Abstract
The apical transmembrane glycoprotein MUC1 is endocytosed to recycle through the trans-Golgi network (TGN) or Golgi complex to the plasma membrane. We followed the hypothesis that not only the known follow-up sialylation of MUC1 in the TGN is associated with this process, but also a remodeling of O-glycan core structures, which would explain the previously described differential core 2- vs core 1-based O-glycosylation of secreted, single Golgi passage and recycling membrane MUC1 isoforms (Engelmann K, Kinlough CL, Müller S, Razawi H, Baldus SE, Hughey RP, Hanisch F-G. 2005. Glycobiology. 15:1111–1124). Transmembrane and secreted MUC1 probes show trafficking-dependent changes in O-glycan core profiles. To address this novel observation, we used recombinant epitope-tagged MUC1 (MUC1-M) and mutant forms with abrogated clathrin-mediated endocytosis (MUC1-M-Y20,60N) or blocked recycling (palmitoylation-defective MUC1-M-CQC/AQA). We show that the CQC/AQA mutant transits the TGN at significantly lower levels, concomitant with a strongly reduced shedding from the plasma membrane and its accumulation in endosomal compartments. Intriguingly, the O-glycosylation of the shed MUC1 ectodomain subunit changes from preponderant sialylated core 1 (MUC1-M) to core 2 glycans on the non-recycling CQC/AQA mutant. The O-glycoprofile of the non-recycling CQC/AQA mutant resembles the core 2 glycoprofile on a secretory MUC1 probe that transits the Golgi complex only once. In contrast, the MUC1-M-Y20,60N mutant recycles via flotillin-dependent pathways and shows the wild-type phenotype with dominant core 1 expression. Differential radiolabeling of protein with [35S]Met/Cys or glycans with [3H]GlcNH2 in pulse-chase experiments of surface biotinylated MUC1 revealed a significantly shorter half-life of [3H]MUC1 when compared with [35S]MUC1, whereas the same ratio for the CQC/AQA mutant was close to one. This finding further supports the novel possibility of a recycling-associated O-glycan processing from Gal1-4GlcNAc1-6(Gal1-3)GalNAc (core 2) to Gal1-3GalNAc (core 1).
Keywords: glycan processing, membrane trafficking, MUC1, O-glycosylation, recycling
Introduction
The correct sorting of newly synthesized proteins for trafficking to their final cellular destination is an important step in the maintenance of cell function and is accomplished by glycan- and protein-specific sorting signals. In polarized epithelia, sorting to the basolateral surface depends on the presence of short peptide sequences in the cytosolic domains of the respective proteins, whereas apical targeting signals are more diverse. Among these apical targeting signals, a variety of post-translational modifications do play a role, such as N- and O-glycans or glycosylphosphatidylinositol anchors (for review, see Potter et al. 2006a; Vagin et al. 2009). For some apically expressed glycoproteins, it is known that their surface expression is only transient and that they traffic back to the trans-Golgi network (TGN) where they can undergo further glycosylation cycles (Naim et al. 1999). One such protein, which is known to follow this recycling pathway, is the type 1 transmembrane glycoprotein MUC1. This heavily O-glycosylated membrane-bound mucin was early demonstrated to recycle several times through the TGN and to gain in this way a maturation of its O-glycans through continued sialylation (Litvinov and Hilkens 1993). The endocytosis and the vesicular transport of the protein were shown to follow a clathrin-mediated pathway (Altschuler et al. 2000), later confirmed to be dependent on specific tyrosine residues within the highly conserved cytosolic domain of MUC1 (Tyr20 and Tyr60) necessary for binding to the adaptor protein complex 2 and Grb2, respectively (Kinlough et al. 2004). The reentry into the secretory pathway of MUC1 from endosomes was revealed as dependent on the Cys-palmitoylation of the CQC motif located at the intersection of the cytosolic and transmembrane domains (Kinlough et al. 2006).
MUC1 can also be shed from the plasma membrane by the action of specific sheddases or by the dissociation of the heterodimeric complex formed by the larger subunit containing the extracellular tandem-repeat domain and the smaller transmembrane subunit (Lillehoj et al. 2003; Thathiah et al. 2003; Thathiah and Carson 2004). Accordingly, the exosome-depleted supernatant of the culture medium from cells transfected with a recombinant MUC1 was shown to contain a soluble form of the fusion protein that lacks the transmembrane and cytosolic domains (Engelmann et al. 2005).
A previous study from our group revealed that secreted (MUC1-S) and membrane-tethered MUC1 (MUC1-M) exhibit dramatic differences in the patterns of their O-linked chains (Engelmann et al. 2005). We observed that not only the degree of sialylation varied as could be expected from the work of Litvinov and Hilkens (1993), but surprisingly also the core structures of the O-glycans (Engelmann et al. 2005). Strikingly, the secreted isoform, which passes through the Golgi complex only once, expressed predominantly neutral core 2-based glycans, whereas the membrane-bound isoform, which recycles through the TGN, expressed almost exclusively sialylated core 1 glycans. Hence, the single-pass isoform MUC1-S was modified with the more complex, branched core structure when compared with the recycling MUC1-M glycoform. This difference in the O-glycan structures observed for MUC1-S and MUC1-M could be either due to the differential glycosylation of soluble vs transmembrane proteins within the biosynthetic pathway or due to the continued processing of core 2 O-glycans on recycling MUC1-M by the concerted action of glycosidases and glycosyltransferases, analogous to the terminal processing of N-linked glycans. Such processing activities for O-linked glycans are not yet known to exist in the Golgi complex or the TGN. It is accordingly important to learn whether the observed changes in O-glycosylation are actually related to the recycling of MUC1.
To test the hypothesis that core 2 O-glycans on newly synthesized MUC1 are remodeled during recycling, we characterized the structures of O-glycans on a recombinant epitope-tagged variant of MUC1 (MUC1-M) and on mutated forms with either reduced clathrin-mediated endocytosis (MUC1-M-Y20,60N) or abrogated recycling from endosomes back to the plasma membrane (MUC1-M-CQC/AQA). However, the mutant isoform MUC1-M-Y20,60N could follow alternative endocytosis pathways based on lipid rafts, since we showed in a proteomic study that MUC1 is sorted into flotillin-2-associated rafts (Staubach et al. 2009). Accordingly, MUC1-M-Y20,60N could still recycle through the Golgi complex or the TGN and could show the partial conversion of the core 2 O-glycans. On the other hand, the MUC1-M-CQC/AQA isoform is predicted to recycle poorly and accumulate in endosomes after its initial endocytosis. If the postulated O-glycan processing by core O-glycan reconstruction or follow-up glycosylation during recycling is a valid assumption, we have to expect that the non-recycling MUC1-M-CQC/AQA will not have the conversion of core 2 O-glycans and would expresses O-glycan profiles similar to those of secreted isoforms. Data in this manuscript support the hypothesis that O-glycans on MUC1 are remodeled from core 2 to core 1 structures during recycling after initial delivery to the cell surface.
