FGFR and promote the formation of a stable FGF–FGFR signal transducing complex at a ratio of 2:2 (REF. 4) (FIGS 1c,7a). In the complex, each ligand interacts with both receptors, and the two receptors also interact directly with each other. These multivalent ligand–receptor and receptor–receptor contacts are fortified by heparan sulphate, which simultaneously engages ligand and receptor in the complex. At the primary ligand–receptor interaction site, an extensive network of hydrogen bonds is formed between the ligand, the receptor D2–D3 linker and the receptor D3 domain4,37,39,82,87–89 (FIG. 7b,c). This hydrogen-bonding network confers structural rigidity on the interface between the ligand and the D3 domain and D2–D3 linker of the receptor, and its composition and geometry thus determine the orientation of the D3 domain and the D2–D3 linker in the receptor dimer. Comparative analysis of the crystal structures of FGF–FGFR complexes has identified three receptor residues and two ligand residues as key constituents of the hydrogen-bonding network (FIG. 7b). The receptor residues, an Arg residue in the D2–D3 linker and an Asp and a Gln residue in the βB′–βC loop of the D3 domain, are conserved among mammalian FGFRs. The ligand residues, a Glu residue in the β8 strand and an Asp in the β9 strand, are nearly completely conserved among the mammalian FGFs. Importantly, these receptor and ligand residues also form entropically favourable intramolecular hydrogen bonds with similarly conserved residues. In ligand–receptor complexes containing this conserved core of hydrogen-bonding interactions at the interface between the ligand and the D3 domain and D2–D3 linker of the receptor, the orientation of the D3 domain and D2–D3 linker is similar. Accordingly, the distance between the C-terminal membrane insertion points of the D3 domains in a dimer of ligand-bound receptors is similar among these complexes (FIG. 7a). . This is the case for complexes of FGFR with FGF1 subfamily ligands or FGF10 (REFS 4,39,82,87–89).
Figure 7. Ligand-dependent differences at the FGF–FGFR interface differentially regulate FGFR dimerization.
a | Superimposition of a 2:2 fibroblast growth factor 8b–FGF receptor 2c (FGF8b–FGFR2c) complex onto a 2:2 FGF2 – FGFR2c complex. The α-carbon traces of the complexes are shown. For the FGF8b–FGFR2c complex model, FGF8b from the FGF8b–FGFR2c structure (Protein Databank identifier (PDB ID): 2FDB)37 was superimposed onto each of the two FGF2 ligands in the 2:2:2 FGF2–FGFR1c–heparan sulphate complex (PDB ID: 1FQ9)4. The FGF2–FGFR2c model was created in a similar manner using FGF2 from the FGF2–FGFR2c structure (PDB ID: 1EV2)82. Note the differences in the orientation of the receptor D3 domain and D2–D3 linker between the two dimers of ligand-bound FGFR2c, which translate into differences in the spatial distance between the carboxy-terminal membrane insertion points of the D3 domains.b | The interface between the ligand and the D3 domain and D2–D3 linker of the receptor in the FGF2–FGFR2c structure (PDB ID: 1EV2)82 is shown. The ligand is coloured orange, the receptor is coloured green (D2–D3 linker and common amino-terminal portion of the D3 domain) and teal (alternatively spliced C-terminal portion of the D3 domain). The network of hydrogen bonds (dashed lines) formed at this interface is highly conserved among complexes of FGFR with FGF1 subfamily ligands or FGF10. Its key constituents comprise Glu105 and Asn113 of FGF2 and Arg251, Asp283 and Gln285 of FGFR2c. Most of these residues also form entropically favourable intramolecular hydrogen bonds with similarly conserved residues, such as Glu105 and Asn113 of FGF2 with Tyr115 of FGF2.c | The interface between the ligand and the D3 domain and D2–D3 linker of the receptor in the FGF8b–FGFR2c structure (PDB ID: 2FDB)37 is shown. Ligand and receptor are coloured as in partb. The composition and geometry of the network of hydrogen bonds (dashed lines) formed at this interface differ from that observed in the FGF2–FGFR2c complex (see, partb). For example, Asn113 and Tyr115 of FGF2 are replaced with Thr and Leu, respectively, in FGF8b. The side chain of Glu131 in FGF8b, the corresponding residue to Glu105 in FGF2, adopts a different rotamer conformation compared to that in the FGF2–FGFR2c complex as it makes an intramolecular hydrogen bond with Lys176, a residue specific to FGF8 subfamily ligands. A total of four hydrogen bonds unique to the FGF8b–FGFR2c complex (dashed red lines) are formed at this interface between FGF8b and FGFR2c. Images in partsb and c are modified, with permission, from REF. 37 © (2006) Cold Spring Harbor Laboratory Press.
