Abstract
The human respiratory tract pathogen Moraxella catarrhalis expresses lipooligosaccharides (LOS), glycolipid surface moieties that are associated with enhanced colonization and virulence. Recent studies have delineated the major steps required for the biosynthesis and assembly of the M. catarrhalis LOS molecule. We previously demonstrated that the glucosyltransferase enzyme Lgt3 is responsible for the addition of at least one glucose (Glc) molecule, at the β-(1-4) position, to the inner core of the LOS molecule. Our data further suggested a potential multifunctional role for Lgt3 in LOS biosynthesis. The studies reported here demonstrate that the Lgt3 enzyme possesses two glycosyltransferase domains (A1 and A2) similar to that of other bifunctional glycosyltransferase enzymes involved in surface polysaccharide biosynthesis in Escherichia coli, Pasteurella multocida and Streptococcus pyogenes. Each Lgt3 domain contains a conserved DXD motif, shown to be involved in the catalytic activity of other glycosyltransferases. To determine the function of each domain, A1 (N-terminal), A2 (C-terminal) and double A1A2 site-directed DAD to AAA mutants were constructed and the resulting LOS phenotypes of these modified strains were analyzed. Our studies indicate that the Lgt3 N-terminal A1 catalytic domain is responsible for the addition of the first β-(1-3) Glc to the first Glc on the inner core. The C-terminal catalytic domain A2 then adds the β-(1-4) Glc and the β-(1-6) Glc, confirming the bifunctional nature of this domain. The results from these experiments demonstrate that Lgt3 is a novel, multifunctional transferase responsible for the addition of three Glcs with differing linkages onto the inner core of M. catarrhalis LOS.
Keywords: glucosyltransferase, lipooligosaccharide, Moraxella catarrhalis, trifunctional
Introduction
Bacterial lipooligosaccharides (LOS) play an important role in virulence and pathogenesis for many Gram-negative mucosal organisms (Jacques 1996; Gronow and Brade 2001; Erridge et al. 2002). Although LOS is often heterologous due to phase and antigenic variation, only three major, nonvariant LOS serotypes (A, B and C) have been described for Moraxella catarrhalis (Edebrink et al. 1994, 1995, 1996; Edwards et al. 2005). Moraxella catarrhalis is an important cause of middle ear infections in children and lower respiratory infections in adults with compromised lung function, thus understanding the role of LOS in disease could provide valuable information for novel drug and vaccine development (Verduin et al. 2002).
A number of enzymes involved in the biosynthesis of the LOS molecule have been described for M. catarrhalis, although the steps involved in assembly are not completely understood (Zaleski et al. 2000; Verduin et al. 2002; Luke et al. 2003; Edwards et al. 2005; Peng, Choudhury et al. 2005; Peng, Hong et al. 2005; Wilson et al. 2006; Peak et al. 2007; Schwingel et al. 2008). Recent reports have described six genes that encode glycosyltransferase enzymes that are critical to the addition of glucose (Glc) (Lgt1, Lgt3 and Lgt6), galactose (Lgt2 and Lgt5) or N-acetylglucosamine (Lgt4) to the growing LOS chain (Edwards, Allen et al. 2005; Edwards, Schwingel et al. 2005; Wilson et al. 2006; Peak et al. 2007; Schwingel et al. 2008). While those studies confirmed the function of Lgt1, Lgt2, Lgt4, Lgt5 and Lgt6, the specific function of Lgt3 in M. catarrhalis LOS biosynthesis was not completely defined. It has been shown that mutations in lgt3 yielded the same LOS structure when constructed in all M. catarrhalis LOS serotypes and that the acceptor substrate for Lgt3 is α-d-Glc-(1-5)-KDO2-lipid A (Edwards et al. 2005; Peng et al. 2007; Schwingel et al. 2009; Faglin et al. 2010). Lgt3 was experimentally shown to function as a β-(1-4) glucosyltransferase; however, the structural data for the LOS synthesized by the Lgt3 mutant suggested three possible functions for this enzyme (Edwards et al. 2005). Moreover, although a report detailing the biochemical characterization of Lgt3 determined the pH and optimal temperature for enzymatic function, the linkage specificity and Glc additions could not be ascertained (Faglin et al. 2010). Therefore, additional studies were conducted to define the specific functions of Lgt3 as a multifunctional β-glucosyltransferase involved in M. catarrhalis LOS biosynthesis.
Glycosyltransferases are a large group of enzymes that catalyze the transfer of a saccharide moiety from an activated sugar donor onto a variety of acceptor molecules specific to the enzyme (Coutinho et al. 2003). Most glycosyltransferase enzymes involved in LOS or lipopolysaccharide (LPS) core biosynthesis are responsible for one type of sugar addition onto the growing chain (Raetz and Whitfield 2002). Some bacterial bifunctional glycosyltransferase enzymes have been described for capsule polysaccharide biosynthesis and belong to the recently classified GT-2 family of modular glycosyltransferases characterized as tandems of two active sites (domains) on one polypeptide (Coutinho et al. 2003). Included in this group are the KfiC of Escherichia coli, a bifunctional processive glycosyltransferase responsible for K5 capsule biosynthesis, as well as the pmHAS of Pasteurella multocida, a nonprocessive bifunctional glycosyltransferase responsible for hyaluronan capsule biosynthesis (Griffiths et al. 1998; Jing and DeAngelis 2000, 2003). Recently, the Mycobacterium tuberculosis GlfT2, a processive glycosyltransferase involved in galactan synthesis, was shown to be a bifunctional enzyme responsible for the catalysis of both β-(1-5) and β-(1-6) linkages with a single active site (May et al. 2012). The active site for many glycosyltransferase enzymes contains highly conserved DXD motif(s) thought to be involved in binding the ribose ring of the UDP-sugar donor and a divalent metal ion coordinated to the phosphate (P) groups (Tarbouriech et al. 2001; Coutinho et al. 2003; Jing and DeAngelis 2003).
The Lgt3 glucosyltransferase enzyme of M. catarrhalis is similar in amino acid sequence to members of the GT-2 family of bifunctional glycosyltransferase enzymes and contains two putative DXD domains, termed A1 (N-terminal) and A2 (C-terminal). Site-directed mutagenesis of the conserved DXD motifs in both putative catalytic domains of Lgt3 suggest that each has a critical role in catalysis for this enzyme, enabling full-length LOS biosynthesis for this organism. Our studies demonstrate that the M. catarrhalis Lgt3 is a unique trifunctional glucosyltransferase responsible for the transfer of three Glc moieties onto the core Glc-Kdo acceptor molecule, with the A1 catalytic domain mediating a β-(1-3) linkage and a bifunctional A2 catalytic domain responsible for catalysis of both β-(1-4) and β-(1-6) linkages.