Results
Generation of MUC1 isoforms with altered cellular trafficking
To address whether MUC1 endocytosis and recycling (through the TGN or the Golgi complex) has an impact on the O-glycan core 2/core 1 expression patterns of the transmembrane glycoprotein, we created mutant isoforms of the previously described fusion protein MUC1-M. MUC1-M exhibits wild-type characteristics, since the small transmembrane subunit is identical to endogenous MUC1, and the large subunit contains a truncated tandem-repeat domain and two N-terminal epitope tags (Engelmann et al. 2005). The mutant isoforms of MUC1-M differ with respect to site-specific mutations in their cytosolic domains (see Figure 1 for MUC1-M-Y20,60N and MUC1-M-CQC/AQA). The double replacement of Tyr20 and Tyr60 by Asn had previously been shown to reduce the clathrin-mediated endocytosis of a Tac-MUC1 chimera by 77% (Kinlough et al. 2004). On the other hand, the two point mutations of Cys to Ala in the Cys-palmitoylation motif CQC located at the intersection of the cytosolic and transmembrane domains had been demonstrated to impair the recycling of the MUC1 probe by blocking reentry of the Cys-palmitoylation-defective probe into the secretory pathway and by causing its accumulation in recycling endosomes (Kinlough et al. 2006). The wild-type MUC1 probe and its mutant isoforms were stably expressed in the MUC1-positive breast carcinoma cell line MCF-7 and in the MUC1-negative Human embryonic kidney-293 (HEK-293) cells. The membrane expression of the epitope-tagged isoforms was secured for the two cell lines by a confocal laser microscopy (Figure 2A). Endocytosis of surface MUC1-M was followed after complexing the mucin with an ectodomain-specific C595 antibody. We demonstrated the effective internalization of MUC1-M and its mutant forms in the cell lines under investigation (Figure 2B). Although the probes MUC1-M and MUC1-M-Y20,60N showed vesicular staining, the probe MUC1-M-CQC/AQA was found to accumulate in larger endosomal compartments. Endocytosed MUC1 co-localized with the lipid-raft marker flotillin in MCF-7 cells consistent with the use of alternative endocytotic pathways for the mucin that are independent of clathrin vesicles (Figure 2C). Co-localization with endocytosed MUC1 was also shown for the raft-stabilizing galectin-3 (Figure 2C), which has been previously found to interact directly with some apical cargo in a glycan-dependent manner (Delacour et al. 2007).
Fig. 1.
The structure of truncated MUC1-M and mutated isoforms with impaired endocytosis or recycling. Schematic presentation of domain topologies of constructs with indicated N-glycosylation sites and amino acid sequences of cytosolic domains in the constructs MUC1-M, MUC1-M-Y20,60N and MUC1-M-CQC/AQA.
Fig. 2.
Surface expression and endocytosis of MUC1-M probes revealed by a confocal laser microscopy. (A) Immunofluorescence microscopy of HEK-293 cell surface expressed MUC1-M and mutant MUC1-M isoforms with anti-MUC1 ectodomain antibody C595 (magnification: ×400). (B) Endocytosis of antibody-tagged MUC1-M, MUC1-M-Y20,60N and MUC1-M-CQC/AQA for 1 h at 37°C in HEK-293 cells (upper panel) and in MCF-7 cells (lower panel; magnification: ×400). (C) Co-localization studies of MUC1-M and lipid-raft marker Flot-2 (flotillin-2) and raft-associated Gal-3 (galectin-3) in MCF-7 cells (magnification: ×400). The white scale bars correspond to 4 µm.
Isolation of shed MUC1-M isoforms
To study the O-glycosylation profiles of MUC1-M isoforms, we decided to isolate the membrane-shed large ectodomain subunits. For this purpose, the constituents of primed culture supernatants were separated into exosome-depleted and exosomal fractions. Western blot analysis of primed exosome-depleted culture medium (50 mL each) revealed much stronger shedding of MUC1-M compared with the mutant isoforms. In particular, the MUC1-M-CQC/AQA mutant was shed at much lower rates by HEK-293 and MCF-7 cells (Figure 3A). The cellular expression on the other hand did not vary significantly for different isoforms (Figure 3B), except for a slight increase observed for the CQC/AQA mutant expressed in both MCF-7 and HEK-293 cells. Yields for shed MUC1-M in HEK-293 cells were ∼1 mg/L of medium. Both MUC1-M and mutant isoforms were demonstrated in immunoblots to lack the cytosolic domain at 31 kDa detectable in exosomal membranes (Figures 4 and 5C). The shed isoforms were isolated from exosome-depleted culture supernatant by nickel chelate affinity chromatography and reversed-phase HPLC and analyzed for purity by sodium dodecyl sulfate (SDS) gel electrophoresis with silver staining (Figure 5A) or by glycan detection after transfer to nitrocellulose (Figure 5B).
Fig. 3.
Comparative analysis of cellular expression rates vs shedding rates of wild-type and mutant isoforms. (A) Semi-quantitative estimation of MUC1-M isoform shedding by HEK-293 and MCF-7 cells. The same volume of culture supernatant (50 mL) was analyzed for shed MUC1-M isoforms after affinity isolation by nickel chelate chromatography using western blotting with the anti-MUC1 (C595) antibody. (B) Comparison of expression levels of the wild-type and mutant MUC1-M isoforms in extracts of HEK-293 and MCF-7 cells. The ectodomains of the MUC1 isoforms were stained with C595 antibody and staining intensity was compared with that of reference protein GAPDH.
Fig. 4.
Cellular release of MUC1-M occurs via shedding. Western blot analysis of exosome-free culture supernatant from HEK-293 and MCF-7 breast cancer cells. Different volumes of culture supernatant were used for the isolation of shed wild-type and mutant isoforms to account for their varying expression rates (Figure 3). Shed MUC1-M is devoid of the small endodomain subunit, which spans the membrane.
Fig. 5.
Purity control of shed MUC1-M isoforms. (A) SDS gel electrophoresis with silver staining of shed samples obtained from culture supernatants of HEK-293 and MCF-7 cells. Equal protein amounts were loaded onto the gel, accordingly the staining intensities do not reflect the differential shedding of the MUC1-M isoforms demonstrated in Figure 3. (B) Western blot for the glycan detection of identical samples described in (A). The band marked with C refers to a minor glycoprotein contaminant from calf serum, which was not separable by affinity chromatography combined with reversed phase chromatography. (C) Western blot of proteins in exosomal membranes stained with either anti-MUC1 ectodomain antibody C595 or with anti-MUC1-endodomain antibody H295.
Non-recycling MUC1 probes exhibit altered O-glycosylation profiles
The shed isoforms of MUC1-M, MUC1-M-Y20,60N and MUC1-M-CQC/AQA were digested in-gel with proteinase K after SDS gel electrophoresis and the eluted (glyco)peptides were analyzed for their O-glycan profiles by reductive β-elimination of the oligosaccharides and mass spectrometric analysis of the permethylated glycans. The relative expression ratios in HEK-293 and MCF-7 cells (of core 2 vs core 1 and of sialylated vs neutral glycans) together with a list of the respective glycan masses and structures are presented in Figure 6.
