Abstract
Background
Cansiliella servadeii (Coleoptera) is an endemic troglobite living in deep carbonate caves in North-Eastern Italy. The beetle constantly moves and browses in its preferred habitat (consisting in flowing water and moonmilk, a soft speleothem colonized by microorganisms) self-preens to convey material from elytra, legs, and antennae towards the mouth. We investigated its inner and outer microbiota using microscopy and DNA-based approaches.
Results
Abundant microbial cell masses were observed on the external appendages. Cansiliella’s midgut is fully colonized by live microbes and culture-independent analyses yielded nearly 30 different 16S phylotypes that have no overlap with the community composition of the moonmilk. Many of the lineages, dominated by Gram positive groups, share very low similarity to database sequences. However for most cases, notwithstanding their very limited relatedness with existing records, phylotypes could be assigned to bacterial clades that had been retrieved from insect or other animals’ digestive traits.
Conclusions
Results suggest a history of remote separation from a common ancestor that harboured a set of gut-specific bacteria whose functions are supposedly critical for host physiology. The phylogenetic and coevolutionary implications of the parallel occurrences of these prokaryotic guilds appear to apply throughout a broad spectrum of animal diversity. Their persistence and conservation underlies a possibly critical role of precise bacterial assemblages in animal-bacteria interactions.
Keywords: Cansiliella servadeii, Gut bacteria, Animal-bacteria coevolution, Cave, Moonmilk, Food web
Background
The associations between microorganisms and insects are widespread in nature [1,2]. Relationships between obligate symbioses and instances of co-evolution have been reported for mealybugs [3], whiteflies [4], weevils [5], tsetse flies [6], cockroaches and termites [7], aphids [8], planthoppers [9], carpenter ants [10]. In previous work of ours we have examined a number of symbiotic occurrences within dipterans, describing the novel species ‘Candidatus Erwinia dacicola’ dwelling in the oesophageal bulb of the olive fly [11,12] and the novel genus Stammerula,[13]; for which we highlighted evidences of joint evolution with the insects [14,15].
Hosting bacteria can result in different benefits for insects, among which a specific nutritional complementation is critical for those living on a markedly imbalanced diet, e.g. aphids [16] or ants. In the latter example trophic metabolism has been recognized as a major contributor of evolutionary shifts [17], as in the case of the Tetraponera ants [18]. In these ants the onset of herbivory has been postulated to be the result of the link with internal bacteria. Further examples include other hymenoptera whereby members of the characteristic bacterial microbiota of the honey bee Apis mellifera were absent from most species outside of the corbiculate bees, and a specific co-evolution between these hymenoptera and a defined bacterial guild was postulated to explain such association [19]. All of these relationships have also been hypothesized to involve oxidative recycling of nitrogen-rich metabolic waste and are encaged in specialized hindgut- or midgut-derived pouches. Stinkbugs host Burkholderia in their midgut crypts [20,21], while the medicinal leech carries Aeromonas and a member of the Rickenellaceae in its intestinal assemblage [22,23].
For invertebrates that permanently live in secluded habitats with little exchange with the external biota, such as cave environments, the importance of microsymbionts can be particularly critical for host adaptation and survival. Some cave-dwelling animals owe their life to symbioses with chemolithoautotrophic bacteria [24,25]. We previously described a novel genus and two species of a troglobitic beetle, Cansiliella tonielloi[26,27] and Cansiliella servadeii (Figure 1a) [28], which are endemic of few karst caves in Northern Italy. The latter has been the object of more detailed studies [29,30], where we further described the physico-chemical features of its environment.
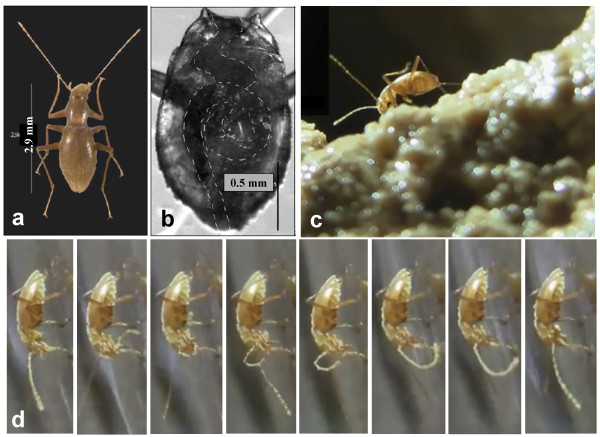
Figure 1.
Cansiliella servadeii and its habitat. a) Top view of the adult insect. b) detail of the abdomen with indication of the gut position and coiling; c) insect browsing on moonmilk in Grotta della Foos cave floor. d) sequence showing C.servadeii on location, preening its left antenna and passing it through mouthparts.
The beetles live in a hygropetric habitat in the presence of a peculiar, soft speleothem called moonmilk, which consists of carbonate minerals that are constantly covered by a thin layer of running water [31]. This habitat type is common in air-filled caves, and is typified by dripwaters or sheetflow that bring allochthonous, surface-derived organic matter [32]. Hydrological isolation for some cave hygropetric habitats may restrict the influx of organic matter, and this can lead to nutritional limitations for troglobites and troglophiles over extended periods of time and be a major driver for evolutionary adaptation for troglobites [32].
Moonmilk usually carries high amounts of microbial biomass [33-38]. In the Grotta della Foos, one of the cave systems being studied, the wet moonmilk contains ~108 microbial cells/ml and ~104 meiofaunal cells/m2 and its bacterial community characterization is described in a parallel study of ours [39]. The insect spends most of its time browsing the moonmilk surface and frequently self-preening. Videos of live C. servadeii in Grotta della Foos (http://www.youtube.com/watch?v=iXF5pDrF2J0) were taken, and its activities and behaviour were recorded. The mouthparts are consistent with reported models of adaptation for browsing/filtering organic particles in semi-aquatic environments [40], and differ markedly from those of the majority of other troglobitic Leptodirini [32,41-43]. The mouthparts have modified hoe-shaped mandibles and spoon-like galeae covered by dense setae spaced 1–1.5 μm. This distance could effectively rake particles of compatible size, such as bacteria [29,30].