By contrast, two of the key residues in the hydrogen- bonding network seen in these complexes are not conserved in FGF8 subfamily ligands, namely the Asp and a Tyr residue in the β9 strand. These substitutions, together with other divergent residues, account for an altered composition and geometry of the hydrogen-bonding network in the FGF8b–FGFR2c complex, and hence for a different orientation of the D3 domain and D2–D3 linker in comparison to the other FGF–FGFR complexes37 (FIG. 7c). As a result of the different domain orientation, the C-terminal membrane insertion points of the D3 domains are closer to each other in a dimer of FGF8b-bound receptors than in the dimers of receptors bound to other FGF ligands37 (FIG. 7a). Such differences in the spatial positioning of the receptor ectodomains among different FGF–FGFR complexes may translate into differences in the juxtapositioning of the intra-cellular receptor kinase domains and distinct signalling outputs. In the case of an FGF8 signalling complex, the kinase domains would be closer to each other than in other FGF signalling complexes. This might generate the more intense or persistent signal that might be required for FGF8 functions such as the organizer activity at signalling centres that drive brain and limb development.
Klotho co-receptors in FGF signalling
Endocrine FGFs depend on Klotho co-receptors for signalling because compared to paracrine FGFs, these ligands have intrinsically low binding affinity for both heparan sulphate and FGFR14,90. Their low affinity for heparan sulphate enables these FGFs to signal in an endocrine fashion, whereas their low affinity for FGFR safeguards against nonspecific off-target signalling. Klotho co-receptors, which include α-Klotho, β-Klotho and γ-Klotho, are members of the Klotho subfamily91–94 of family 1 β-glycosidases95–97. Klotho co-receptors contain an extracellular domain composed of 1 (γ-Klotho) or 2 (α-Klotho and β-Klotho) β-glycosidase-like domains, a single-pass trans-membrane domain and a short cytoplasmic tail91–93 (FIG. 1b). α-Klotho is required for FGF23 signalling19,20,23 and β-Klotho is required for signalling by FGF19 and FGF21 (REFS 17,18,21,22). γ-Klotho may function as an additional co-receptor in FGF19 signalling98, but in vivo evidence for such a role is lacking. Expression of α-Klotho and β-Klotho co-receptors is restricted to the target tissues of endocrine FGFs98, where these co-receptors constitutively associate with cognate FGFRs of endocrine FGFs18,19,21,23. When bound to α-Klotho or β-Klotho co-receptors, FGFRs exhibit high affinity for endocrine ligands18,19,21,23,35,90. Although this role of α-Klotho and β-Klotho is fairly well established, recent findings have provided new insight into the mechanism by which these co-receptors convert FGFRs into specific receptors for endocrine FGFs99.
Similarly to ligands of the FGF4, FGF8 and FGF9 subfamilies2,74, α-Klotho and β-Klotho preferentially bind to the FGFRc splice isoforms of FGFR1, FGFR2 and FGFR3 (REFS 19,21,23,99,100). This raised the possibility that the binding site for α-Klotho and β-Klotho co-receptors on FGFR may overlap with the binding site for these paracrine FGFs. Indeed, it was shown that α-Klotho and β-Klotho engage the conserved hydrophobic groove in the D3 domain of FGFRc isoforms99, which is also used by FGF8 subfamily ligands for receptor binding37 (FIG. 5c). Thus, the complex formation of α-Klotho and β-Klotho co-receptors with FGFRc iso-forms and FGFR4, which also contains the hydrophobic groove in its D3 domain, obscures the binding site for FGF8 subfamily ligands and, as a result, inhibits signalling by these ligands99. This mechanism is likely to be biologically relevant, as α-Klotho and β-Klotho co-receptors and FGF8 subfamily ligands are co-expressed in several adult tissues, such as the liver, kidney and mature ovarian follicles18,23,101–103. Notably, α-Klotho and β-Klotho co-receptors are also co-expressed with other paracrine FGFs in adult tissues25,104 and might also inhibit signalling by these FGFs by binding to FGFRs at a site that overlaps with that of these ligands. Together, the findings support the concept of a dual mechanism by which α-Klotho and β-Klotho convert FGFRs into specific receptors for endocrine FGFs; these co-receptors not only enhance the binding affinity of FGFRs for endocrine ligands but concomitantly suppress the binding of paracrine FGFs, such as FGF8 subfamily ligands, to FGFRs. In essence, Klotho co-receptors modify the ligand-binding specificity of FGFRs in favour of endocrine FGFs. This co-receptor function probably evolved to dedicate FGFRs in target cells of endocrine FGFs exclusively to endocrine FGFs, avoiding any interference from paracrine FGFs.