Fig. 6.
Profiling of O-glycosylation on shed MUC1-M isoforms and their mutant isoforms expressed in HEK-293 and MCF-7 cells. (A) Schematic presentation of O-linked glycans with core 1 and core 2 structures and the molecular masses of sodium adducts of methylated glycan alditols found on recombinantly expressed MUC1-M probes. a, core 1 refers to glycans with one HexNAc residue (core-GalNAc) in their calculated monosaccharide compositions, core 2 to glycans with more than one HexNAc. HEK-293 express non-elongated core 1 and GlcNAc-containing core 2 glycans according to previous studies (Müller and Hanisch, 2002; Engelmann et al. 2005). b, the masses refer to pseudo-molecular ions M + Na. c, the proposed structures are based on published evidence from mass spectrometry of methylated glycan alditols and partially methylated alditol acetates derived from MUC1-M (Engelmann et al. 2005). (B) Representative MALDI-TOF mass spectra registered for samples of permethylated glycan alditols from MUC1-M (upper panel) and MUC1-M-CQC/AQA (lower panel). The ion signals were annotated with respect to the relative masses of molecular ions (m/z) detected as sodium adducts and by assignment of the respective core structure (1 or 2). F refers to the primary fragment NeuAc1Hex1HexNAc1 induced during laser ionization. (C) The bars correspond to the relative amounts of glycans (%) with core 1 (gray shaded) vs core 2 (open) structures or to sialylated (black) vs neutral glycans (gray-shaded). 1, 2 and 3 refer to MUC1-M, MUC1-M-Y20,60N and MUC1-M-CQC/AQA, respectively.
The shed MUC1 probes revealed changes in the relative expression ratios of core 1 vs core 2 O-glycans and with respect to the ratio of sialylated vs neutral glycans (Figure 6B). Whereas the profiles changed only slightly in the Y20,60N mutant when compared with the wild-type analogous MUC1-M, there was a dramatic shift in the ratios toward core 2 O-glycan expression for the CQC/AQA mutant. Impaired clathrin-mediated endocytosis of the Y20,60N mutant could hence be compensated by alternative routes of vesicular transport as revealed in a recent proteomic study (Staubach et al. 2009). On the other hand, the abrogation of recycling due to defective Cys-palmitoylation and accumulation in endosomes apparently has a much stronger effect on the glycosylation of the transmembrane protein. Expectedly, the ratio of sialylated vs neutral glycans decreased when comparing the wild-type profile to that of the CQC/AQA mutant, but the effect was weaker than the shift in core expression.
Comparing the half-lives of MUC1 metabolically labeled with [3H]GlcNAc or [35S]Met/Cys
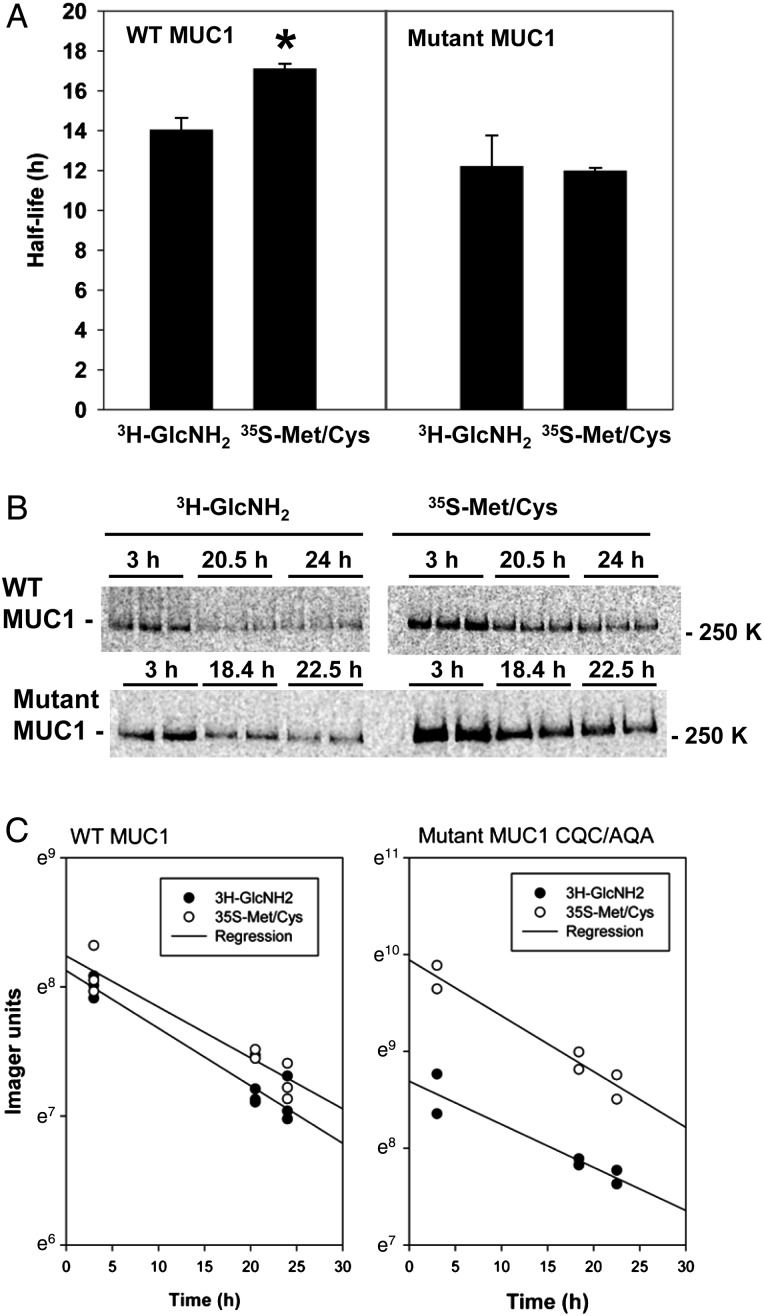
To test the possibility that core 2 O-glycans added to MUC1 in the biosynthetic pathway are remodeled during recycling by the removal of GlcNAc to reveal a core 1 glycan structure, we compared the half-lives of MUC1 metabolically labeled with either [3H]GlcNAc or [35S]Met/Cys (Figure 7). In preliminary experiments, we found that metabolic labeling of proteins with [3H]GlcNAc was considerably more efficient in MDCK cells when compared with HEK-293 and MCF-7 cells, and [3H]GlcNAc incorporation into MUC1 with 22 tandem repeats was notably higher when compared with its incorporation into MUC1-M with only six tandem repeats. MDCK cells stably transfected with MUC1, or MUC1 with the mutation CQC/AQA, were pulse labeled with either [3H]GlcNAc or [35S]Met/Cys for 1 h, then chased for 1 h in the absence of radioactive metabolites, a time frame that allows newly synthesized MUC1 to reach the cell surface (Kinlough et al. 2011; Hanisch et al. 2012). Cells were moved to ice and treated with sulfo-NHS-SS-biotin to tag all surface proteins, then the cells were returned to culture for 3–24 h in order to follow the disappearance of metabolically labeled and surface-biotinylated MUC1. Biotinylated MUC1 was recovered from immunoprecipitates obtained at each time point with avidin-conjugated beads and analyzed with a BioRad phosphoimager after SDS–polyacrylamide gel electrophoresis (PAGE). The half-lives (mean and SEM) were determined from three independent experiments and revealed that MUC1 metabolically labeled with [3H]GlcNAc turned over significantly faster than MUC1 labeled with [35S]Met/Cys (14.0 ± 0.6 vs 17.1 ± 0.2 h, respectively, with P < 0.05; Figure 7). In contrast, MUC1 with the mutation CQC/AQA that prevents recycling had no significant difference in its half-lives determined in three separate experiments whether labeled with [3H]GlcNAc or [35S]Met/Cys (12.2 ± 1.5 vs 12.0 ± 0.2 h, respectively). Altogether, the data support the hypothesis that GlcNAc in core 2 O-glycans added within the biosynthetic pathway are subsequently removed during the recycling of MUC1 after it reaches the cell surface.