Our previous stable isotope investigations [30] demonstrated that C. servadeii derives its nutritional requirements from the moonmilk and from dissolved organic matter in the percolating waters. To our knowledge, there are no molecular studies of the gut microbiota of cave invertebrates. The current project aimed at characterizing the feeding behaviour of C. servadeii from Grotta della Foos and the nature of its gut microbiota. The results provided insights pointing towards the existence of a universal guild of bacteria which appears to be common to many animal digestive systems and that suggests to have shared ancestors established prior to their hosts evolution.
Methods
Sampling site, specimen observation and collection
The Grotta della Foos cave system formed within Monte Ciaurlec located in north-eastern Italy, and is underlain by Cretaceous and Triassic limestone units [44] The cave contains over 2600 m of passages. Ten sampling locations within the cave were used for the investigations of behaviour and insect collection. the sites covered altogether 13.3, square meters, which is the whole area which Cansiliella is regularly found in Grotta de la Foos cave. The density monitored varied from 1.4 to 1.8 specimens per square meter. Examined specimen were all adults and included both sexes. Live C. servadeii were collected in sterile falcon tubes and transported to the laboratory.
Microscopy, insect dissection, and gut content evaluation
Insects external teguments were stained with DAPI (5 μg/ml) and observed in visible light and in epifluorescence using a Leica DM4000 inverted microscope equipped with a DFC300 FX camera. Images were acquired by using the LAS software.
Insects were dissected to remove the midgut to analyze the intestinal microflora. Before dissection, specimens were stunned by keeping vials at 4°C for 20 min. To extract the midgut, the insect’s abdomen was opened under a stereomicroscope (Figure 1b) in a laminar flow hood using sterile equipment and sterile water. The midgut was transferred in a sterile Eppendorf tube and used for both bacterial culturability tests and bacterial DNA extraction and amplification, and was stored at −20°C until extraction.
A segment of each midgut was observed under microscopy after staining with the LIVE/DEAD® BacLight Bacterial Viability Kit (Molecular Probes, California, USA). Slides were also prepared for Gram staining and morphological characterization, which was performed under an Olympus BX60 microscope.
Bacterial cultivation
In order to examine external bacteria adhering to the insect exoskeletal tegument, live specimens collected with cave water in falcon tubes were handled with sterile forceps and gently touched over the surface of Plate Count Agar (PCA) (Oxoid) plates.
The possible culturability of the microorganisms hosted in the insect midgut was verified by plating aliquots of resuspended, dissected gut material extracted onto PCA plates.
All plates were incubated in the dark at 20°C for up to 10 days.
DNA extraction, 16S rRNA gene amplification, cloning, and sequencing
DNA was extracted from the content of the midguts, as previously described [45]. PCR amplification targeting the 16S rRNA gene was carried out in 20 μl, 1x PCR GoTaqFlexi Buffer (Promega), 2.5 mM MgCl2, 0.1 mM dNTPs, 0.5 μM of each primer, 1 U of GoTaq Flexi DNA polymerase (Promega), and 1 μl of a 1:30 dilution of the DNA extraction. The universal bacterial 16S rRNA primers used were 63f and 1389r [46] to yield an expected amplicon of ~1300 bp. The cycling program consisted of a 95°C 2 min step followed by 35 cycles at 96°C for 30 s, 56°C for 30 s, 72°C for 90 s, and a final extension at 72°C for 10 min. PCR products were checked by 1.0% agarose gel stained with SYBR®Safe (Invitrogen) and purified with the ExoSAP-IT kit (Amersham Biosciences) before sequencing.
Amplicons (1300 bp) were cloned into JM109 competent cells using the P-GEM-T Easy vectors (Promega), following the manufacturer’s recommendations. Thirty clones from each of the three gut specimen samples were picked. Transformation was verified using PCR assays with the M13-T7 universal primers pair. The amplification products were sequenced by capillary electrophoretic sanger sequencing using M13 and T7 primers at the BMR Genomics service (Padova, Italy). Restriction enzyme (BsaI, Euroclone) digestion patterns of the amplified 16S gene (ARDRA) were used as a parallel clone screening in addition to nucleotide sequencing.
Sequence analysis
Sequence chromatograms were visually inspected and sequences were edited and aligned by using MEGA 4.0.2 (http://www.megasoftware.net/).
Chimeras were searched with the CHIMERA CHECK program of the Ribosomal Database Project II (http://rdp.cme.msu.edu).
A BLASTN GenBank analysis of all the sequences was done through the NCBI website (http://www.ncbi.nlm.nih.gov/) and closely related sequences from the databases were retrieved and added to the alignment. Phylogenetic relationships among newly retrieved gut microbiota sequences to close relatives were estimated using a maximum likelihood analysis (ML) with a GTR+I+G model.
The software package DOTUR [47] was used to assign sequences to operational taxonomic units (OTUs) for the bacterial identities found in the midgut of C. servadeii. This program assigns sequences to OTUs based on sequence data by using values that are less than the cut-off level, which were at the 97% and 95% identity thresholds. The Chao1 richness estimator [48] was also calculated using DOTUR. The richness estimates are reported for 3% difference between sequences.
16S rRNA gene sequences of clones from the guts of C. servadeii are accessible under numbers JQ308110 to JQ308155 and from JX463074 to JX463100.
The sequences from the culturable microbial community from the midgut and the external tegument are accessible under numbers JQ308156 to JQ308165.