Results
Sequence analysis of Lgt3 suggests two putative catalytic domains
An NCBI BLAST search, using the predicted amino acid sequence of Lgt3 of M. catarrhalis 7169 (GenBank accession number AY789049), revealed two conserved domains, each with homology to members of the GT-2 family of glycosyltransferase enzymes. Each separate domain exhibited homology to full-length glycosyltransferase enzymes involved in LOS biosynthesis for other Gram-negative bacteria, including LgtF of Haemophilus ducreyi, a β-(1-4) glucosyltransferase (42% identity/56% similarity), and WaaV of E. coli, a β-(1-3) glucosyltransferase (14% identity/29% similarity). Figure 1 depicts a ClustalW alignment of the N-terminal domain (amino acids 13–250) with the C-terminal domain (amino acids 287–526) of Lgt3, which revealed 41% identity and 58% similarity to each other. Additional sequence analyses identified two conserved DXD motifs, one in each domain (asterisks). The two domains of Lgt3 were designated A1 (N-terminal) and A2 (C-terminal) due to their homology with other bifunctional glycosyltransferase enzymes that possess multiple “domain A” motifs, including the pmHAS of P. multocida (Saxena et al. 1995; Jing and DeAngelis 2000, 2003).
Fig. 1.
(A) The relative positions of the two putative catalytic DAD motifs in the Lgt3 polypeptide and the two GT-2 domains as predicted by sequence homology to known β-glycosyltransferases and bifunctional glycosyltransferase enzymes. (B) Alignment of the amino acid sequences of the two domains; the aspartic acid residues of the DAD motifs mutated to alanines are marked with asterisks. Shading denotes identical (dark) and similar (light) amino acid residues.
Table I depicts the conserved DXD motifs, predicted to be important for substrate binding, found in a number of different glycosyltransferase enzymes from multiple strains of bacteria compared with those motifs found in the Lgt3 enzyme from M. catarrhalis (Saxena et al. 1995; Griffiths et al. 1998; Jing and DeAngelis 2000; Coutinho et al. 2003; Jing and DeAngelis 2003). The LgtF of H. ducreyi and the WaaV of E. coli transfer a single Glc molecule to their respective LOS and LPS molecules, whereas the KfiC, KfoC, pmHAS, HasA and GlfT2 all transfer multiple sugars to surface polysaccharides of their respective strains (Saxena et al. 1995; Griffiths et al. 1998; Filiatrault et al. 2000; Jing and DeAngelis 2000, 2003; Ninomiya et al. 2002; Coutinho et al. 2003; May et al. 2012). The aspartic acid residues in this conserved motif are important for donor substrate binding for domain A of the KfiC of E. coli, both domain As of pmHAS of P. multocida and the single bifunctional catalytic domain of the M. tuberculosis GlfT2 (Griffiths et al. 1998; Jing and DeAngelis 2000, 2003; May et al. 2012). Each domain of the Lgt3 enzyme for M. catarrhalis possesses homology with these enzymes in the conserved regions. Thus, these two DAD motifs of Lgt3 were chosen for site-directed mutagenesis to investigate their contribution to the function of Lgt3 in M. catarrhalis LOS biosynthesis.
Table I.
Primary amino acid sequences of the conserved domain A DXD motif in GT-2 family β-glycosyltransferases
| Organism | Enzyme | Function | Sequence (DXD) motifa |
|---|---|---|---|
| M. catarrhalis | Lgt3—Domain A1 | β-glucosyltransferase; LOS biosynthesis Edwards et al. (2005) | 77..YATGDWILALDADEVVTP..94 |
| M. catarrhalis | Lgt3—Domain A2 | β-glucosyltransferase; LOS biosynthesis Edwards et al. (2005) | 351..YATGDYVLVLDADERLDQ..368 |
| H. ducreyi | LgtF | β (1-4) glucosyltransferase; LOS biosynthesis Filiatrault et al. (2000) | 72..YVTSDYVLWLDADERVTP..89 |
| E. coli | WaaV | β (1-3) glucosyltransferase; rLPS biosynthesis Heinrichs et al. (1998) | 83..KAQGQFICFMDDDDEIDP..100 |
| E. coli | KfiC—Domain A | β (1-4) GlcA transferase; K5 capsule biosynthesis Griffiths et al. (1998) | 342..KAHGNFITFQDADDLSHP..359b |
| E. coli | KfoC—Domain A1 | β (1-3) GlcA transferase; K4 chondroitin capsule biosynthesis Ninomiya et al. (2002) | 229..AAKYNYVAILDCDMAPNP..246 |
| E. coli | KfoC—Domain A2 | β (1-4) GalNAc transferase; K4 chondroitin capsule biosynthesis Ninomiya et al. (2002) | 509..LCRGFYIGQLDSDDFLEPD..527 |
| P. multocida | pmHAS—Domain A1 | β (1-3) GlcNAc transferase; hyaluronan capsule synthesis Jing and DeAngelis (2000), Jing and DeAngelis (2003) | 237..LAKYDFIFLLDCDMAPNP..254b |
| P. multocida | pmHAS—Domain A2 | β (1-4) GlcUA transferase; hyaluronan capsule synthesis Jing and DeAngelis (2000), Jing and DeAngelis (2003) | 517..FAKGYYIGQLDSDDYLEP..534b |
| M. tuberculosis | GlfT2 | Bifunctional β(1-5)/β(1-6) UDP-galactofuranosyl transferase; galactan synthesis May et al. (2012) | 246.. NTDCQQILFMDDDIRLEP..263b |
| Streptococcus pyogenes | HasA | Bifunctional β(1-3)/β(1-4) GlcUA/GlcNAc transferase; hyluronic acid capsule synthesis DeAngelis et al. (1993), Dougherty and van de Rijn (1994) | 148..RSDADVFLTVDSDTYIYP..165 |
aConserved aspartic acid residues are underlined; the putative DXD catalytic sites of Lgt3 selected for mutagenesis studies are in boldface type.
bThese conserved aspartic acid residues have demonstrated catalytic activity for their respective enzymes.
Alteration of the DAD motifs affects Lgt3 function
Three different site-directed mutant strains were constructed that encoded Lgt3 enzymes with the following amino acid changes: Lgt3A1 represents an N-terminal DAD mutant with aspartic acid residues 87 and 89 mutated to alanine, Lgt3A2 represents a C-terminal DAD mutant with aspartic acid residues 361 and 363 mutated to alanine and the double DAD mutant Lgt3A1A2 had all the four aspartic acid residues from both domains (amino acids 87/89/361/363) mutated to alanine (Figure 1, Table II).