Fig. 7.
MUC1 labeled with [3H]GlcNH2 has a shorter half-life than MUC1 labeled with [35S]Met/Cys. MDCK cells stably expressing either wild-type (WT) MUC1 or mutant MUC1 (CQC/AQA) that does not recycle from endosomes, were metabolically labeled with [3H]GlcNH2 or [35S]Met/Cys (1 h pulse and 1 h chase, in duplicate or triplicate) prior to cell surface biotinylation. Cells were returned to culture for 3–24 h as indicated in each experiment, and biotinylated MUC1 or mutant MUC1 were recovered with avidin-conjugated beads from immunoprecipitates from cell extracts and subjected to SDS–PAGE and analysis with a BioRad phosphoimager. Radioactive bands were quantified and half-lives from three independent experiments are presented as the mean and SEM (A). Note that the half-life for [3H]MUC1 is significantly shorter than the half-life for [35S]MUC1 (*P < 0.05), whereas no statistical difference was found between [3H]mutant and [35S]mutant (see text for details). The radioactive bands on the SDS gels (B) and semi-log plots of the data (C) from one representative experiment are shown.
Internalized wild-type MUC1 traffics to the TGN more efficiently than the CQC/AQA mutant
To test whether wild-type MUC1 and CQC/AQA mutant differentially access the TGN during recycling, transiently transfected HEK-293 cells expressing the TGN marker mCherry-GalT were incubated for 1 h at 37°C with an anti-MUC1 antibody. Residual surface-bound antibody was stripped and the cells were fixed and processed for indirect immunofluorescence. Confocal image stacks were scored by two blinded observers for co-localization between internalized B27.29 antibody and mCherry-GalT (Figure 8). Both observers scored 100% of the MUC1 expressing cells (n = 7) as having some co-localization of the two markers. In contrast, fewer than 50% of the cells expressing MUC1 mutant CQC/AQA were scored as having co-localization (1–3 of seven; Figure 8). Though semi-quantitative, these results are consistent with the interpretation that internalized MUC1 traffics to the TGN from the cell surface more efficiently than the CQC/AQA mutant.
Fig. 8.

Internalized anti-MUC1 antibodies traffic to the TGN. HEK-293 cells expressing wild-type MUC1 or the mutant MUC1 CQC/AQA and the TGN fluorescent marker mCherry-GalT (red) were incubated with anti-MUC1 tandem repeat antibody B27.29 for 1 h at 37°C to allow antibody internalization. Residual surface antibody was removed by stripping and the coverslips were processed for immunofluorescence as described in Materials and methods to detect internalized anti-MUC1 antibody (green). Confocal sections and merged images are shown. Arrows point to areas where co-localization (yellow) is evident. Scale bar: 7.5 μm.
Discussion
There are many open questions surrounding the trafficking routes of apical cell surface membrane glycoproteins, like MUC1. We know that MUC1 recycles at the plasma membrane, but not how it is sorted for either rapid recycling back to the cell surface or slowly through the TGN or why this sorting takes place at all. We also know that some follow-up glycosylation occurs during recycling through the TGN, and thereby secreted isoforms exhibit distinct O-glycan core patterns compared with recycling membrane isoforms. These latter, quite dramatic changes in the O-glycan core profiles (Engelmann et al. 2005) correspond to similar findings for N-linked glycans on native MUC1 from human skim milk (secreted mucin) or human milk fat globule (HMFG) membranes (Parry et al. 2006). In the latter study, it was shown that a single passage of the glycosylation machinery resulted in preponderant high-mannose-type N-glycosylation of secreted MUC1, whereas the HMFG-MUC1 was nearly exclusively modified with complex-type chains. Altogether, these previous findings indicate that changes of MUC1 glycosylation during recycling may not be restricted to simply continued sialylation in the TGN. Instead, it now appears that a remodeling of both N- and O-linked glycans takes place during recycling.
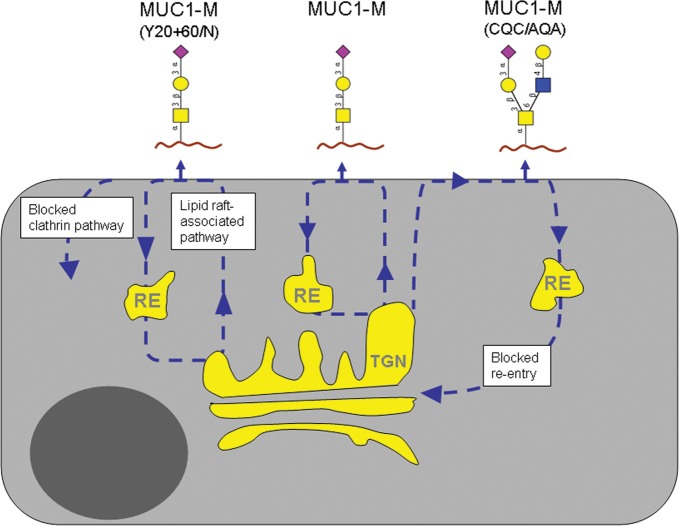
In this study, we obtained experimental evidence that the observed dramatic changes in O-glycosylation profiles are clearly associated with recycling of MUC1. Recycling transmembrane probes exhibit a distinct O-glycosylation pattern (sialylated core 1) compared with the secreted probe (largely core 2-based; Figure 9). Contrasting to this, non-recycling mutant forms of MUC1 (CQC/AQA) transmembrane probes with impaired TGN reentry expressed O-glycan profiles similar to the secreted MUC1 probe (Figure 9). A mutant with impaired clathrin-mediated endocytosis, however, expressed O-glycan profiles similar to the wild-type probe (Figure 9). This latter finding may suggest alternative, clathrin-independent routes of endocytosis as indicated by proteomic data on MUC1 and flotillin-2 positive lipid rafts from MCF-7 cells (Staubach et al. 2009).
Fig. 9.