Results
Observations of insect behaviour
Live activities were monitored for C. servadeii individuals within Grotta della Foos on six different expeditions (Figure 1). Consistent behavioural patterns could be defined from a continuous 24-hour period from eight specimens. The insect spends 44% of the time at a depth between 4 and 20 mm under the water that flows over the moonmilk speleothem. During this activity, the mouthparts and head are engaged in a prolonged browsing to rubbing motion (Figure 1c). Nearly half of the time was dedicated to self-preening of the head, legs, elytra and antennae; this behaviour is suggestive of a feeding activity as it moves organic particulates from the body towards the mouth. Typically, during preening, the insect passed the posterior legs over the elytra, then the middle legs brushed the posterior ones, the forelegs brushed the middle ones, each antenna, and then the forelegs passed between the mandibles and galeae. Antennae were combed for their entire length, as shown by the consecutive frames of the sequential series (Figure 1d), taken from footage available at http://www.youtube.com/watch?v=iXF5pDrF2J0. The observed aquatic and semi-aquatic movement actively displaced superficial sediment granules and disrupted moonmilk into trenches ~0.2 to 3 mm long.
In support of the hypothesis that the browsing and preening activities are related to feeding, possibly to acquire organic matter or cellular material from the wet moonmilk, the DAPI fluorescent stain shows that the hair-covered upper underside and interior legs of the insect body parts, that are continuously rubbed during preening, are covered by masses of bacteria-containing material (Figure 2). Crawling across the soft moonmilk, and passing the antennae tightly by the mouthparts, as shown by the sequence in Figure 1d, contributes to scooping up abundant organic material visible on the ventral segment of the body (Figure 2c).
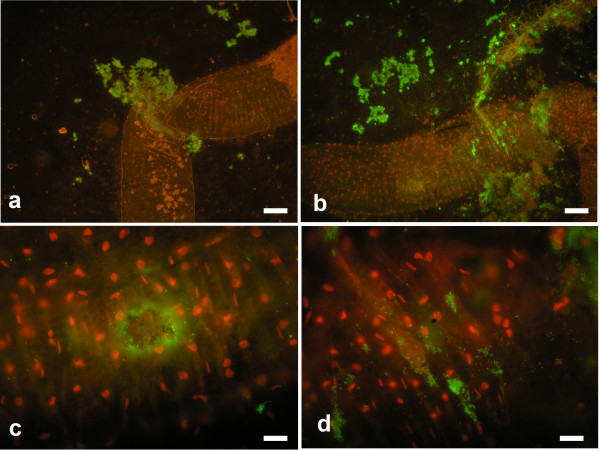
Figure 2.
Cansiliella servadeii observation under epifluorescence stereomicroscope after staining with the DNA-specific DAPI fluorochrome. a), c): head and torso view; b), d) detail of foreleg underside. a), b): white illumination; c), d): UV illumination. The presence of masses of bacteria staining with DAPI on the insect head, limbs, antennae and ventral side of body is visible. Scale bars: a), c): 250 μm; b), d): 50 μm.
Presence and viability of midgut bacteria
We explored C. servadeii midgut (Figure 1b) by pulling it out gently from dissected specimens and staining it with the Bac/Light live-dead bacterial stain. The results shown in Figure 3, reveal that abundant alive (green-staining), prevailingly rod-shaped, bacterial cells fill the lumen of the gut. In the images, in which the nuclei of the insect epithelial layers stain in red, profuse live bacterial content is seen oozing out from the gut tube in correspondence of its ruptures. In Figure 3c a hole was pierced with forceps on the gut wall, through which a lump of bacterial cells are consequently pouring out. The data indicate that this cave beetle hosts live prokaryotes in its digestive tract. In order to investigate their identities we proceeded with both culture-dependent and independent approaches as follows.
Figure 3.
BacLight staining of dissected Cansiliella servadeii midgut resuspended material. Live bacterial cells stain in green while insect epithelial nuclei stain in red. In a) clumps of bacteria are seen flowing out from the rupture of the bent gut tract. In b) a different portion is shown and the abundant masses of extracted bacteria. In c) individual bacterial cells are released from the gut epithelium through a hole pierced with forceps. In d) a region of the gut from which several distinct bacterial cells can be seen along with others in more clustered formations. Scale bars: a),b): 350 μm,c),d): 50 μm.
Culturable microbial community from the external tegument and midgut
Touching the external tegument of wet live specimens onto PCA plates resulted in colonies that belonged to four 16S phylotypes representing three lineages (Gammaproteobacteria, Actinobacteria, and Firmicutes) (Table 1).
Table 1.
Taxonomical assignment based on 16S rRNA gene sequencing of culturable isolates from the external exoskeleton of Cansiliella servadeii (non-surface sterilized specimens) or from its midgut content (surface-sterilized specimens)
| Taxonomy | Isolate, GenBank code | Top database similarities (%)1 | Habitat of subject2 | |
|---|---|---|---|---|
| Tegument |
γ-Proteobacteria |
InGrP, (JQ308165) |
(100) Pseudomonas sp. EU182834 |
Soil |
| Actinobacteria |
InGrG, (JQ3081649) |
(99.4) Streptomyces sp. JF292927 |
Endophyte in Lobularia sp. |
|
| Actinobacteria |
InGrA3, (JQ308163) |
(99.4) Rhodococcussp. HQ256783 |
Cloud water from mountain summit |
|
| Firmicutes |
InGrA1, (JQ308162) |
(96.8) Unc.bacterium JF107304 |
Human skin, antecubital fossa |
|
| Midgut | γ-Proteobacteria |
CP1a, (JQ308158) |
(100) Pseudomonas sp. AB569967 |
Chitinolitic biota in rhizosphere soil |
| γ-Proteobacteria |
CP1b, CP2b, (JQ308159) |
(100) Pseudomonas sp. AJ243602 |
Lumbricus rubellus gut (Annelida) |
|
| Actinobacteria |
CP2a, CP3aL, (JQ308160) |
(100) Streptomyces champavatii HQ143637 |
Soil |
|
| γ-Proteobacteria |
CP3a, (JQ308161) |
(100) Unc. Pseudomonas sp. JF500897 |
Rye grass rhizosphere |
|
| Firmicutes |
CP4.1, CP4.2, (JQ308156) |
(99.4) Unc. Firmicutes EU005283 |
Inert surfaces immersed in marine water |
|
| Firmicutes | CP4.3, (JQ308157) | (98.6) Unc.bacterium DQ860054 | Anchovy intestinal microflora |
1Description of GenBank subjects displaying the top-scoring BLAST alignment results of sequence similarity.