Table II.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | Host strain used for cloning | Stratagene |
| M. catarrhalis | ||
| 7169 | Wild-type strain, pediatric middle ear isolate | Luke et al. (1999) |
| 7169::lgt3KH | lgt3 isogenic mutant | Edwards et al. (2005) |
| 7169::lgt1KB | lgt1 isogenic mutant | Edwards et al. (2005) |
| Lgt3A1 | 7169 lgt3 N-terminal DAD site-directed mutant [D87A/D89A] | This study |
| Lgt3A2 | 7169 lgt3 C-terminal DAD site-directed mutant [D361A/D363A] | This study |
| Lgt3A1A2 | 7169 lgt3 double DAD site-directed mutant [D87A/D89A/D361A/D363A] | This study |
| Lgt3A1::lgt1K | Lgt1-null Lgt3A1 mutant | This study |
| Lgt3A2::lgt1K | Lgt1-null Lgt3A2 mutant | This study |
| Lgt3A1A2::lgt1K | Lgt1-null Lgt3A1A2 mutant | This study |
| Plasmids | ||
| pGEM-T easy | Commercially available cloning vector | Promega |
| pWW115 | M. catarrhalis in trans complementation plasmid | Wang and Hansen (2006) |
| plgt3A1 | Site-directed lgt3 A1 mutant (A2 catalytic motif functional) subcloned into pWW115 | This study |
| plgt3A2 | Site-directed lgt3 A2 mutant (A1 catalytic motif functional) subcloned into pWW115 | This study |
| plgt3 | lgt3 subcloned into pWW115 | This study |
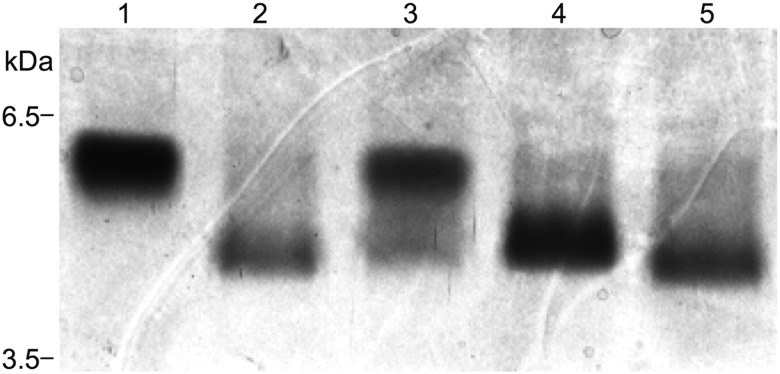
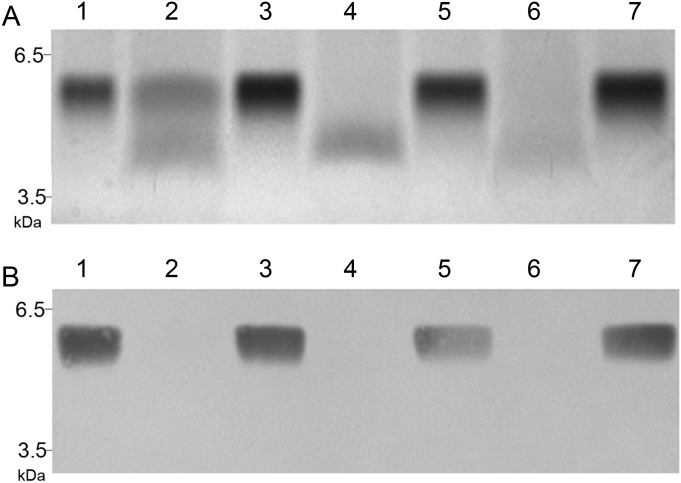
The Lgt3A1 site-directed construct produced a LOS with two major glycoforms by silver-stained gel, one that migrated slightly faster than the wild-type LOS molecule and one that migrated similar to that of the previously described lgt3-null mutant (Figure 2, lanes 3, 1 and 2, respectively) (Edwards et al. 2005). The LOS molecule synthesized by Lgt3A2, the C-terminal DAD mutant (lane 4), migrated slightly slower than the lgt3 mutant glycoform (lane 2) but faster than wild-type (lane 1). The double site-directed DAD mutant Lgt3A1A2 synthesized an LOS molecule that was consistent with the LOS of the lgt3 mutant (lane 5 compared with lane 2). To evaluate whether the LOS phenotypes exhibited by the mutant strains were solely the result of Lgt3 and not the result of compensatory transferase activity, in trans complementation was performed on all strains. Complementation of Lgt3A1 with lgt3A2 subcloned into the M. catarrhalis shuttle vector pWW115 restored the LOS phenotype of the site-directed strain to wild-type (Figure 3A, lanes 1–3). Likewise, complementation of both Lgt3A2 with lgt3A1 in trans and Lgt3A1A2 with the wild-type lgt3 in trans restored the LOS phenotype to the wild-type glycoform (lanes 4–7). The reversion to the wild-type phenotype was confirmed by immunoblot using monoclonal antibodies (MAbs) 3F7 (Figure 3B) and 4G5 (data not shown), which react to full-length M. catarrhalis serotype B LOS (Edwards et al. 2005). These data indicated that the catalytic domains A1 and A2 were responsible for the function of Lgt3 and that each domain can function independently.
Fig. 2.
A silver-stained SDS–PAGE gel depicting the LOS profiles of wild-type 7169 (lane 1), 7169::lgt3KH (lane 2) and the site-directed mutant constructs Lgt3A1 (lane 3), Lgt3A2 (lane 4) and Lgt3A1A2 (lane 5).
Fig. 3.
Composite of a silver-stained SDS–PAGE gel (A) and the corresponding western blot probed with MAb 3F7 (B) evaluating LOS profiles of wild-type strain 7169 (lane 1), Lgt3A1 (lane 2), Lgt3A1 + plgt3A2 (lane 3), Lgt3A2 (lane 4), Lgt3A2 + plgt3A1 (lane 5), Lgt3A1A2 (lane 6), Lgt3A1A2 + plgt3 (lane 7) demonstrating restoration of wild-type phenotype following in trans complementation of the site-directed mutants with the corresponding functional catalytic domain(s).
To further confirm that Lgt3 was responsible for the β-(1-3), β-(1-4) and β-(1-6) Glc additions to the lipid A-KDO2-(1-5)-d-Glc core, Lgt1 was mutated in each of the site-directed Lgt3 mutant constructs. Lgt1, although an α-(1-2) glucosyltransferase, is the only other glucosyltransferase involved in M. catarrhalis 7169 LOS assembly and therefore the only likely transferase with the potential to exhibit compensatory activity (Edwards et al. 2005). Phenotypic analyses of the resulting Lgt1-site-directed DAD mutants indicated that there was no compensatory activity by this glucosyltransferase (data not shown), further confirming that Lgt3 alone exhibited trifunctional activity. Mass spectrometry (MS) structural analyses of the LOS assembled by all mutant constructs were performed to delineate each glycolipid and confirm the function of Lgt3.
Structural analyses
Capillary electrophoresis electrospray MS (CE-ES-MS) analysis of LPS-OH from the mutant strains (Table III) revealed a range of glycoforms from 1Hex to 8Hex for the Lgt3A1 mutant, whereas the Lgt3A2 mutant elaborated just a single 2Hex glycoform. To simplify the interpretation of the MS data, the corresponding lgt1 double mutants were examined so that further elongation of inner core glycoforms due to the action of the Lgt1 glycosyltransferase was precluded. While the Lgt3A2::lgt1K double mutant still elaborated a 2Hex glycoform, the Lgt3A1::lgt1K double mutant now elaborated a 1Hex-to-3Hex range of glycoforms. Finally, the MS spectrum of the Lgt3A1A2 double mutant revealed a single 1Hex glycoform consistent with no extension beyond the initial Glc residue attached to the Kdo molecule. These results also suggested that the A1 region of the Lgt3 glycosyltransferase, functional in the LgtA2 mutant background, was able to add a single hexose (Hex) to the Glc-Kdo stub, whereas the A2 region of the Lgt3 glycosyltransferase, functional in the A1 mutant background, was able to add two Hex residues to the Glc-Kdo acceptor. These data were corroborated by MS analysis of the fully deacylated (KOH treated) LPS from the Lgt3A1::lgt1K double mutant, which also revealed a range of 1Hex-to-3Hex glycoforms. These glycoforms were fractionated by anion-exchange chromatography resulting in the isolation of predominantly a 1Hex-containing oligosaccharide and a 3Hex-containing oligosaccharide.