Correlation between MUC1 trafficking and glycosylation. Trafficking pathways of mutant MUC1 probes with impaired clathrin-mediated endocytosis (MUC1-M-Y20,60N) or impaired reentry into the secretory pathway and blocked recycling (MUC1-M-CQC/AQA). Although MUC1-M-Y20,60N may recycle via alternative lipid raft-associated pathways and shows similar O-glycosylation as the wild-type MUC1-M, the CQC/AQA mutant accumulates in endosomal compartments and resembles the non-recycling MUC1-S with respect to O-glycan core expression. Light squares refer to GalNAc, light circles to Gal, dark squares to GlcNAc, rhombs to sialic acid. RE, recycling endosome; TGN, trans-Golgi network.
Although a follow-up sialylation during the recycling of MUC1 has been demonstrated two decades ago (Litvinov and Hilkens 1993), there is no evidence yet, which would support a remodeling of O-linked glycans similar to the trimming of endoplasmic reticulum N-glycoforms of the high-mannose-type and the construction of complex-type chains in the Golgi complex and the TGN. The processing of extended core 2 O-glycans [Gal1-4GlcNAc1-6(Gal1-3)GalNAc] would require the action of at least two glycosidases, a β-galactosidase and a β-N-acetylglucosaminidase, which would trim the predominant core 2-tetrasaccharide down to a core 1-disaccharide, which then forms the substrate of sialyltransferases in the TGN. Although these trimming glycosidases were not yet identified, we could obtain evidence that at least a partial removal of [3H]GlcNAc that had been incorporated into MUC1 glycans on the first passage of the Golgi complex is associated with recycling of MUC1. The shorter half-life of MUC1-bound [3H]GlcNAc compared with the MUC1 [35S]protein is consistent with the partial removal of the core 2-characteristic sugar during recycling. The observed difference in half-lives may be regarded as small; however, the processing of N-linked chains on MUC1 to complex-type glycans carrying additional GlcNAc residues should be considered to partially compensate for the removal of GlcNAc from O-linked chains.
Taken together, the expression of different O-glycoforms was shown to be associated with the cellular recycling of MUC1 in two different cell lines (HEK-293 and MCF-7), indicating a functional link between these phenomena. Vice versa, we have already published that MUC1 apical targeting is glycan-dependent. We initially observed that MUC1 traffics to the apical surface via apical recycling endosomes in polarized renal epithelial cells, a route used by transmembrane proteins such as endolyn with an N-glycan-dependent apical targeting signal (Cresawn et al. 2007; Mattila et al. 2009; Mo et al. 2012). Subsequently, we did establish that core-glycosylated mucin-like repeats from the N terminus of MUC1 represent an apical targeting signal (Kinlough et al. 2011). This finding was consistent with the earlier studies that observed the accumulation of immature glycoforms of proteins including MUC1 in endosomal compartments after the treatment of epithelial cells with benzyl-GalNAc which perturbs the extension of O-glycans beyond the initial formation of GalNAc-Ser/Thr (Delacour et al. 2003).
Further support for the involvement of glycans in membrane sorting came from the finding that galectins (Gal) can play a role in this process (Schuck and Simons 2004; Delacour et al. 2005, 2006, 2007; Delacour and Jacob 2006; Mo et al. 2012). In particular, the knockdown of Gal-3 in MDCK cells resulted in the non-polarized expression of the normally apical lactase-phlorizin hydrolase, gp114 and p75 neurotrophin receptor, likely by disruption of galectin-dependent cross-linking. Interestingly, MUC1 in ZR-75-1 cells is also reported to interact with Gal-3 through the single N-glycan on its small transmembrane subunit, but we found that the efficient knockdown of Gal-3 in MDCK cells did not perturb MUC1 apical targeting (Ramasamy et al. 2007; Kinlough et al. 2011). We previously reported that MUC1 expressed in MDCK cells preferentially binds to Gal-3 and both carbohydrate recognition domains of Gal-9 (Gal-9N and Gal-9C) in pull-down assays with all of the GST-tagged canine galectins (Poland et al. 2011). More recently, we established that the O-glycan-dependent apical targeting signal of MUC1 preferentially binds to GST-Gal-9C in pull-down assays with all the canine galectins, and future studies utilizing Gal-9 knockdown in MDCK cells will investigate any role for this galectin in MUC1 apical targeting (Kinlough, Poland and Hughey, unpublished data/in press). Interestingly, similar pull-down assays with GST-tagged canine galectins and endolyn showed a kifunensine-sensitive interaction of endolyn with Gal-3, Gal-4, Gal-8 and Gal-9N, but only the knockdown of Gal-9 in MDCK cells blocked apical targeting of this protein with established N-glycan-dependent targeting (Mo et al. 2012). As similar N-glycan sorting determinants are required for the apical delivery of endolyn along both the biosynthetic and the postendocytic pathways, it is also likely that MUC1 utilizes O-glycan-dependent targeting for both apical biosynthetic and postendocytic delivery as well (Potter et al. 2006b). It is not clear yet whether the reentry of recycling MUC1 into the secretory pathway is restricted to late compartments in the TGN or whether also earlier compartments, like the cis- to trans-Golgi compartments could be involved as shown for DPPIV (Volz et al. 1995).
In summary, accumulating evidence supports a dependency of trafficking/recycling and O-glycosylation in a way that proteins following different pathways exhibit distinct glycosylation patterns (this paper) and vice versa that differential glycosylation determines trafficking routes of some glycoproteins (reviewed by Potter et al. 2006a). Both are true for MUC1, as the O-glycans direct sorting processes and the trafficking determines the glycan processing. Core 2 glycans could enhance MUC1 delivery to the plasma membrane from both the biosynthetic and recycling pathways, whereas the conversion to core 1 glycans could reduce the efficiency endocytic trafficking directed by the cytoplasmic Tyr-motifs and thereby enhance MUC1 degradation or accumulation and release through exosomes.
Materials and methods
Cells and cell culture
The breast cancer cell line MCF-7 and the human epithelial kidney cell line HEK-293 were obtained from the American Type Culture Collection and were cultured in 75 or 150 cm2 flasks at 37°C in the presence of 5% CO2. Minimal essential medium, supplemented with glutamax I, 10% fetal calf serum, 0.1 mM sodium pyruvate, 100 IU penicillin and 100 µg/mL streptomycin was used for MCF-7 and HEK-293. After transfection with plasmids encoding MUC1 variants, 0.1 µg/mL puromycin was added to all media. Medium was exchanged every 3–4 days and the cells were passaged when they reached 80–90% confluency using 0.1% trypsin and 0.2 mM EDTA. Puromycin was from Sigma, Taufkirchen, Germany, all other cell culture reagents were obtained from Life Technologies, Karlsruhe, Germany.
MDCK cells were transfected with pcDNA3(neo) encoding either MUC1 or mutant MUC1 CQC/AQA (both with 22 tandem repeats), and clonal cell lines were selected by cell growth in G418 (0.5 mg/mL) in Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 (1:1) with 5% fetal bovine serum (DMEM/F-12/5% FBS) at 37°C in a humidified incubator with 5% CO2 (all reagents from Invitrogen Life Technologies, Karlsruhe, Germany).