2Animal host or other environment in which the subject having homology with the present sequence s described in GenBank records.
From the extracted insect guts, there were sparse colonies that grew on PCA plates, and the most frequent morphological colony type resulted in isolate CP4.1. Sequences obtained from the external tegument, as well as from the culturable fraction of midgut bacteria, had high homology values (in most cases 99-100%) to bacterial taxa featuring multiple occurrencies in the nucleotide sequence database (for references consult the GenBank accession numbers given in the fourth column of Table 1), although there was no close affinity to sequences previously retrieved from insect guts.
Culture-independent analysis of the midgut microbial community
Under the limitations posed by working with a rare endemic and protected species with minimum sampling allowed, we analyzed three specimens from which separate clone libraries of 16S rRNA gene amplicons were generated and 87 clones screened. Sequences from the three different guts are labeled with the suffixes A, B, and C, respectively, on Table 2. At this resolution level the number of Dotur-defined species was 29 and the Chao1 estimator [48] predicted a total number of species of 51,7. We also calculated the estimated coverage by applying the Good’s index [49] which, at species level, resulted 81.6 %. In order to check with an independent method whether the sampling size had been truly effective in yielding an adequate representation of the community, we compared the cluster analysis dendrogram obtained with the first 46 clones screened (Additional file 1: Material S1 and Additional file 2: Material S2) with those generated with the whole set of 87 (Figures 4 and 5), from whose comparison it can be observed that the community structure was already fully delineated from the first stepwise subset of randomly selected clones. Further, considering the phylum rank as a more functional assessment of population diversity we run rarefaction curves with OTUs defined at a phylum level similarity threshold (81%). The result obtained indicated a saturating curve and is shown in the supplementary Additional file 3: Figure S3.
Table 2.
Taxonomical assignment of cloned 16S rRNA amplicons from the midgut content of Cansiliella servadeii
| OTU # | Taxonomy | Clone and GenBank Code | Top database similarities (%)1 | Habitat2 |
|---|---|---|---|---|
| 1 |
Firmicutes |
7B, (JQ308118) |
(92.4) Unc3. bacterium AB606297 |
Mouse faeces |
| (92.1) Unc. Clostridiaceae AB088980 |
Reticulitermes speratus gut (Isoptera: Termitidae) |
|||
| 43A;14B; 9B; 33C, (JQ308112, JQ308119, JQ308111, JQ308113) |
(92.6) Unc. bacterium AB606297 |
Mouse faeces |
||
| (92.4) Unc. bacterium DQ815954 |
Mouse cecum |
|||
| (92.3) Unc. Clostridiaceae AB088980 |
R. speratus gut |
|||
| 19B; 23C; 25C; 28C; 39C, 50B, 53B, 57B, 73A, 74A (JQ308115, JQ308116, JQ308110, JQ308114, JQ308117, JX463078, JX463086, JX463088, JX463089), JX463090 |
(92.9) Unc. bacterium AB606297 |
Mouse faeces |
||
| (92.6) Unc. Clostridiaceae AB088980 |
R. speratus gut |
|||
| 41A, (JQ308120) |
(93.1) Unc. bacterium AB606297 |
Mouse faeces |
||
| (92.9) Unc. bacterium DQ815954 |
Mouse cecum |
|||
| (92.8) Unc. Clostridiaceae AB088980 |
R. speratus gut |
|||
| 49B (JX463074) |
(92.9) Unc. bacterium AB606297 |
Mouse faeces |
||
| (92.6) Unc. bacterium DQ815954 |
Mouse cecum |
|||
| (92.5) Unc. Clostridiaceae AB088980 |
R. speratus gut |
|||
| 2 |
Firmicutes |
10B, (JQ308121) |
(92.3) Unc. bacterium EF602946 |
Mouse cecum |
| 3 |
Firmicutes |
4A; 42A, (JQ308123, JQ308124) |
(95.9) Unc. Clostridiales AB088981 |
R. speratus gut |
| (94.4) Unc. bacterium GU451010 |
Tipula abdominalis gut (Diptera: Tipulidae) |
|||
| 67A, 72A (JX463084, JX463085) |
(94.8) Unc. Clostridiales AB088981 |
R. speratus gut |
||
| 8B, (JQ308122) |
(95.5) Unc. bacteriumEF608549 |
Poecilus chalcites gut (Coleoptera: Carabidae) |
||
| 4 |
Firmicutes |
32C, (JQ308126) |
(95.2) Unc. Clostridiaceae AB192046 |
Microcerotermes spp. gut (Isoptera: Termitidae) |
| 48A, 68A, 75A (JQ308127, JX463080, JX463091) |
(95.7) Unc. bacterium AJ852374 |
Melolontha melolontha gut (Coleoptera: Scarabaeidae) |
||
| 5 |
Firmicutes |
21C, (JQ308125) |
(94,5) Unc. bacterium FJ374218 |
Pachnoda spp. gut (Coleoptera: Scarabaeidae) |
| 6 |
Firmicutes |
2A;12B, (JQ308128, JQ308129) |
(97.1) Unc. Clostridiaceae AB192046 |
Microcerotermes spp. gut (Isoptera: Termitidae) |
| 6B, (JQ308130) |
(96.9) Unc. bacterium FJ374218 |
Pachnoda spp. larval gut (Coleoptera: Scarabaeidae) |
||
| 46A, 63A (JQ308131, JX463079) |
(94.5) Unc. bacterium FJ374218 |