Table III.
Negative ion CE-ESI-MS data and proposed compositions of O-deacylated LPS and KOH-treated LPS from M. catarrhalis strains as indicated
| Preparation/strain | Observed ions (m/z) |
Molecular mass (Da)a |
Relative intensity | Proposed composition | ||
|---|---|---|---|---|---|---|
| (M–H)− | (M–2H)2− | Observed | Calculated | |||
| De-O-acylated LPS | ||||||
| LgtA1 | — | 749.5 | 1501.0 | 1499.5 | 1.0 | Hex, 2Kdo, Lipid A-OH |
| — | 830.5 | 1663.0 | 1661.7 | 0.3 | 2Hex, 2Kdo, Lipid A-OH | |
| — | 1073.5 | 2149.0 | 2148.1 | 0.4 | 5Hex, 2Kdo, Lipid A-OH | |
| — | 1155.0 | 2312.0 | 2310.3 | 0.2 | 6Hex, 2Kdo, Lipid A-OH | |
| — | 1235.5 | 2473.0 | 2472.4 | 0.6 | 7Hex, 2Kdo, Lipid A-OH | |
| — | 1317.0 | 2636.0 | 2634.6 | 0.5 | 8Hex, 2Kdo, Lipid A-OH | |
| LgtA2 | — | 830.5 | 1663.0 | 1661.7 | 1.0 | 2Hex, 2Kdo, Lipid A-OH |
| LgtA1::lgt1K | — | 749.5 | 1501.0 | 1499.5 | 0.8 | Hex, 2Kdo, Lipid A-OH |
| — | 830.5 | 1663.0 | 1661.7 | 0.6 | 2Hex, 2Kdo, Lipid A-OH | |
| — | 911.0 | 1824.0 | 1823.9 | 1.0 | 3Hex, 2Kdo, Lipid A-OH | |
| LgtA2::lgt1K | — | 830.5 | 1663.0 | 1661.7 | 1.0 | 2Hex, 2Kdo, Lipid A-OH |
| LgtA1A2 | — | 749.5 | 1501.0 | 1499.5 | 1.0 | Hex, 2Kdo, Lipid A-OH |
| KOH-treated LPS | ||||||
| LgtA1::lgt1K | 1101.6 | 550.5 | 1102.8 | 1102.8 | 1.0 | Hex, 2Kdo, 2HexN, 2P, H2O |
| 1263.6 | 631.8 | 1265.1 | 1264.9 | 0.5 | 2Hex, 2Kdo, 2HexN, 2P, H2O | |
| 1425.6 | 712.8 | 1427.1 | 1427.1 | 0.6 | 3Hex, 2Kdo, 2HexN, 2P, H2O | |
| LgtA1::lgt1K (Fr. 1) | 1426.2 | 712.5 | 1427.1 | 1427.1 | 1.0 | 3Hex, 2Kdo, 2HexN, 2P, H2O |
| LgtA1::lgt1K (Fr. 2) | 1101.6 | 550.2 | 1102.5 | 1102.8 | 1.0 | Hex, 2Kdo, 2HexN, 2P, H2O |
| — | 631.5 | 1265.0 | 1264.9 | 0.1 | 2Hex, 2Kdo, 2HexN, 2P, H2O | |
aAverage mass units were used for calculation of molecular mass based on proposed composition as follows: Hex, 162.15; Kdo, 220.18; HexN, 161.19; P, 79.95; O-deacylated lipid A (Lipid A-OH), 897.00.
Methylation analyses were performed on the core oligosaccharides (OS) from the two single mutants and the two corresponding double mutants in the lgt1K background to determine the linkage pattern of the OS. Analysis of the core OS from the Lgt3A1 single mutant revealed the presence of terminal Glc, terminal Gal, 2-substituted Glc, 4-substituted Glc, 4-substituted Gal and 4,6-disubstituted Glc, consistent with the MS analyses. The absence of a 3-linked Glc inferred that the A1 region of the Lgt3 glycosyltransferase was responsible for this modification of the inner core OS. Analysis of the core OS from the Lgt3A2 single mutant and the Lgt3A2::lgt1K double mutant revealed the presence of terminal Glc and 3-substituted Glc, consistent with the MS data corroborating that the A1 region of the Lgt3 glycosyltransferase adds a Glc residue to the 3-position of the Glc-Kdo acceptor and inferring that the A2 region of the Lgt3 glycosyltransferase is involved in the addition of Glc residues to the 4- and 6-positions. Methylation analysis of the core OS from the Lgt3A1::lgt1K double mutant revealed the presence of terminal Glc and 4,6-disubstituted Glc, consistent with the MS analyses and corroborating that the A2 region of the Lgt3 glycosyltransferase adds Glc residues to the 4- and 6-positions of the Glc-Kdo acceptor.
To elucidate the exact locations and linkage patterns of the oligosaccharide, NMR studies were performed on the fractionated OS from the Lgt3A1::lgt1K double-mutant strain following complete deacylation. MS analyses had revealed that fraction 1 (Fr. 1) elaborated a 3Hex glycoform and fraction 2 (Fr. 2) a predominantly 1Hex glycoform. As detailed in Table IV and consistent with the methylation analyses, the NMR analysis confirmed that the 3-linked Glc was absent in this mutant background by virtue of 13C, 1H chemical shifts for the 3-position of residue E at 72.3, 3.94 compared with 75.2, 4.32 for the 3-position of this same residue in the fully substituted oligosaccharide. Furthermore, evidence of substitution at the 4-position was evident from 13C, 1H chemical shifts for the 4-position of residue E at 79.0, 3.88 compared with 74.0, 4.04 for the 4-position of this same residue in the fully substituted oligosaccharide, and evidence of substitution at the 6-position was evident from 13C, 1H chemical shifts for the 6-position of residue E at 68.2, 4.08/4.23 compared with 67.2, 4.16/4.22 for the 6-position of this same residue in the fully substituted oligosaccharide (Cox et al. 2011).
Table IV.