Yet another role for Klotho co-receptors in endocrine FGF signalling is suggested by a recent study demonstrating that heparan sulphate is dispensable for the metabolic activity of endocrine FGFs90. This study showed that endocrine FGF mutants devoid of residual heparan sulphate binding have the same capacity as the native ligands to activate FGFR in the presence of α-Klotho or β-Klotho co-receptor and to elicit metabolic effects in vivo90. Thus, heparan sulphate is not a component of the endocrine FGF signal transduction unit (FIG. 1d), which in turn implies that Klotho co-receptors fulfil the same role in the formation of the endocrine FGF signalling complex as heparan sulphate does in the formation of the paracrine FGF signalling complex. This suggests that Klotho co-receptors promote FGFR dimerization upon endocrine FGF binding, which is required for FGFR kinase activation.
Conclusion and perspective
Recent advances have improved our understanding of the structural basis of FGF signalling and provided a glimpse at the complexity of molecular control that is in place for this essential signalling system. Importantly, the structural findings suggest that there may be no functional redundancy among FGF ligands. This is supported by genetic studies that demonstrate that the gene knockout of one FGF ligand cannot be compensated for by any other member of the same FGF subfamily. For example, FGF10-deficient mouse embryos show agenesis of the lungs78,79, aplasia of the salivary glands105 and hypoplasia of the mammary glands106, whereas mouse embryos lacking FGF7 exhibit defects in ureteric bud growth and branching in the developing kidney107. The distinct phenotypes caused by these gene knockouts reflect that, although FGF7 and FGF10 are closely related members of the same subfamily and both branching morphogens, these ligands cannot substitute for one another. Perhaps even more strikingly, FGF7 and its subfamily member FGF22 both promote the differentiation of presynaptic terminals on dendrites of CA3 pyramidal neurons in the hippocampus108. However, the two ligands promote the formation of functionally distinct synapses108, and neither ligand can replace the other. Hence, future studies should be directed at identifying novel ligand-specific functions of FGF signalling.
Although it is established that Klotho co-receptors convert FGFRs into specific receptors for endocrine FGFs, a full understanding of the molecular basis for this functional conversion awaits the determination of the crystal structure of a ternary complex of endocrine FGF with FGFR and Klotho co-receptor. A ternary complex structure may also unveil how Klotho co-receptors promote FGFR dimerization upon endocrine FGF binding and provide clues as to why an endocrine FGF signalling complex sends a weaker signal than a paracrine FGF signalling complex18,23. Importantly, this crystal structure would also offer a template for the structure-based design of endocrine FGF agonists or antagonists for the treatment of metabolic disorders.
Moreover, although it is known that paracrine FGFs cooperate with one another to direct developmental processes, it has yet to be explored how paracrine FGF signalling is integrated with endocrine FGF signalling to regulate metabolism. The latest discoveries on the roles of FGF1 and FGF21 in the dynamic remodelling and metabolic activity of adipose tissue may set the stage for future investigations in this direction. Both FGF1 and FGF21 are induced postprandially in gonadal white adipose tissue by the nuclear hormone receptor PPARγ (peroxisome proliferator activated receptor-γ)26,109, which is a major regulator of adipocyte differentiation and metabolic function110. FGF1 is required for the remodelling of adipose tissue to adjust to fluctuations in nutrient availability26, and this process is influenced by FGF21 (REFS 109,111). Notably, as part of a positive feedback loop, FGF21 stimulates PPARγ activity in adipocytes109, raising the intriguing possibility that FGF21 regulates FGF1 signalling in adipose tissue through PPARγ. Nuclear hormone receptor signalling might therefore serve as a mechanism that integrates the functions of endocrine FGFs and paracrine FGFs in the control of metabolic homeostasis.