Generation and expression of mutant constructs
Preparation of MUC1-M (with N-terminal His6-myc tags and six tandem repeats) in pCEP-PU was previously described (Engelmann et al. 2005). Mutations were introduced into the cytoplasmic tail of MUC1-0TR (MUC1 with zero tandem repeats) in pBS-SK(-) (Stratagene, Cedar Creek, TX) using PCR-based, site-directed mutagenesis using primers with specific nucleotide changes to produce double mutants CQC/AQA or Y20N,Y60N. Nucleotide sequences encoding the SEA domain, transmembrane domain and cytoplasmic domain of the two MUC1-0TR mutants were amplified by PCR (sense primer 5′-ACA CTC GAG CAC AGC ACT TTT CCC CAG TTG-3¢ and reverse primer 5′-ATT GGA TCC CTA CAA GTT GGC AGA AGC GGC-3¢). Underlined nucleotides are the MUC1 sequence. The amplified DNA was cut with Xho1 and BamH1 and ligated into MUC1-M in pCEP-PU that was similarly cut with Xho1 and BamH1, using standard techniques. MUC1-M-CQC/AQA and MUC1-M-Y20,60N in pCEP-PU were sequenced for accuracy. Preparation of MUC1 mutant CQC/AQA with 22 tandem repeats was described previously (Kinlough et al. 2006).
Isolation of fusion proteins from culture supernatant
FCS was pre-depleted of calf exosomes by centrifugation in three steps: 1200 × g for 10 min, 10,000 × g for 30 min and 100,000 × g for 1 h. The exosomes contained in cell culture supernatants were isolated in four centrifugation steps: 1200 × g for 10 min to remove cells, 10,000 × g for 30 min to remove cellular debris, 10,000 × g for 30 min to remove remaining cellular debris and 100,000 × g for 1 h to pellet the exosomes. The exosomal pellet was washed three times in phosphate-buffered saline at 100.000 × g for 1 h.
To inhibit proteases, PMSF in DMSO was added to the exosome-depleted supernatant (final concentration: 1 mM) and the pH was adjusted with 0.5 M NaH2PO4. Ni Sepharose 6 Fast flow column (Amersham Pharmacia Biotech, Nümbrecht, Germany) was washed with sterile deionized water and with phosphate-buffered saline (PBS), pH 7.0, for equilibration. The cleared protein sample was then passed through the Ni column at 4°C. Afterwards, the column was washed with PBS followed by a stepwise elution with 10, 20, 40, 80, 150 and finally 250 mM imidazole in PBS, 10 mL each. The eluted samples were concentrated via (Amicon Centriprep Ultracel YM-10 Da, Millipore, Schwalbach, Germany) then analyzed by SDS–PAGE and western blotting. MUC1-positive fractions were subjected to HPLC purification on a reversed-phase column (Vydac 208TP1015, MZ Analysentechnik, Mainz, Germany). Samples were injected in 500 µL aliquots and the column was eluted with a gradient of 2–80% acetonitril in 0.1% TFA over 30 min. A flow rate of 1 mL/min was used and the chromatogram was registered spectrophotometrically at 214 nm. Peaks were collected manually and the quality of the preparations was analyzed by SDS–PAGE. The gels were stained with silver or transferred to nitrocellulose membranes and the fusion proteins were detected with an anti-MUC1 ectodomain-specific monoclonal antibody (C595; gift from Dr. Graem Denton, University of Nottingham, UK) or an anti-MUC1 endodomain-specific polyclonal antibody H-295 (Santa Cruz Biotechnology, Inc., Heidelberg, Germany).
Purified fusion proteins were checked for contaminating protein (silver-stained gel) or glycoprotein (DIG Glycan Detection Kit, Roche, Mannheim, Germany) by gel electrophoresis and western blotting as directed by the manufacturer.
Endocytosis studies by confocal laser scanning microscopy
Cells were detached from culture flasks by trypsin treatment, resuspended in complete media and plated on Permanox chamber slide (sterile plastic, Nunc, Langenselbold, Germany) overnight at 37°C, 5% CO2. Surface exposed MUC1 was stained with a primary mouse monoclonal antibody (C595: 5 µg/mL) and a polyclonal anti-MUC1 rabbit antibody (10 µg/mL) in cell culture medium and the immune complex was allowed to follow endocytosis for 1 h at 37°C, 5% CO2. The cell culture supernatant was removed, cells were rinsed three times in sterile PBS for 5 min and fixed in 2% paraformaldehyde (Roth, Karsruhe, Germany) at RT for 10 min. Surface charges were blocked by addition of 2% glycine (Roth) in PBS, pH 7.5, for 30 min at RT followed by three washing steps and permeabilization with 0.1% Triton X-100 (Fluka, Taufkirchen, Germany). Non-specific protein binding was blocked with 3% BSA in PBS for 1 h at RT followed by rinsing three times with sterile PBS and in 0.1% Tween-20/PBS (buffer A), each for 5 min at RT. As secondary antibody, the fluorescently labeled anti-mouse IgG Alexa Fluor 488 and the fluorescently labeled anti-rabbit IgG Alexa Fluor 555 (Molecular Probes, Göttingen, Germany; diluted 1:1000 in buffer A with 0.1% BSA) were incubated for 1 h at RT. Cells were washed three times in buffer A for 5 min at RT in the dark. Slides were mounted in fluorescent mounting medium (Dako, Hamburg, Germany). The microscopic analysis was performed on a confocal laser-scanning microscope Leica TCS SL (Leica, Heidelberg, Germany).
TGN co-localization studies
HEK-293 cells grown on glass coverslips in DMEM supplemented with 10% FBS were transfected using Lipofectamine 2000 with plasmid cDNA encoding MUC1 WT or mutant MUC1 CQC/AQA (both with 22 tandem repeats) along with the TGN fluorescent marker mCherry-GalT (galactosyltransferase). The following day, cells were incubated with 5 μg/mL mouse monoclonal anti-MUC1 antibody B27.29 (directed against the extracellular tandem repeats) for 1 h at 37°C. The coverslips were washed with PBS three times and surface-bound antibody was stripped by incubation with 150 mM glycine buffer, pH 2.3, for 15 min at 4°C. Cells were rinsed with PBS and then fixed with 2% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min and blocked with 2% BSA for 1 h. Cells were labeled with a goat anti-mouse secondary antibody (IgG Alexa Fluor 488; 1:500; Invitrogen Life Technologies) for 30 min, washed and mounted in ProLong Gold Antifade Reagent (Invitrogen Life Technologies). Confocal images were acquired using a Leica TCS SP5 microscope and processed using Adobe Photoshop.