NMR data for Lgt3A1::lgt1k KOH treatment-derived oligosaccharides 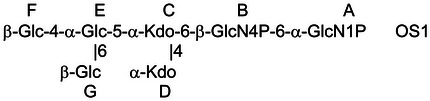 ,
, 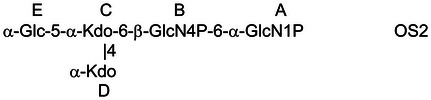
| H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 | |
|---|---|---|---|---|---|---|
| E, OS1 | 5.28 | 3.58 | 3.94 | 3.88 | 4.32 | 4.08; 4.23 |
| 100.4 | 73.1 | 72.3 | 79.0 | 70.6 | 68.2 | |
| E, OS2 | 5.29 | 3.53 | 3.83 | 3.55 | 4.07 | 3.86; 3.89 |
| 100.7 | 73.4 | 73.8 | 70.3 | 73.0 | 61.2 | |
| F, OS1 | 4.61 | 3.32 | 3.52 | 3.43 | 3.44 | 3.74; 3.91 |
| 103.6 | 74.3 | 77.0 | 70.8 | 77.0 | 61.9 | |
| G, OS1 | 4.51 | 3.34 | 3.51 | 3.43 | 3.48 | 3.74; 3.92 |
| 103.6 | 74.2 | 77.0 | 70.8 | 77.1 | 61.9 |
Taken together, these data indicate that Lgt3 is a novel, trifunctional glucosyltransferase. In the Lgt3A2 mutant, only the N-terminal DAD motif is functional. The β-(1-3) is added to the Glc-KDO2-lipid A core by the A1 catalytic domain, but the LOS chain cannot be further extended. Conversely, only the A2 catalytic domain is functional in the Lgt3A1 mutant, resulting in the β-(1-4) and β-(1-6) Glc additions. However, both phenotypic and structural analyses suggest that the preferred sequence of addition is the β-(1-3) Glc by the A1 domain, followed by the addition of the β-(1-4) and β-(1-6) Glc by the A2 domain. With the β-(1-4) and β-(1-6) Glcs linked to the core Glc-Kdo acceptor in the Lgt3A1 mutant, although this catalysis does not occur with full-efficiency, Lgt1 is able to extend the chain. Importantly, the double Lgt3A1A2 site-directed construct exhibits the same phenotype and LOS structure as the Lgt3-null mutant. Further, the deletion of Lgt1 in the site-directed mutants and resulting analyses support the trifunctional enzymatic activity of Lgt3 and proves that Lgt1, the only other glucosyltransferase, does not exhibit compensatory activity. The three linkage functions of Lgt3 in the biosynthesis of M. catarrhalis LOS are summarized in Figure 4.
Fig. 4.
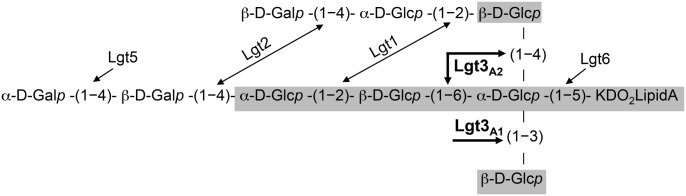
Schematic depicting the biochemical structure of the wild-type M. catarrhalis serotype B LOS molecule. All the three LOS serotypes contain the common core OS indicated in gray. The functions of the glycosyltransferases involved in LOS biosynthesis are indicated. The trifunctional catalysis mediated by Lgt3 was determined in this study.
Discussion
Prior to this study, the specific function of Lgt3 in M. catarrhalis LOS biosynthesis remained undefined. Although Lgt3 demonstrated the ability to function as a β-(1-4) glucosyltransferase using in trans complementation of an H. ducreyi LOS mutant defective in endogenous β-(1-4) glucosyltransferase activity; the structural data for the LOS synthesized by the lgt3 mutant suggested two additional functions for this enzyme (Edwards et al. 2005). This current report confirms the ability of Lgt3 to function as a β-(1-4) glucosyltransferase and further supports its additional functions as a β-(1-3) and β-(1-6) glucosyltransferase critically involved in M. catarrhalis LOS biosynthesis.
Analyses of the predicted amino acid sequence of Lgt3 revealed two conserved domains, each with homology to members of the GT-2 family of glycosyltransferase enzymes, whose members are characterized as enzymes with tandem glycosyltransferase active sites (domains) on one polypeptide (Coutinho et al. 2003). Each Lgt3 domain exhibited homology to full-length glycosyltransferase enzymes involved in LOS biosynthesis and possessed homology to the domain As of other bifunctional glycosyltransferase enzymes. This type of domain is responsible for transferring UDP-sugar substrates to the nonreducing end of sugar-acceptor molecules found in the surface polysaccharides of other bacteria (Saxena et al. 1995; Jing and DeAngelis 2000, 2003). Based on primary amino acid sequence similarities, Lgt3 did not appear to possess a domain B, which is normally associated with processive polymerization that involves alternating α and β linkages in polysaccharide biosynthesis, catalyzed by some multifunctional glycosyltransferase enzymes (Saxena et al. 1995; Griffiths et al. 1998; Jing and DeAngelis 2000).
Additional analyses of the amino acid sequence for each Lgt3 domain revealed two highly conserved DXD motifs, which are associated with binding to the ribose ring of the UDP-sugar donor and a divalent metal ion coordinated to the P groups for other glycosyltransferase enzymes (Tarbouriech et al. 2001; Coutinho et al. 2003; Jing and DeAngelis 2003). To determine whether these DXD motifs were important for the function of each Lgt3 glycosyltransferase domain, site-directed DAD mutants were constructed. The N-terminal DAD mutant synthesized two LOS glycoforms and our analyses suggested a defect in the addition of the first Glc residue onto the Glc-2-keto-3-deoxyoctulosonic acid (KDO2) acceptor molecule for this mutant. Mutagenesis of the C-terminal DAD motif resulted in a truncated LOS glycoform which was slightly larger that the previously constructed lgt3 mutant LOS glycoform (Edwards et al. 2005). Structural analyses of the LOS moieties produced by these constructs indicated that the N-terminal DAD motif adds the β-(1-3) Glc to the Glc-KDO core, whereas the C-terminal DAD motif is critical to the β-(1-4) and β-(1-6) Glc additions to the growing LOS chain. The double DAD mutant synthesized a LOS glycoform identical to the previously constructed lgt3 mutant, supporting the function of the A1 and A2 DAD motifs as the critical catalytic domains essential and sufficient for Lgt3 function. Moreover, these data demonstrate that the C-terminal domain has a broader acceptor-substrate specificity, which has been demonstrated for some LOS/LPS glycosyltransferase enzymes from other organisms and that the single A2 catalytic site exhibits bifunctional enzymatic activity (Rocchetta et al. 1998; Blixt et al. 1999; Piekarowicz and Stein 2002; May et al. 2012).
In conclusion, the data presented in this report describe the characterization of Lgt3, a novel, trifunctional glucosyltransferase critically involved in the assembly of M. catarrhalis LOS. Our data indicate that the A1 catalytic domain of Lgt3 functions as a β-(1-3) glucosyltransferase and that the Lgt3 A2 catalytic domain exhibits bifunctional catalysis by mediating the addition of both the β-(1-4) and β-(1-6) Glc to the Glc-KDO LOS core.
Materials and methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table II. M. catarrhalis and E. coli XL1-blue were cultured as described previously using Mueller-Hinton or Luria-Bertani broth and agar plates, respectively (Luke et al. 2004; Edwards et al. 2005). For NMR structural analyses, cells were grown in a 24-L fermenter as described (Cox et al. 2011). Antibiotics were supplemented as necessary at 15 μg per mL spectinomycin (Spec), 20 μg per mL kanamycin (Kan) and 100 μg per mL ampicillin (Amp).