Lastly, key questions remain in regard to the molecular basis of FGF signal transduction specificity. For example, what determines the speed, extent and duration of transphosphorylation of FGFR kinases in a ligand-bound FGFR dimer? Moreover, how do the speed, extent and duration of kinase transphosphorylation contribute to the generation of distinct signalling outputs? Future studies addressing these questions and more are poised to unravel new layers of molecular control, which operate to fine-tune FGF signalling. Answering these questions may not only advance our understanding of how FGFs generate specific signalling outputs in enormously diverse biological contexts but also pave the way for new treatments for disorders caused by dysregulated FGF signalling.
Acknowledgments
The authors thank J. Ma for help with preparing the structures. Studies on the structural basis of FGF signalling in the Mohammadi laboratory are funded by the U.S. National Institutes of Health grant DE13686.
Glossary
- Paracrine
Refers to a mode of signalling in which the cell responding to a signalling molecule is near the cell secreting the molecule
- Endocrine
Refers to a mode of signalling in which the cell responding to a signalling molecule is far away from the cell secreting the molecule
- Transphosphorylation
The process by which one kinase molecule in a kinase dimer phosphorylates the other (also referred to as autophosphorylation)
- Proteoglycans
Protein–glycan conjugates that consist of a core protein to which one or more glycosaminoglycan chains are attached
- Morphogenesis
Process of cell movement during embryonic development that controls the size, shape and patterning of tissues and organs
- β-trefoil
Portion of a protein that consists of 12 β-strands arranged into 3 similar sets of 4-stranded β-sheets
- Epimerization
Process by which an epimer of a molecule is converted into its stereoisomeric counterpart
- Submandibular gland
Gland located under the mandible bone that secretes saliva into the mouth
- Elbow knee synostosis
(Eks). Mouse skeletal phenotype characterized by bone fusion at elbow and knee joints
- Lacrimal gland
Gland of the eye that produces tear fluid to keep the eye lubricated
- Idiopathic hypogonadotropic hypogonadism
Hereditary disorder characterized by the failure of sexual maturation and infertility due to deficiency of gonadotropin-releasing hormone
- Renal phosphate wasting disease
Inherited or acquired condition characterized by excessive renal excretion of phosphate due to impaired tubular reabsorption
- Hyperphosphatemia
Increase in blood phosphate levels above normal
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Moosa Mohammadi’s homepage: http://www.med.nyu.edu/mohammadi
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Regina Goetz, Email: Regina.Goetz@nyumc.org.
Moosa Mohammadi, Email: Moosa.Mohammadi@nyumc.org.
References
- 1.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J, et al. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. Reveals the structural basis by which heparan sulphate promotes ligand-induced FGFR dimerization. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, et al. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc Natl Acad Sci USA. 2008;105:19660–19665. doi: 10.1073/pnas.0807752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter G, Ji Q. Phospholipase C-γ as a signal-transducing element. Exp Cell Res. 1999;253:15–24. doi: 10.1006/excr.1999.4671. [DOI] [PubMed] [Google Scholar]
- 9.Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 10.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nature Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 12.Asada M, et al. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim Biophys Acta. 2009;1790:40–48. doi: 10.1016/j.bbagen.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 14.Goetz R, et al. Molecular insights into the Klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. Reveals the structural basis for the endocrine mode of action of FGF19 subfamily ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 16.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 17.Ding X, et al. βKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab. 2012;16:387–393. doi: 10.1016/j.cmet.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurosu H, et al. Tissue-specific expression of βKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–26695. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurosu H, et al. Regulation of fibroblast growth factor-23 signaling by Klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatani T, Ohnishi M, Razzaque MS. Inactivation of klotho function induces hyperphosphatemia even in presence of high serum fibroblast growth factor 23 levels in a genetically engineered hypophosphatemic (Hyp) mouse model. FASEB J. 2009;23:3702–3711. doi: 10.1096/fj.08-123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa Y, et al. βKlotho is required for metabolic activity of fibroblast growth factor 21. Proc Natl Acad Sci USA. 2007;104:7432–7437. doi: 10.1073/pnas.0701600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomiyama K, et al. Relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc Natl Acad Sci USA. 2010;107:1666–1671. doi: 10.1073/pnas.0913986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 24.Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart AW, Baeza N, Apelqvist A, Edlund H. Attenuation of FGF signalling in mouse β-cells leads to diabetes. Nature. 2000;408:864–868. doi: 10.1038/35048589. Identifies a crucial role for FGF signalling in pancreatic β-cell function and glucose homeostasis in the adult. [DOI] [PubMed] [Google Scholar]