Western blot analysis
Samples were run on SDS–polyacrylamide gels (3% stacking gel, 10% running gel) in a Mini Protean 3 cell (BioRad, Munich, Germany). The gel was equilibrated in transfer buffer (20 mM glycine, 24 mM Trizma-base, 20% methanol) before it was wet-blotted (BioRad) onto a nitrocellulose membrane (Schleicher and Schull, Einbeck, Germany) at 90 mA overnight. Thereafter, the membrane was blocked in Tris-buffered saline containing 5% non-fat dried milk and 0.1% Tween-20 for 1 h at room temperature before incubation with the primary antibody (1 h at room temperature). Immunoblots were incubated with rabbit anti-mouse IgG, or pig anti-rabbit HRP-conjugated secondary antibody (Dako) and MUC1 was detected using a Lumilight Kit (Roche) and super RX film (Fujifilm Europe, Düsseldorf, Germany). Between each incubation step, the membrane was washed three times with TBST (20 mM Tris–HCl, 137 mM NaCl, pH 7.6, 0.1% Tween-20). The large ectodomain of MUC1 probes was stained with C595 antibody.
Mass spectrometric O-glycan analysis
Due to a contaminating glycoprotein detectable at ∼126 kDa, O-glycosylation profiles were determined after electrophoretic separation by in-gel proteolysis of the glycoproteins, elution of glycopeptides from the gel and reductive β-elimination of the O-linked chains. In detail, the region covering proteins with apparent molecular masses from 65 to 85 kDa in Coomassie-stained gels was cut and washed in two 1 mL aliquots of acetonitrile/water (for 1 h each time). Gel slices were dried by vacuum rotation prior to addition of 50 µL of 20 mM Tris, pH 7.5, 1 mM CaCl2 containing 1 µg proteinase K (Qiagen, Hilden, Germany). After overnight incubation at 56°C, the resulting protease-stable glycopeptides were eluted consecutively in 200 µL aliquots of water (2 aliquots) and 50% acetonitrile/water each by rotation for 1 h. The combined aliquots were dried by vacuum rotation and the glycans were β-eliminated in 50 µL NaBH4 (1 M in 50 mM NaOH) under argon by overnight incubation at 50°C. After desalting with Dowex50 × 8 (H+) in a batch procedure and the removal of borate by codistillation of the methylester from acidified methanol, the dried residues were methylated according to the Ciucanu and Kerek (1984) method. Glycan profiles were measured by performance of MALDI-TOF mass spectrometry according to previously published protocols (Müller and Hanisch 2002; Engelmann et al. 2005).
Metabolic labeling experiments with surface biotinylated MUC1 in MDCK cells
MDCK cells stably transfected with MUC1 or MUC1 mutant CQC/AQA (both with 22 tandem repeats) were plated on 12-well size plates (Costar) in DMEM/F-12/5% FBS. The following day, cells in one-half of the wells were washed with 1 mL Dulbecco's PBS (DPBS) without calcium and magnesium (Cellgro, Mediatech, Inc., Manassas, VA), starved for 15 min in 0.5 mL medium lacking Met/Cys (Sigma) in the CO2 incubator, and metabolically labeled for 1 h in 0.5 mL of the same medium containing [35S]Met/Cys (20 μCi/mL Easy Tag Express-[35S]Protein Labeling Mixture, Perkin Elmer Life Sciences, Baesweiler, Germany).
The other one-half of the wells were washed twice with 1 mL DPBS and metabolically labeled for 1 h with [3H]GlcNH2 in 0.5 mL of “low glucose medium” prepared with DMEM-no glucose (Invitrogen), 1 mM sodium pyruvate (Sigma), 0.1 mg/mL D-(+)glucose (Sigma), 5% FBS (GIBCO) and [3H]GlcNH2 (50 μCi/mL D-[6-3H(N)-glucosamine HCl, Perkin Elmer Life Sciences).
After 1 h of metabolic labeling, radioactive medium was removed, cells were washed with 1 mL DPBS, incubated for 1 h in DMEM/F-12/5% FBS in the CO2 incubator, and then moved to ice for biotinylation of cell surface proteins with sulfo-NHS-SS-biotin as described previously (Kinlough et al. 2006; Hanisch et al. 2012). Cells were then returned to culture in DMEM/F-12/5% FBS for varying times from 3 to 24 h. At each time point, MUC1 immunoprecipitates were prepared from cell extracts of duplicate or triplicate wells, and biotinylated MUC1 was recovered with avidin-conjugated beads for analysis with a BioRad phosphoimager after SDS–PAGE. [35S]MUC1 and [3H]MUC1 levels were analyzed with Quantity One software and the half-life was calculated from the slope of the linear regression of a semi-log plot of the data using Sigma Plot.
Funding
This study was supported by a Deutsche Forschungsgemeinschaft Grant HA 2092/17-1 (to F.-G.H.), NIH DK054787 (to R.P.H.) and DK54407 (to O.A.W.).
Conflict of interest
None declared.
Abbreviations
BSA, bovine serum albumine; DMEM, Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; DPBS, Dulbecco's PBS; EDTA, ethylene diamine tetraacetic acid; FCS, fetal calf serum; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathion S transferase; HEK, Human embryonic kidney; HMFG, human milk fat globule; HPLC, high-performance liquid chromatography; HRP, horse-raddish peroxidase; MALDI-TOF, matrix-assisted laser dissociation ionization-time-of-flight; MDCK cells, madin-darby canine kidney cells; MUC1-M, membrane-tethered MUC1; MUC1-S, secreted MUC1; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PMSF, phenylmethyl sulfonylfluorid; RT, room temperature; SDS, sodium dodecyl sulfate; TBST, tris-buffered saline-tween; TFA, trifluoro acetic acid; TGN, trans-Golgi network.