Nucleic acid manipulations and analyses
Standard molecular biology reagents were obtained from New England Biolabs, Inc. (Beverly, MA) or Invitrogen (Carlsbad, CA). Chromosomal DNA was isolated using standard methods. PCR amplifications were performed for 25 cycles with the GeneAMP PCR System 9700 (P.E. Applied Biosystems, Foster City, CA) using Platinum Taq High-Fidelity DNA Polymerase (Invitrogen) and primer-set-dependent annealing temperatures and extension times. PCR amplicons were purified with the MinElute Reaction Clean-up Kit (Qiagen, Santa Clarita, CA) and ligated into the TA cloning vector pGEM®-T Easy (Promega, Madison, WI) for further modifications or used for natural transformation. Site-directed mutant constructs were complemented in trans with the M. catarrhalis shuttle vector pWW115 containing the wild-type DAD motif that was mutated in the chromosome (Wang and Hansen 2006). Lgt1 mutants were constructed exactly as we described previously (Edwards et al. 2005). DNA nucleotide sequences for all constructs were obtained via automated DNA sequencing (RPCI Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, NY) and analyzed with MacVector 12 and the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, WI).
Site-directed mutagenesis
Mutagenesis was achieved using the QuickChange® II Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA) and mutagenic primers 2476, 2477, 2478 and 2479 (Table V) to introduce defined mutations in lgt3. The previously reported plasmid pLGT-3KE was used as the template for mutagenic reactions, which were performed according to the manufacturer's protocol (Edwards et al. 2005). Following DpnI-digestion and electroporation into XL1-blue competent cells, plasmid DNA was isolated from the resulting transformants using a QIAPrep Spin Mini Kit (Qiagen) and subjected to DNA sequence analysis to confirm the presence of the desired mutations. Purified amplicons, generated by previously designed primers 698 and 699, were used to naturally transform the formerly constructed Kan-resistant Lgt3 mutant M. catarrhalis 7169::lgt3KM (Luke et al. 2004; Edwards et al. 2005). Kan-sensitive transformants were selected by replica plating (Luke et al. 2002; Furano and Campagnari 2004; Schwingel et al. 2009). The presence of the desired site-directed mutation(s) was confirmed by sequence analysis of chromosomal DNA isolated from each transformant using primers 418 and 419, which were designed to flank the site of recombination. The resulting M. catarrhalis strains were designated 7169::lgt3A1 (D87A/D89A; the N-terminal A1 domain DAD to AAA mutant), 7169::lgt3A2 (D361A/D363A, the C-terminal A2 domain DAD to AAA mutant) and 7169::lgt3A1A2 (D87A/D89A/D361A/D363A double mutant containing both A1 (N-terminal) and A2 (C-terminal) DAD domains mutated to AAA).
Table V.
Sequences of oligonucleotide primers
| Primer | Sequence (5′-3′)a | Description |
|---|---|---|
| Pr418F | TTGTTGAGAGTCATTCCCC | Sequence analysis |
| Pr419R | TGATGTTGATACAGCAGGTTC | Sequence analysis |
| Pr2476F | TGGATTTTGGCATTGGCCGCTGCTGAAGTGGTGACA | D87A/D89A |
| Pr2477R | TGTCACCACTTCAGCAGCGGCCAATGCCAAAATCCA | D87A/D89A |
| Pr2478F | GCGATTATGTGCTTGTGCTTGCTGCTGCTGAACG | D361A/D363A |
| Pr2479R | CGTTCAGCAGCAGCAAGCACAAGCACATAATCGC | D361A/D363A |
aNucleotide changes engineered for D to A site-directed mutagenesis are underlined.
Isolation of M. catarrhalis LOS for phenotypic analysis
LOS was prepared from proteinase-K-treated whole cell lysates, resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) on a 16% acrylamide separating gel with a bilayer stacker, and visualized by silver staining or western blot analysis with MAbs 4G5 or 3F7 as previously described (Tsai and Frasch 1982; Campagnari et al. 1991; Inzana et al. 1999; Edwards et al. 2005; Schwingel et al. 2009).
Preparation of M. catarrhalis LOS for structural analyses
For the initial, rapid profiling of LOS glycoforms by ES-MS, plate-grown bacterial cells were treated with proteinase K followed by successive DNase and RNase treatments to release the LOS, which was O-deacylated in situ with anhydrous hydrazine (Li et al. 1998; Schwingel et al. 2009). For linkage analysis, cell pellets from 5 L of stationary-phase bacteria were treated with 1% phenol (final concentration), successively washed with ethanol, acetone and petroleum ether and subjected to the hot phenol–water LOS extraction procedure as described in detail (Westphal and Jann 1965; Masoud et al. 1994; Schwingel et al. 2008, 2009). The core OS were isolated by treating the LPS (∼20–40 mg) with 1% acetic acid (10 mg mL−1, 100°C, 1.5 h) with subsequent removal of the insoluble lipid A by centrifugation (5000 × g). The lyophilized OS samples were desalted on a Sephadex G-25 column. To prepare sufficient material for NMR analyses, strain Lgt3A1::lgt1K was grown in a 24L fermenter and LPS was isolated as described previously except that it was necessary to isolate the LPS from the phenol phase following the Westphal extraction (Cox et al. 2011). To prepare completely deacylated LPS (LPS-KOH), purified LPS (180 mg) isolated from the phenol phase of the fermenter grown cells was treated with 4N KOH at 125°C for 30 h, cooled and neutralized with 4N HCl followed by a low speed spin. The supernatant was desalted on a Sephadex G-25 column and lyophilized. NMR analyses were performed on the purified fully deacylated oligosaccharide, following anion-exchange chromatography as described previously (Russo et al. 2013). Briefly, the fully deacylated LPS was separated on a Carbopac PA100 column using a gradient of 20–100% 1 M NaOAc in 0.1 M NaOH, at a flow rate of 2 mL/min. The resulting fractions were desalted on a Sephadex G-25 column and lyophilized.
Glycoform profiling and linkage analyses
Glycoform profiling of O-deacylated LOS samples and linkage analyses of core OS were performed as described (Masoud et al. 1994; Schwingel et al. 2008, 2009). In brief, lyophylized O-deacylated LOS samples were dissolved in 1 M ammonium acetate solution and analyzed directly on a Crystal Model 310 CE instrument (ATI Unicam, Boston, MA) coupled to a 4000 QTRAP mass spectrometer (Applied Biosystems/MDS Sciex, Canada). Molecular mass calculations of proposed compositions were determined using the following average mass units: Hex, 162.15; KDO, 220.18; phosphoethanolamine, 123.05; P, 79.95; O-deacylated lipid A (Lipid A-OH), 897.00. For linkage delineation, methylated core OS samples were hydrolyzed with 2 M trifluoroacetic acid followed by reduction of the liberated glycoses with NaBD4 and acetylation by acetic anhydride (Ac2O); partially methylated alditol acetates were separated by gas–liquid chromatography and identified by electron impact-MS on a Varian Saturn 2000 as previously detailed (Ciucanu and Kerek 1984; Masoud et al. 1994; Schwingel et al. 2009). NMR experiments were performed as described previously (Cox et al. 2011).
Funding
This work was supported by the National Institutes of Health (grant numbers AI46422, DC005837 to A.A.C.).
Conflict of interest
None declared.