- 26.Jonker JW, et al. A PPARγ–FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485:391–394. doi: 10.1038/nature10998. Identifies a crucial role for FGF1 in the remodelling of adipose tissue in response to fluctuating nutrient availability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M, et al. Fibroblast growth factor 2 control of vascular tone. Nature Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaue H, et al. Requirement of fibroblast growth factor 10 in development of white adipose tissue. Genes Dev. 2002;16:908–912. doi: 10.1101/gad.983202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quarles LD. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nature Rev Endocrinol. 2012;8:276–286. doi: 10.1038/nrendo.2011.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razzaque MS. The FGF23–Klotho axis: endocrine regulation of phosphate homeostasis. Nature Rev Endocrinol. 2009;5:611–619. doi: 10.1038/nrendo.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long YC, Kharitonenkov A. Hormone-like fibroblast growth factors and metabolic regulation. Biochim Biophys Acta. 2011;1812:791–795. doi: 10.1016/j.bbadis.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nature Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkie AO. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Goetz R, et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23–FGFR–Klotho complex formation. Proc Natl Acad Sci USA. 2010;107:407–412. doi: 10.1073/pnas.0902006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalinina J, et al. Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Mol Cell Biol. 2009;29:4663–4678. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen SK, et al. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. Identifies the molecular mechanism by which N-terminal splicing regulates the biological activity of FGF8 and provides structural evidence for ligand-induced differences in FGFR dimerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plotnikov AN, et al. Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J Biol Chem. 2001;276:4322–4329. doi: 10.1074/jbc.M006502200. [DOI] [PubMed] [Google Scholar]
- 39.Yeh BK, et al. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc Natl Acad Sci USA. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. Reveals the molecular basis by which alternative splicing in the D3 domain of FGFR2 regulates the ligand-binding specificity of this receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher JT. Heparan sulfate: growth control with a restricted sequence menu. J Clin Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Mohammadi M, Flanagan JG. Graded levels of FGF protein span the midbrain and can instruct graded induction and repression of neural mapping labels. Neuron. 2009;62:773–780. doi: 10.1016/j.neuron.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harada M, et al. FGF9 monomer–dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nature Genet. 2009;41:289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarenkova HP, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu X, et al. Lacrimal gland development and Fgf10–Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 2011;286:14435–14444. doi: 10.1074/jbc.M111.225003. Demonstrates that a specific fine structure of the heparan sulphate produced by the cell responding to an FGF ligand is required for FGF signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu X, et al. Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development. 2012;139:2730–2739. doi: 10.1242/dev.079236. Demonstrates that the heparan sulphate produced by the cell secreting an FGF ligand mediates the formation of an FGF gradient in the extracellular matrix and that its fine structure is a crucial determinant in this process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami H, et al. Elbow knee synostosis (Eks): a new mutation on mouse chromosome 14. Mamm Genome. 2002;13:341–344. doi: 10.1007/s00335-001-2143-6. [DOI] [PubMed] [Google Scholar]
- 47.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 48.Gemel J, Gorry M, Ehrlich GD, MacArthur CA. Structure and sequence of human FGF8. Genomics. 1996;35:253–257. doi: 10.1006/geno.1996.0349. [DOI] [PubMed] [Google Scholar]
- 49.MacArthur CA, et al. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development. 1995;121:3603–3613. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- 50.Xu J, Lawshe A, MacArthur CA, Ornitz DM. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev. 1999;83:165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 51.Sato T, Araki I, Nakamura H. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development. 2001;128:2461–2469. doi: 10.1242/dev.128.13.2461. [DOI] [PubMed] [Google Scholar]