References
- Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol Biol Cell. 2000;11:819–831. doi: 10.1091/mbc.11.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. doi:10.1016/0008-6215(84)85242-8. [Google Scholar]
- Cresawn KO, Potter BA, Oztan A, Guerriero CJ, Ihrke G, Goldenring JR, Apodaca G, Weisz OA. Differential involvement of endocytic compartments in the biosynthetic traffic of apical proteins. EMBO J. 2007;26:3737–3748. doi: 10.1038/sj.emboj.7601813. doi:10.1038/sj.emboj.7601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacour D, Cramm-Behrens CI, Drobecq H, Le Bivic A, Naim HY, Jacob R. Requirement for galectin-3 in apical protein sorting. Curr Biol. 2006;16:408–414. doi: 10.1016/j.cub.2005.12.046. doi:10.1016/j.cub.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Delacour D, Gouyer V, Zanetta JP, Drobecq H, Leteurte E, Grard G, Moreau-Hannedouche O, Maes E, Pons A, Andre S, et al. Galectin-4 and sulfatides in apical membrane trafficking in enterocyte-like cells. J Cell Biol. 2005;169:491–501. doi: 10.1083/jcb.200407073. doi:10.1083/jcb.200407073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacour D, Goyer V, Leteurte E, Ait-Slimane T, Drobecq H, Lenoir C, Moreau-Hannedouche O, Trugnan G, Huet G. 1-benzyl-2-acetamido-2-deoxy-alpha-D-galactopyranoside blocks the apical biosynthetic pathway in polarized HT-29 cells. J Biol Chem. 2003;278:37799–37809. doi: 10.1074/jbc.M305755200. doi:10.1074/jbc.M305755200. [DOI] [PubMed] [Google Scholar]
- Delacour D, Greb C, Koch A, Salomonsson E, Leffler H, Le Bivic A, Jacob R. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic. 2007;8:379–388. doi: 10.1111/j.1600-0854.2007.00539.x. doi:10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- Delacour D, Jacob R. Apical protein transport. Cell Mol Life Sci. 2006;63:2491–2505. doi: 10.1007/s00018-006-6210-8. doi:10.1007/s00018-006-6210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann K, Kinlough CL, Müller S, Razawi H, Baldus SE, Hughey RP, Hanisch F-G. Transmembrane and secreted MUC1 probes show trafficking-dependent changes in O-glycan core profiles. Glycobiology. 2005;15:1111–1124. doi: 10.1093/glycob/cwi099. doi:10.1093/glycob/cwi099. [DOI] [PubMed] [Google Scholar]
- Hanisch FG, Kinlough CL, Staubach S, Hughey RP. MUC1 membrane trafficking: Protocols for assessing biosynthetic delivery, endocytosis, recycling, and release through exosomes. Methods Mol Biol. 2012;842:123–140. doi: 10.1007/978-1-61779-513-8_7. doi:10.1007/978-1-61779-513-8_7. [DOI] [PubMed] [Google Scholar]
- Kinlough CL, McMahan RJ, Poland PA, Bruns JB, Harkleroad KL, Stremple RJ, Kashlan OB, Weixel KM, Weisz OA, Hughey RP. Recycling of MUC1 is dependent on its palmitoylation. J Biol Chem. 2006;281:12112–12122. doi: 10.1074/jbc.M512996200. doi:10.1074/jbc.M512996200. [DOI] [PubMed] [Google Scholar]
- Kinlough CL, Poland PA, Bruns JB, Harkleroad KL, Hughey RP. MUC1 membrane trafficking is modulated by multiple interactions. J Biol Chem. 2004;279:53071–53077. doi: 10.1074/jbc.M409360200. doi:10.1074/jbc.M409360200. [DOI] [PubMed] [Google Scholar]
- Kinlough CL, Poland PA, Gendler SJ, Mattila PE, Mo D, Weisz OA, Hughey RP. Core-glycosylated mucin-like repeats from MUC1 are an apical targeting signal. J Biol Chem. 2011;286:3972–3981. doi: 10.1074/jbc.M111.289504. doi:10.1074/jbc.M111.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj EP, Han F, Kim KC. Mutagenesis of a Gly-Ser cleavage site in MUC1 inhibits ectodomain shedding. Biochem Biophys Res Commun. 2003;307:743–749. doi: 10.1016/s0006-291x(03)01260-9. doi:10.1016/S0006-291X(03)01260-9. [DOI] [PubMed] [Google Scholar]
- Litvinov SV, Hilkens J. The epithelial sialomucin, episialin, is sialylated during recycling. J Biol Chem. 1993;268:21364–21371. [PubMed] [Google Scholar]
- Mattila PE, Kinlough CL, Bruns JR, Weisz OA, Hughey RP. MUC1 traverses apical recycling endosomes along the biosynthetic pathway in polarized MDCK cells. Biol Chem. 2009;390:551–556. doi: 10.1515/BC.2009.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo D, Costa SA, Ihrke G, Youker RT, Pastor-Soler N, Hughey RP, Weisz OA. Sialylation of N-linked glycans mediates apical delivery of endolyn in MDCK cells via a galectin-9-dependent mechanism. Mol Biol Cell. 2012;23:3636–3646. doi: 10.1091/mbc.E12-04-0329. doi:10.1091/mbc.E12-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Hanisch F-G. Recombinant MUC1 probe authentically reflects cell-specific O-glycosylation profiles of endogenous breast cancer mucin: High density and prevalent core 2-based glycosylation. J Biol Chem. 2002;277:26103–26112. doi: 10.1074/jbc.M202921200. doi:10.1074/jbc.M202921200. [DOI] [PubMed] [Google Scholar]
- Naim HY, Joberty G, Alfalah M, Jacob R. Temporal association of the N- and O-linked glycosylation events and their implication in the polarized sorting of intestinal brush border sucrase-isomaltase, aminopeptidase N, and dipeptidyl peptidase IV. J Biol Chem. 1999;274:17961–17967. doi: 10.1074/jbc.274.25.17961. doi:10.1074/jbc.274.25.17961. [DOI] [PubMed] [Google Scholar]
- Parry S, Hanisch F-G, Leir S-H, Sutton-Smith M, Morris HR, Dell A, Harris A. N-Glycosylation of the MUC1 mucin in epithelial cells and secretions. Glycobiology. 2006;16:623–634. doi: 10.1093/glycob/cwj110. doi:10.1093/glycob/cwj110. [DOI] [PubMed] [Google Scholar]
- Poland PA, Rondanino C, Kinlough CL, Heimburg-Molinaro J, Arthur CM, Stowell SR, Smith DF, Hughey RP. Identification and characterization of endogenous galectins expressed in Madin Darby canine kidney cells. J Biol Chem. 2011;286:6780–6790. doi: 10.1074/jbc.M110.179002. doi:10.1074/jbc.M110.179002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter BA, Hughey RP, Weisz OA. Role of N- and O-glycans in polarized biosynthetic sorting. Am J Physiol Cell Physiol. 2006a;290:C1–C10. doi: 10.1152/ajpcell.00333.2005. doi:10.1152/ajpcell.00333.2005. [DOI] [PubMed] [Google Scholar]
- Potter BA, Weixel KM, Bruns JR, Ihrke G, Weisz OA. N-Glycans mediate apical recycling of the sialomucin endolyn in polarized MDCK cells. Traffic. 2006b;7:146–154. doi: 10.1111/j.1600-0854.2005.00371.x. doi:10.1111/j.1600-0854.2005.00371.x. [DOI] [PubMed] [Google Scholar]
- Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. doi:10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. doi:10.1002/pmic.200890093. [DOI] [PubMed] [Google Scholar]
- Staubach S, Razawi H, Hanisch F-G. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics. 2009;9:1–16. doi: 10.1002/pmic.200800793. doi:10.1002/pmic.200890093. [DOI] [PubMed] [Google Scholar]
- Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382:363–373. doi: 10.1042/BJ20040513. doi:10.1042/BJ20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386–3394. doi: 10.1074/jbc.M208326200. doi:10.1074/jbc.M208326200. [DOI] [PubMed] [Google Scholar]
- Vagin O, Kraut JA, Sachs G. Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am J Physiol Renal Physiol. 2009;296:F459–F469. doi: 10.1152/ajprenal.90340.2008. doi:10.1152/ajprenal.90340.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz B, Orberger G, Porwoll S, Hauri HP, Tauber R. Selective reentry of recycling cell surface glycoproteins to the biosynthetic pathway in human hepatocarcinoma HepG2 cells. J Cell Biol. 1995;130:537–551. doi: 10.1083/jcb.130.3.537. doi:10.1083/jcb.130.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]