Pachnoda spp. gut (Coleoptera: Scarabaeidae) |
||
| 7 |
Firmicutes |
15B, (JQ308133) |
(91.7) Unc. bacterium EU465991 |
African elephant faeces |
| (90.5) Unc. bacterium AY654956 |
Chicken gut |
|||
| 29C, (JQ308132) |
(91.9) Unc. bacterium EU465991 |
African elephant faeces |
||
| (90.7) Unc. bacterium AY654956 |
Chicken gut |
|||
| 8 |
Firmicutes |
5A, (JQ308134) |
(93.8) Unc. Clostridiales AB231035 |
Hodotermopsis sjoestedti gut (Isoptera: Termitidae) |
| 9 |
Firmicutes |
69A (JX463081) |
(94.7) Unc. bacterium AB088973 |
R. speratus gut |
| 10 |
Firmicutes |
71A(JX463087) |
(92.7) Unc. bacterium AB088973 |
R. speratus gut |
| 11 |
Firmicutes |
24C, 30C, (JQ308135, JQ308136) |
(92.6) Unc. Firmicutes GQ275112 |
Leptogenys spp. gut (Hymenoptera: Formicidae) |
| 12 |
Actinobacteria |
61A (JX463076) |
(93.2) Unc. Bacterium FR687129 |
Paddy soil |
| 13 |
Actinobacteria |
22C; 36C, 51B, 54B (JQ308137, JQ308138, JX463075, JX463083) |
(97.2) Unc. bacterium DQ521505 |
Lake Vida ice cover |
| (96.9) Unc. bacterium AM940404 |
Rhagium inquisitor gut (Coleoptera: Cerambycidae) |
|||
| 52B (JX463077) |
(96.7) Unc. bacterium DQ521505 |
Lake Vida, ice cover |
||
| (96.5) Unc. bacterium AM940404 |
Rhagium inquisitor gut (Coleoptera: Cerambycidae) |
|||
| 65A (JX463082) |
(97) Unc. bacterium DQ521505 |
Lake Vida, ice cover |
||
| (96.7) Unc. bacterium AM940404 |
Rhagium inquisitor gut (Coleoptera: Cerambycidae |
|||
| 14 |
Actinobacteria |
45A, (JQ308139) |
(99.5) Sanguibacter inulinus HQ326836 |
Thorectes lusitanicus gut (Coleoptera: Geotrupidae) |
| 15 |
α-Proteobacteria |
13B, (JQ308142) |
(96.2) Unc. α-proteobacterium CU920098 |
Mesophilic anaerobic digester treating wastewater sludge |
| |
(93.7) Unc. bacterium FN659093 |
Lumbricus terrestris gut |
||
| 16 |
α-Proteobacteria |
58B (JX463098) |
(100) Brevundimonas sp.JQ316297 |
Soil |
| 17 |
α-Proteobacteria |
44A (JQ308143) |
(92.5) Unc. bacterium EF667926 |
Epithelium Hydra vulgaris |
| (88.2) Unc. bacterium HM779996 |
Adult zebrafish gut |
|||
| (87.9) Unc. bacterium EU148629 |
Agrilus planipennis gut (Coleoptera: Buprestidae) |
|||
| 18 |
δ-Proteobacteria |
3A; 20A, 62A (JQ308144, JQ308145, JX463096) |
(94.3) Unc. δ-proteobacterium DQ307712 |
Macrotermes michaelsenigut (Isoptera: Termitidae) |
| 19 |
δ-Proteobacteria |
60B (JX463100) |
(96) Unc. Desulfovibrionaceae JN653048 |
Gut of millipede Tachypodoiulus niger |
| 20 |
δ-Proteobacteria |
66A, 70A (JX463092, JX463093) |
(94.1) Unc. bacterium FJ374259 |
P. ephippiata gut (Coleoptera: Scarabaeidae) |
| 21 |
β-Proteobacteria |
27C, (JQ308141) |
(95.2) Unc.bacterium AJ852369 |
Melolontha melolontha gut (Coleoptera: Scarabaeidae) |
| 22 |
β-Proteobacteria |
26C, (JQ308140) |
(96.5) Burkholderiales bacterium EU073950 |
Dermolepida albohirtum gut (Coleoptera: Scarabaeidae) |
| 23 |
Bacteroidetes |
11B, (JQ308146) |
(91.9) Unc. bacterium AJ576327 |
Pachnoda ephippiata gut (Coleoptera: Scarabaeidae) |
| 18B, (JQ308147) |
(92.1) Unc. bacterium HQ728219 |
Microbial fuel cell |
||
| (91.9) Unc. bacterium AJ576327 |
P. ephippiata gut (Coleoptera: Scarabaeidae) |
|||
| 24 |
Bacteroidetes |
16B, (JQ308148) |
(92.5) Unc. bacterium FJ674429 |
Cattle feedlot |
| (91.9) Unc. Bacteroidetes AB522123 |
R. santonensis gut (Isoptera: Termitidae) |
|||
| (89.2) Unc. bacterium EF176896 |
Tipula abdominalis gut (Diptera: Tipulidae) |
|||
| 25 |
Bacteroidetes |
35C, (JQ308149) |
(96.2) Unc. bacterium AJ576327 |
P. ephippiata gut (Coleoptera: Scarabaeidae) |
| 26 |
Bacteroidetes |
64A (JX463097) |
(94.2) Unc. bacterium HQ728219 |
Anode of a glucose-fed microbial fuel cell |
| (93.7) Unc. bacterium AJ576361 |
P. ephippiata gut (Coleoptera: Scarabaeidae) |
|||
| 27 |
Bacteroidetes |
31C, (JQ308150) |
(93.1) Unc. bacterium DQ447343 |
Urban biowaste |
| (89.3) Elizabethkingia sp. GU45829 |
R. speratus gut (Isoptera: Termitidae) |
|||
| 40C, (JQ308151) |
(92.8) Unc. bacterium DQ447343 |
Urban biowaste |
||
| (89.2) Unc. Bacteroidetes HM215036 |
Bumble bee gut (Hymenoptera: Apidae) |
|||
| 28 |
Bacteroidetes |
17B; 37C; 34C, 59B (JQ308154, JQ308155, JQ308153, JX463099) |
(94.9) Unc. Bacteroidetes DQ837639 |
Apis mellifera gut (Hymenoptera: Apidae) |
| 55B (JX463095) |
(94.6) Unc. Bacteroidetes DQ837639 |
Apis mellifera gut (Hymenoptera: Apidae) |
||
| 56B (JX463094) |
(94.8) Unc. Bacteroidetes DQ837639 |
Apis mellifera gut (Hymenoptera: Apidae) |
||
| 29 | Bacteroidetes | 38C, (JQ308152) | (94.3) Unc. Bacteroidetes DQ837639 | Apis mellifera gut (Hymenoptera: Apidae) |
1Description of GenBank subjects displaying the top-scoring BLAST alignment results of sequence similarity.