- 52.Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- 53.Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126:4827–4838. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- 54.Christen B, Slack JM. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–466. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- 55.Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- 56.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. Implicates decreased FGF8 signalling in the deficiency of gonadotropin-releasing hormone that underlies idiopathic hypogonadotropic hypogonadism and presents an example for the role of N-terminal alternative splicing in regulating the biological activity of FGF8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimada T, et al. Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology. 2002;143:3179–3182. doi: 10.1210/endo.143.8.8795. [DOI] [PubMed] [Google Scholar]
- 58.White KE, et al. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60:2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 59.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nature Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 60.Gram Schjoldager KT, et al. A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J Biol Chem. 2011;286:40122–40132. doi: 10.1074/jbc.M111.287912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White KE, et al. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nature Genet. 2000;26:345–348. doi: 10.1038/81664. Identifies FGF23 as a phosphaturic hormone and missense mutations at the proteolytic cleavage site of FGF23 as the cause for the renal phosphate wasting disease autosomal dominant hypophosphatemic rickets. [DOI] [PubMed] [Google Scholar]
- 62.Frishberg Y, et al. Hyperostosis-hyperphosphatemia syndrome: a congenital disorder of O-glycosylation associated with augmented processing of fibroblast growth factor 23. J Bone Miner Res. 2007;22:235–242. doi: 10.1359/jbmr.061105. [DOI] [PubMed] [Google Scholar]
- 63.Kato K, et al. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- 64.Hsu YR, et al. Human keratinocyte growth factor recombinantly expressed in Chinese hamster ovary cells: isolation of isoforms and characterization of post-translational modifications. Protein Expr Purif. 1998;12:189–200. doi: 10.1006/prep.1997.0840. [DOI] [PubMed] [Google Scholar]
- 65.Kim GY, et al. HtrA1 is a novel antagonist controlling FGF signaling via cleavage of FGF8. Mol Cell Biol. 2012 Sep 4; doi: 10.1128/MCB.00872-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avivi A, Yayon A, Givol D. A novel form of FGF receptor-3 using an alternative exon in the immunoglobulin domain III. FEBS Lett. 1993;330:249–252. doi: 10.1016/0014-5793(93)80882-u. [DOI] [PubMed] [Google Scholar]
- 67.Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–11627. [PubMed] [Google Scholar]
- 68.Johnson DE, Lu J, Chen H, Werner S, Williams LT. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miki T, et al. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orr-Urtreger A, et al. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 71.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)–FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol. 1993;13:4513–4522. doi: 10.1128/mcb.13.8.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yayon A, et al. A confined variable region confers ligand specificity on fibroblast growth factor receptors: implications for the origin of the immunoglobulin fold. EMBO J. 1992;11:1885–1890. doi: 10.1002/j.1460-2075.1992.tb05240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wuechner C, Nordqvist AC, Winterpacht A, Zabel B, Schalling M. Developmental expression of splicing variants of fibroblast growth factor receptor 3 (FGFR3) in mouse. Int J Dev Biol. 1996;40:1185–1188. [PubMed] [Google Scholar]
- 74.Zhang X, et al. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alarid ET, et al. Keratinocyte growth factor functions in epithelial induction during seminal vesicle development. Proc Natl Acad Sci USA. 1994;91:1074–1078. doi: 10.1073/pnas.91.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 77.De Moerlooze L, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal–epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 78.Min H, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sekine K, et al. Fgf10 is essential for limb and lung formation. Nature Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 80.Xu X, et al. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. Provides genetic evidence for the crucial role of FGFR2 in bidirectional paracrine FGF signalling between the epithelium and the mesenchyme that initiates the outgrowth of limb buds. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X, et al. Reciprocal epithelial–mesenchymal FGF signaling is required for cecal development. Development. 2006;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF–FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 83.Wang F, Kan M, Xu J, Yan G, McKeehan WL. Ligand-specific structural domains in the fibroblast growth factor receptor. J Biol Chem. 1995;270:10222–10230. doi: 10.1074/jbc.270.17.10222. [DOI] [PubMed] [Google Scholar]