Abbreviations
Amp, ampicillin; CE-ES-MS, capillary electrophoresis electrospray mass spectrometry; Fr. 1, fraction 1; Fr. 1, fraction 2; Gal, galactose; Glc, glucose; Hex, hexose; Kan, kanamycin; KDO, 2-keto-3-deoxyoctulosonic acid; lgt, lipooligosaccharide glycosyltransferase gene; LOS, lipooligosaccharides; LPS, lipopolysaccharides; MS, mass spectrometry; MAbs, monoclonal antibodies; NMR, nuclear magnetic resonance; OS, oligosaccharides; P, phosphate; PCR, polymerase chain reaction; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; Spec, spectinomycin; UDP, uridine diphosphate.
Acknowledgements
We thank Perry Fleming for large-batch bacterial growth for structural analyses and Jacek Stupak for ES-MS.
References
- Blixt O, van Die I, Norberg T, van den Eijnden DH. High-level expression of the Neisseria meningitidis lgtA gene in Escherichia coli and characterization of the encoded N-acetylglucosaminyltransferase as a useful catalyst in the synthesis of GlcNAc beta 1–>3Gal and GalNAc beta 1–>3Gal linkages. Glycobiology. 1999;9:1061–1071. doi: 10.1093/glycob/9.10.1061. doi:10.1093/glycob/9.10.1061. [DOI] [PubMed] [Google Scholar]
- Campagnari AA, Wild LM, Griffiths GE, Karalus RJ, Wirth MA, Spinola SM. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. doi:10.1016/0008-6215(84)85242-8. [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. doi:10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Cox AD, St Michael F, Cairns CM, Lacelle S, Filion AL, Neelamegan D, Wenzel CQ, Horan H, Richards JC. Investigating the potential of conserved inner core oligosaccharide regions of Moraxella catarrhalis lipopolysaccharide as vaccine antigens: Accessibility and functional activity of monoclonal antibodies and glycoconjugate derived sera. Glycoconj J. 2011;28:165–182. doi: 10.1007/s10719-011-9332-7. doi:10.1007/s10719-011-9332-7. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Papaconstantinou J, Weigel PH. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993;268:19181–19184. [PubMed] [Google Scholar]
- Dougherty BA, van de Rijn I. Molecular characterization of hasA from an operon required for hyaluronic acid synthesis in group A streptococci. J Biol Chem. 1994;269:169–175. [PubMed] [Google Scholar]
- Edebrink P, Jansson PE, Rahman MM, Widmalm G, Holme T, Rahman M. Structural studies of the O-antigen oligosaccharides from two strains of Moraxella catarrhalis serotype C. Carbohydr Res. 1995;266:237–261. doi: 10.1016/0008-6215(94)00276-l. doi:10.1016/0008-6215(94)00276-L. [DOI] [PubMed] [Google Scholar]
- Edebrink P, Jansson PE, Rahman MM, Widmalm G, Holme T, Rahman M, Weintraub A. Structural studies of the O-polysaccharide from the lipopolysaccharide of Moraxella (Branhamella) catarrhalis serotype A (strain ATCC 25238) Carbohydr Res. 1994;257:269–284. doi: 10.1016/0008-6215(94)80040-5. doi:10.1016/0008-6215(94)80040-5. [DOI] [PubMed] [Google Scholar]
- Edebrink P, Jansson PE, Widmalm G, Holme T, Rahman M. The structures of oligosaccharides isolated from the lipopolysaccharide of Moraxella catarrhalis serotype B, strain CCUG 3292. Carbohydr Res. 1996;295:127–146. doi: 10.1016/s0008-6215(96)90132-9. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Allen S, Gibson BW, Campagnari AA. Characterization of a cluster of three glycosyltransferase enzymes essential for Moraxella catarrhalis lipooligosaccharide assembly. J Bacteriol. 2005;187:2939–2947. doi: 10.1128/JB.187.9.2939-2947.2005. doi:10.1128/JB.187.9.2939-2947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Schwingel JM, Datta AK, Campagnari AA. Multiplex PCR assay that identifies the major lipooligosaccharide serotype expressed by Moraxella catarrhalis clinical isolates. J Clin Microbiol. 2005;43:6139–6143. doi: 10.1128/JCM.43.12.6139-6143.2005. doi:10.1128/JCM.43.12.6139-6143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4:837–851. doi: 10.1016/s1286-4579(02)01604-0. doi:10.1016/S1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- Faglin I, Tiralongo J, Wilson JC, Collins PM, Peak IR. Biochemical analysis of Lgt3, a glycosyltransferase of the bacterium Moraxella catarrhalis. Biochem Biophys Res Commun. 2010;393:609–613. doi: 10.1016/j.bbrc.2010.02.028. doi:10.1016/j.bbrc.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Filiatrault MJ, Gibson BW, Schilling B, Sun S, Munson RS, Jr, Campagnari AA. Construction and characterization of Haemophilus ducreyi lipooligosaccharide (LOS) mutants defective in expression of heptosyltransferase III and beta1,4-glucosyltransferase: Identification of LOS glycoforms containing lactosamine repeats. Infect Immun. 2000;68:3352–3361. doi: 10.1128/iai.68.6.3352-3361.2000. doi:10.1128/IAI.68.6.3352-3361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano K, Campagnari AA. Identification of a hemin utilization protein of Moraxella catarrhalis (HumA) Infect Immun. 2004;72:6426–6432. doi: 10.1128/IAI.72.11.6426-6432.2004. doi:10.1128/IAI.72.11.6426-6432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Cook NJ, Gottfridson E, Lind T, Lidholt K, Roberts IS. Characterization of the glycosyltransferase enzyme from the Escherichia coli K5 capsule gene cluster and identification and characterization of the glucuronyl active site. J Biol Chem. 1998;273:11752–11757. doi: 10.1074/jbc.273.19.11752. doi:10.1074/jbc.273.19.11752. [DOI] [PubMed] [Google Scholar]
- Gronow S, Brade H. Lipopolysaccharide biosynthesis: Which steps do bacteria need to survive? J Endotoxin Res. 2001;7:3–23. [PubMed] [Google Scholar]
- Heinrichs DE, Yethon JA, Amor PA, Whitfield C. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharides of Escherichia coli. The R1 core-specific beta-glucosyltransferase provides a novel attachment site for O-polysaccharides. J Biol Chem. 1998;273:29497–29505. doi: 10.1074/jbc.273.45.29497. doi:10.1074/jbc.273.45.29497. [DOI] [PubMed] [Google Scholar]
- Inzana TJ, Apicella MA Virginia-Maryland Regional College of Veterinary Medicine VPI, State University BUSAtve. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis. 1999;20(3):462–465. doi: 10.1002/(SICI)1522-2683(19990301)20:3<462::AID-ELPS462>3.0.CO;2-N. doi:10.1002/(SICI)1522-2683(19990301)20:3<462::AID-ELPS462>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Jacques M. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 1996;4:408–409. doi: 10.1016/0966-842X(96)10054-8. doi:10.1016/0966-842X(96)10054-8. [DOI] [PubMed] [Google Scholar]
- Jing W, DeAngelis PL. Dissection of the two transferase activities of the Pasteurella multocida hyaluronan synthase: Two active sites exist in one polypeptide. Glycobiology. 2000;10:883–889. doi: 10.1093/glycob/10.9.883. doi:10.1093/glycob/10.9.883. [DOI] [PubMed] [Google Scholar]
- Jing W, DeAngelis PL. Analysis of the two active sites of the hyaluronan synthase and the chondroitin synthase of Pasteurella multocida. Glycobiology. 2003;13:661–671. doi: 10.1093/glycob/cwg085. doi:10.1093/glycob/cwg085. [DOI] [PubMed] [Google Scholar]