2Animal host or other environment in which the subject having homology with the present sequence is described in GenBank records.
3Unc. = ‘Uncultured’.
OTUs are defined at 97% similarity threshold. Clones ID are followed by letters A,B or C to identify the three insect guts specimens.
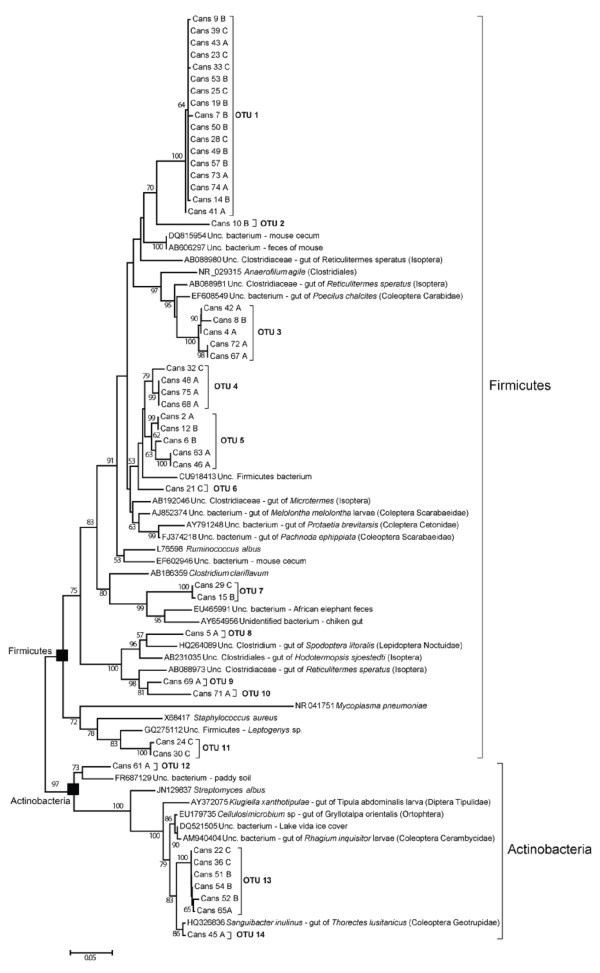
Figure 4.
Maximum likelihood tree of 16S rRNA gene clone sequences recovered of the midgut of Cansiliella servadeii affiliated with gram-positive bacteria. The sequences of GenBank dataset showing the closest similarity levels have been added. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap value shown next to the branches. Only values greater than 50 are indicated. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option).
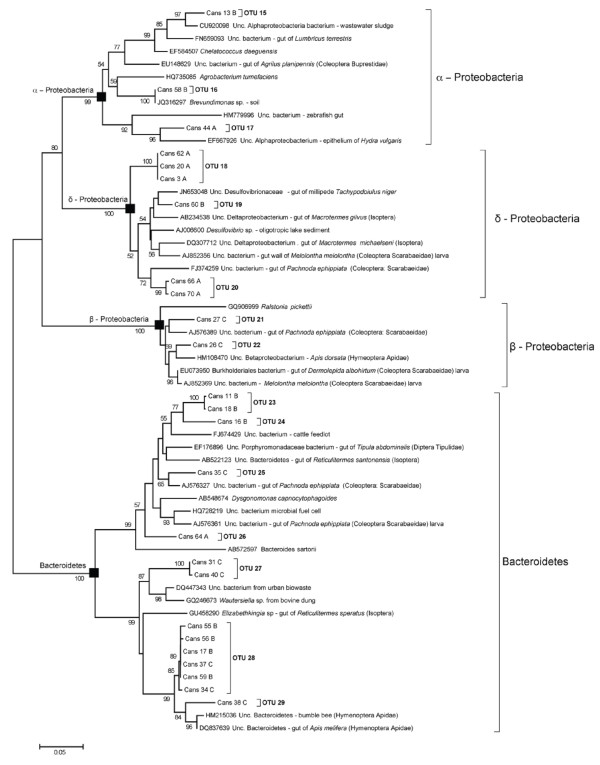
Figure 5.
Maximum likelihood tree of 16S rRNA gene clone sequences recovered of the midgut of Cansiliella servadeii affiliated with Proteobacteria and Bacteriodetes. Sequences from GenBank dataset showing the closest similarity levels have been added. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. Only values higher than 50 are indicated. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option).
Phylogenetic analyses revealed the presence of six distinct major phylogenetic groups from the sequenced clones.
The sequences showed a range of homology values with the GenBank database records that for most cases was remarkably low (Table 2).
Considering the totality of the 87 clones, the Firmicutes phylum represented 58,6% of all retrieved sequences, and over 60% of the clones showed homologies as low as 92-94% with existing database subjects. Bacteroidetes represented 16.1% of the sequences, with homologies 89-94% to GenBank entries. Only few clones of the Actinobacteria (whose phylum represented 11.5% of the retrieved sequences) displayed similarity values qualifying for species level relatedness (≥97%) with described records.
The remainder of the clones were affiliated with the Deltaproteobacteria (8.0%) and with the Alpha- and Betaproteobacteria, classes (<5% each). Although culturable strains affiliated to the Gammaproteobacteria were obtained from the gut (Table 1), no clone sequences affiliated with this class were retrieved, presumably due to their rarity within the total community.