- 84.Lajeunie E, et al. FGFR2 mutations in Pfeiffer syndrome. Nature Genet. 1995;9:108. doi: 10.1038/ng0295-108. [DOI] [PubMed] [Google Scholar]
- 85.Nagase T, Nagase M, Hirose S, Ohmori K. Mutations in fibroblast growth factor receptor 2 gene and craniosynostotic syndromes in Japanese children. J Craniofac Surg. 1998;9:162–170. doi: 10.1097/00001665-199803000-00015. [DOI] [PubMed] [Google Scholar]
- 86.Ibrahimi OA, et al. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum Mol Genet. 2004;13:2313–2324. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olsen SK, et al. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc Natl Acad Sci USA. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 89.Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci USA. 2000;97:49–54. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goetz R, et al. Conversion of a paracrine fibroblast growth factor into an endocrine fibroblast growth factor. J Biol Chem. 2012;287:29134–29146. doi: 10.1074/jbc.M112.342980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ito S, Fujimori T, Hayashizaki Y, Nabeshima Y. Identification of a novel mouse membrane-bound family 1 glycosidase-like protein, which carries an atypical active site structure. Biochim Biophys Acta. 2002;1576:341–345. doi: 10.1016/s0167-4781(02)00281-6. [DOI] [PubMed] [Google Scholar]
- 92.Ito S, et al. Molecular cloning and expression analyses of mouse βKlotho, which encodes a novel Klotho family protein. Mech Dev. 2000;98:115–119. doi: 10.1016/s0925-4773(00)00439-1. [DOI] [PubMed] [Google Scholar]
- 93.Kuro-o M, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 94.Yahata K, et al. Molecular cloning and expression of a novel Klotho-related protein. J Mol Med (Berl) 2000;78:389–394. doi: 10.1007/s001090000131. [DOI] [PubMed] [Google Scholar]
- 95.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fon Tacer K, et al. Research resource: comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goetz R, et al. Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Mol Cell Biol. 2012;32:1944–1954. doi: 10.1128/MCB.06603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kharitonenkov A, et al. FGF-21/FGF-21 receptor interaction and activation is determined by βKlotho. J Cell Physiol. 2008;215:1–7. doi: 10.1002/jcp.21357. [DOI] [PubMed] [Google Scholar]
- 101.Li SA, et al. Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct Funct. 2004;29:91–99. doi: 10.1247/csf.29.91. [DOI] [PubMed] [Google Scholar]
- 102.Machado MF, et al. Regulation and action of fibroblast growth factor 17 in bovine follicles. J Endocrinol. 2009;202:347–353. doi: 10.1677/JOE-09-0145. [DOI] [PubMed] [Google Scholar]
- 103.Portela VM, et al. Expression and function of fibroblast growth factor 18 in the ovarian follicle in cattle. Biol Reprod. 2010;83:339–346. doi: 10.1095/biolreprod.110.084277. [DOI] [PubMed] [Google Scholar]
- 104.Gabrielsson BG, et al. Depot-specific expression of fibroblast growth factors in human adipose tissue. Obes Res. 2002;10:608–616. doi: 10.1038/oby.2002.83. [DOI] [PubMed] [Google Scholar]
- 105.Jaskoll T, et al. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev Biol. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mailleux AA, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129:53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- 107.Qiao J, et al. FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development. 1999;126:547–554. doi: 10.1242/dev.126.3.547. [DOI] [PubMed] [Google Scholar]
- 108.Terauchi A, et al. Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature. 2010;465:783–787. doi: 10.1038/nature09041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dutchak PA, et al. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148:556–567. doi: 10.1016/j.cell.2011.11.062. Identifies FGF21 as a mediator of the metabolic actions of PPARγ and hence provides new insight into the mechanism of action of the thiazolidinedione class of antidiabetic drugs, which are PPARγ agonists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARγ. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 111.Hotta Y, et al. Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150:4625–4633. doi: 10.1210/en.2009-0119. [DOI] [PubMed] [Google Scholar]
- 112.Ming AY, et al. Dynamics and distribution of Klothoβ (KLB) and fibroblast growth factor receptor-1 (FGFR1) in living cells reveal the fibroblast growth factor-21 (FGF21)-induced receptor complex. J Biol Chem. 2012;287:19997–20006. doi: 10.1074/jbc.M111.325670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kouhara H, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 114.Lamothe B, et al. The docking protein Gab1 is an essential component of an indirect mechanism for fibroblast growth factor stimulation of the phosphatidylinositol 3-kinase/Akt antiapoptotic pathway. Mol Cell Biol. 2004;24:5657–5666. doi: 10.1128/MCB.24.13.5657-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 116.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 117.Hartwig JH, et al. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 118.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ye S, et al. Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry. 2001;40:14429–14439. doi: 10.1021/bi011000u. [DOI] [PubMed] [Google Scholar]