- Li J, Thibault P, Martin A, Richards JC, Wakarchuk WW, van der Wilp W. Development of an on-line preconcentration method for the analysis of pathogenic lipopolysaccharides using capillary electrophoresis-electrospray mass spectrometry. Application to small colony isolates. J Chromatogr A. 1998;817:325–336. doi: 10.1016/s0021-9673(98)00341-0. doi:10.1016/S0021-9673(98)00341-0. [DOI] [PubMed] [Google Scholar]
- Luke NR, Allen S, Gibson BW, Campagnari AA. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect Immun. 2003;71:6426–6434. doi: 10.1128/IAI.71.11.6426-6434.2003. doi:10.1128/IAI.71.11.6426-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke NR, Howlett AJ, Shao J, Campagnari AA. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect Immun. 2004;72:6262–6270. doi: 10.1128/IAI.72.11.6262-6270.2004. doi:10.1128/IAI.72.11.6262-6270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke NR, Karalus RJ, Campagnari AA. Inactivation of the Moraxella catarrhalis superoxide dismutase SodA induces constitutive expression of iron-repressible outer membrane proteins. Infect Immun. 2002;70:1889–1895. doi: 10.1128/IAI.70.4.1889-1895.2002. doi:10.1128/IAI.70.4.1889-1895.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke NR, Russo TA, Luther N, Campagnari AA. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect Immun. 1999;67:681–687. doi: 10.1128/iai.67.2.681-687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud H, Perry MB, Brisson JR, Uhrin D, Richards JC. Structural elucidation of the backbone oligosaccharide for the lipopolysaccharide of Moraxella catarrhalis serotype A. Can J Chem. 1994;72:1466–1477. doi:10.1139/v94-182. [Google Scholar]
- May JF, Levengood MR, Splain RA, Brown CD, Kiessling LL. A processive carbohydrate polymerase that mediates bifunctional catalysis using a single active site. Biochemistry. 2012;51:1148–1159. doi: 10.1021/bi201820p. doi:10.1021/bi201820p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya T, Sugiura N, Tawada A, Sugimoto K, Watanabe H, Kimata K. Molecular cloning and characterization of chondroitin polymerase from Escherichia coli strain K4. J Biol Chem. 2002;277:21567–21575. doi: 10.1074/jbc.M201719200. doi:10.1074/jbc.M201719200. [DOI] [PubMed] [Google Scholar]
- Peak IR, Grice ID, Faglin I, Klipic Z, Collins PM, van Schendel L, Hitchen PG, Morris HR, Dell A, Wilson JC. Towards understanding the functional role of the glycosyltransferases involved in the biosynthesis of Moraxella catarrhalis lipooligosaccharide. FEBS J. 2007;274:2024–2037. doi: 10.1111/j.1742-4658.2007.05746.x. doi:10.1111/j.1742-4658.2007.05746.x. [DOI] [PubMed] [Google Scholar]
- Peng D, Choudhury BP, Petralia RS, Carlson RW, Gu XX. Roles of 3-deoxy-d-manno-2-octulosonic acid transferase from Moraxella catarrhalis in lipooligosaccharide biosynthesis and virulence. Infect Immun. 2005;73:4222–4230. doi: 10.1128/IAI.73.7.4222-4230.2005. doi:10.1128/IAI.73.7.4222-4230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Hong W, Choudhury BP, Carlson RW, Gu XX. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. 2005;73:7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. doi:10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Hu WG, Choudhury BP, Muszynski A, Carlson RW, Gu XX. Role of different moieties from the lipooligosaccharide molecule in biological activities of the Moraxella catarrhalis outer membrane. FEBS J. 2007;274:5350–5359. doi: 10.1111/j.1742-4658.2007.06060.x. doi:10.1111/j.1742-4658.2007.06060.x. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A, Stein DC. Biochemical properties of Neisseria gonorrhoeae LgtE. J Bacteriol. 2002;184:6410–6416. doi: 10.1128/JB.184.23.6410-6416.2002. doi:10.1128/JB.184.23.6410-6416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. doi:10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta HL, Burrows LL, Pacan JC, Lam JS. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: A fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. doi:10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- Russo TA, Beanan JM, Olson R, MacDonald U, Cox AD, St Michael F, Vinogradov EV, Spellberg B, Luke-Marshall NR, Campagnari AA. The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect Immun. 2013;81:915–922. doi: 10.1128/IAI.01184-12. doi:10.1128/IAI.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Jr, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of beta-glycosyl transferases: Implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingel JM, Edwards KJ, Cox AD, Masoud H, Richards JC, St Michael F, Tekwe CD, Sethi S, Murphy TF, Campagnari AA. Use of Moraxella catarrhalis lipooligosaccharide mutants to identify specific oligosaccharide epitopes recognized by human serum antibodies. Infect Immun. 2009;77:4548–4558. doi: 10.1128/IAI.00294-09. doi:10.1128/IAI.00294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingel JM, St Michael F, Cox AD, Masoud H, Richards JC, Campagnari AA. A unique glycosyltransferase involved in the initial assembly of Moraxella catarrhalis lipooligosaccharides. Glycobiology. 2008;18:447–455. doi: 10.1093/glycob/cwn021. doi:10.1093/glycob/cwn021. [DOI] [PubMed] [Google Scholar]
- Tarbouriech N, Charnock SJ, Davies GJ. Three-dimensional structures of the Mn and Mg dTDP complexes of the family GT-2 glycosyltransferase SpsA: A comparison with related NDP-sugar glycosyltransferases. J Mol Biol. 2001;314:655–661. doi: 10.1006/jmbi.2001.5159. doi:10.1006/jmbi.2001.5159. [DOI] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. doi:10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- Verduin CM, Hol C, Fleer A, van Dijk H, van Belkum A. Moraxella catarrhalis: From emerging to established pathogen. Clin Microbiol Rev. 2002;15:125–144. doi: 10.1128/CMR.15.1.125-144.2002. doi:10.1128/CMR.15.1.125-144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hansen EJ. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid. 2006;56:133–137. doi: 10.1016/j.plasmid.2006.03.002. doi:10.1016/j.plasmid.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of procedure. In: Whistler R, editor. Methods in Carbohydrate Chemistry. vol. V. New York, London: Academic Press; 1965. pp. 83–91. [Google Scholar]
- Wilson JC, Collins PM, Klipic Z, Grice ID, Peak IR. Identification of a novel glycosyltransferase involved in LOS biosynthesis of Moraxella catarrhalis. Carbohydr Res. 2006;341:2600–2606. doi: 10.1016/j.carres.2006.07.009. doi:10.1016/j.carres.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Zaleski A, Scheffler NK, Densen P, Lee FK, Campagnari AA, Gibson BW, Apicella MA. Lipooligosaccharide P(k) (Galalpha1-4Galbeta1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect Immun. 2000;68:5261–5268. doi: 10.1128/iai.68.9.5261-5268.2000. doi:10.1128/IAI.68.9.5261-5268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]