The taxonomical groups resulted homogeneously distributed through the samples analyzed. There was no statistical difference in the distribution of the phylogenetic groups of Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes from the different midgut samples (Fisher’s exact test, P = 0.22). All guts had an outstanding majority of OTUs belonging to the Firmicutes.
Although the BLAST analysis gave similarities that in most cases were below the species and even genus limit (respectively for the 89.04% and 63% of the samples), nevertheless the best matches of a vast majority of clones corresponded to bacteria occurring in different insects gut, including ants, termites, and beetles (Table 2). It is worth adding that more than 80% of these hosts spend at least part of their life cycle in the soil, and ~46% of them belong to the Coleoptera order (Carabidae, Scarabaeidae and Geotrupidae). Another key finding is the fact that groups of taxonomically distinct clones from C. servadeii have their respective GenBank matches in sequences that were found also in the same insect host species. For example, three non-identical Clostridiales clones are closely related to three different bacteria that all come from the coleopteran Pachnoda epipphiata, [50] which also hosts the closest relatives to some of the Bacteroidetes clones (Table 2). Also, closest sequences to the clones affiliated with the Clostridiales and some Proteobacteria have been retrieved from the gut of the Melolontha melolontha beetle, while several Clostridiaceae clones and one Bacteroidetes clone were closely related to sequences that were all retrieved from the same dipteran host Tipula abdominalis gut (GU451010). Given the low homologies and the recurring multiple instances it appears highly unlikely that these occurrences could be coincidental, constituting a significant element in favour of distant but conserved host-bacteria interactive relationships, in which given subsets of bacterial taxa seem to co-occur in a number of parallel situations hosted by very different insects.
In order to better visualize the distribution of bacterial phyla found in C. servadeii along with that of the hosts/habitats where their closest GenBank relatives had been found, in Figure 6 we plotted these across the span of 16S homology at which the BLAST match was found for each clone or isolate. Interestingly, for the midgut clones, the identity levels show a bimodal distribution. Figure 6a shows the distribution of the bacterial taxonomical divisions found within Cansiliella’s gut assemblages. When the same are inspected as regards the habitat of the nearest database subject (Figure 6b), a distinction arises separating the insect-related cases (higher homology region, peaking at 95%) from the rest of non-insect environments including mammal guts/faeces, etc., (more distant homology region peaking at 93%). The two peaks (93% and 95%) are significantly different (Wilcoxon Mann–Whitney test, p<0.01) (Figure 6b). The fraction of culturable bacteria instead (Figure 6c) displays high levels of similarity shared in all cases with non-insect GenBank subjects.
Figure 6.
Phylotype and host partitioning in GenBank subjects with similarity to Cansiliella-associated bacteria. a) Abundance of 16S rDNA phylotypes found from the midgut using a culture-independent approach and respective GenBank homology percentage classes. b) Proportions of insects orders or other environments hosting bacterial subjects resulting in different degrees of sequence homology (x axis) with clones of the non-culturable microbial community from the midgut. The smaller diagram in the upper right corner shows the same data as line graphs and by pooling the insect orders together to put in evidence the separation from the cases found in non-insect environments. c) Proportions of insects orders or other environments hosting bacterial subjects resulting in different degrees of sequence homology (x axis) with culturable microbial community isolates from the midgut and external tegument. The definition ‘other’ includes all non-insect guts, faeces, and other habitats as reported in Table 2.
Discussion
Cansiliella spp. mouthparts are distinct from other cave beetles, in general and from the large majority of the Leptodirini, and show features uncommon to beetles with more saprophagous diets [28]. The beetles in Grotta della Foos have also a semi-aquatic lifestyle associated with moonmilk, which is a rich microbiological substrate mixed with carbonate minerals. Our previous stable isotope investigations, and observations of moonmilk particles in beetle mouths, reveal that C. servadeii from Grotta della Foos derives nutrition from moonmilk and habitat waters which contain dissolved organic carbon at a concentration of 10.11 mg/l [30]. The present data show that the insect midgut hosts a bacterial community whose members, as far as it can be judged from the sequenced clones, appear to belong to heterotrophic guilds. The midgut of the insect contains live bacterial cells whose culture-independent analysis yielded a bacterial assemblage dominated by the phyla Firmicutes and featuring presences of Bacteoridetes, Actinobacteria, together with Alpha-, Beta- and Deltaproteobacteria. A possible role of these bacteria in nutritional physiology with activities within the nitrogen metabolism could be postulated on the basis of parallel examples in other gut systems.
The sampling depth proved suitable as this community structure was already fully outlined in terms of phyla and their proportions from the first round of 46 clones. Upon nearly doubling the number, the whole set of 87 clones maintained the same pattern as the new sequences merged into groups which had already appeared. (Additional file 1: Material S1 and Additional file 2: Material S2 vs. Figure 4 and Figure 5).
Interestingly, as seen from each of the subject score lists of the BLAST analysis, the identities of the C. servadeii gut bacteria did not overlap with any of the sequences already obtained from our parallel project targeting the bacteria in the moonmilk of the very same cave [39]. In that work, 169 sequences are described (and are available in GenBank under the accession numbers from EU431666 to EU431834). Although moonmilk biota encompassed phyla belonging to the Bacteriodetes, Firmicutes, and Betaproteobacteria, there was no OTU overlap (no BLAST identity nor close similarity) between the potentially ingested moonmilk bacteria and the gut-hosted community described in the present report.
These findings confirm the presence of a gut microbiota specificity in C. servadeii similarly to what is found in the gut of some insects such as soil or humus-feeding termites [51], european cockchafer larvae (Melolontha melolontha) [52] and scarab beetle larvae (Pachnoda spp.) [50,53]. For these insects no correspondence has been found either between the gut community and the microbiota of their soil-related diet. On the contrary in insects having a more diverse and richer diet such as crickets and cockroaches higher correspondence between diet and gut bacterial flora has been identified in culture-dependent studies [54,55].
While the uncultured clone library community had such far divergence from known database entries, the culturable bacteria isolated from external tegument and midgut showed a much higher sequence similarity to previously retrieved sequences available in GenBank. Approximately 86% of these sequences have close or equal to 100% sequence similarity (average 97%) (Table 1). In contrast, the uncultured gut clone sequences have lower homology to any previously described bacterial species or environmental sequences, with some as low as 92% (Table 2, Figure 6). Among the dominant OTUs groups, belonging mostly to Firmicutes and Bacteriodetes phyla, sequence similarity with described taxa is ~92% and 94%, respectively, which suggests novel bacterial lineages at the genus-level, if not higher taxonomic ranks. Such result is nowadays an unusual occurrence as the GenBank database contains a large, ever-expanding number of sequences obtained from many different microbiological environments, and it is therefore no longer common to find such low sequence homology, especially when working with a set of several different sequences, all of which turned out consistently distant from known records. Except for two clones corresponding to OTU 14 and OTU 16 that show 100% identity with the Actinobacteria Sanguibacter inulinus isolated from the gut of Thorectes lusitanicus (Coleoptera Geotrupidae) and Brevundimonas sp. isolated from the soil, the rest of the bacterial communities isolated from the gut of C. servadeii are highly different from bacteria typical of other gut systems studied until now by culture-independent methods.
Noteworthy, for a number of different groups of taxonomically distinct bacteria hosted by the cave beetle, the insect hosting the closest relatives of each case turned out to be the same (Table 2). For example, the sequences of given firmicutes, bacteroidetes and betaproteobacteria happen to have their top matching GenBank subjects all occurring within the Melolontha scarab. Others, also encompassing different phyla have their relatives coinciding within a coleopteran of the Pachnoda genus, other clusters co-occur in the Dipteran Tipula abdominalis, others within the termite Reticulitermes speratus. Given the peculiarity of the sequences, these repeated occurrences appear non-coincidental and support the hypothesis of a selection ensuring the maintenance of a given microbial assemblage for a relevant physiological scope.
Because of the semi-aquatic feeding behaviour of C. servadeii, it has been speculated that its ancestor, like that of other hygropetric coleopterans, may have been aquatic [32]. Neverthelesss, considering that the C. servadeii gut microbiota having the highest degrees of homology (95-98%) to previously retrieved sequences from invertebrate gut bacteria that spend at least a part of their biological cycle in the soil (Table 2, Figure 4), and mainly to insects belonging to the Isoptera and Coleoptera orders, one could in alternative speculate that the C. servadeii ancestor had a terrestrial origin. However in available databases, bacteria from aquatic insects could be still poorly represented to enable a thorough assessment in this regard. About these aspects, a survey of microbial phylotypes from the guts of the other species in the genus, and a barcoding comparison of the insect genes are envisaged as parts of future research.
Considering the evolutionary history of the C. servadeii and its gut symbiont system, a long history of separation from other invertebrates and microorganisms appears to have occurred. At the same time its situation reveals the existence of phylogenetic similarities across the digestive tracts of many different hosts (Table 2). It is conceivable that there may be a common ancestry involving a functional guild of bacteria that has endured the host lineage separation, as well as the erosion of sequence identities, through the paths of independent evolution. The dual pattern of homology among clone sequences from gut bacteria in Cansiliella to other insects further suggests this scenario (Figure 6b); a progressive phenomenon of divergence from common ancestries is suggested by the double-peaking instance of homology existing between C. servadeii’s sequence queries and GenBank subjects, that set the insect-dwelling cases separated from the general intestinal/faecal cases. It is noteworthy that, while the hosts are set apart by sequence homology thresholds, the taxonomical groups of the bacteria found in Cansiliella are rather evenly represented across the different homology span (Figure 6a). It can be seen that Firmicutes, Bacteroidetes and Proteobacteria are almost equally present throughout the sequence similarity gradient, underscoring the need of the whole functional assemblage to be conserved both in distantly- as well as in recently-diverged hosts. This emphasizes a supposedly crucial role of a well-defined set of prokaryotic taxa that appear to have remained in charge within the alimentary tract of animals in spite of ages of separation of their hosts.
More recent acquisitions across different hosts appear to correspond to higher degrees of homology for bacterial symbionts, while acquisitions and symbiotic associations that are older would correspond to lower degrees of homology (Figure 6). The evidences depicted in Figure 6 appear to fit the contour of an evolutionary path of separation of the midgut bacteria from those of other insects; it appears that matching bacteria that are hosted in other insects (i.e. hosts that are closer to Cansiliella) share higher homology with its symbionts (peak at 95%), while those living in animals which are evolutionarily more distant from the beetle, or in other habitats, have undergone a correspondingly higher divergence from them (peak at 93%). These instances support the existence of a group of common ancestors for a set of different bacteria and a history of isolation and coevolution within the hosts. The same analysis performed with the culturable biota isolated from the external tegument or, as a minority, from the midgut, shows the opposite scenario (Figure 6c) i.e. a high level of similarity with non-insect environments (Table 1), suggesting that plate-culturable taxa are also more prone to spread/reproduce and be part of a more diffuse cosmopolitanism.
Conclusions
The insects hereby examined feature a defined gut community of bacteria suggesting a long history of inheritance and a coevolution.with their hosts. Corresponding, but genetically diverged, microbial assortments appear to exist, in parallel, in a series of other animals’ digestive systems. It appears that the reproductive boundaries arisen between the hosts at their speciation stages, have, at the same pace, prevented the exchange of their gut bacteria. The conservation of these sets of prokaryotic taxa suggests a relevant role in animal physiology.
The evidence of such patterns casts light on their biology at both physiological and evolutionary scales. Elucidating, in future studies, the details of the bacterial transmission in C. servadeii will offer useful insights to further interpret bacterial evolution and the critical roles of prokaryotes in the animal-microbe interactions ecology.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MGP defined the whole experimental plan of the research, organized the fieldwork and identified the zoological samples; LM, MS and IMS performed the gut microscopy and the cloning and sequencing of microbial 16S genes and constructed the phylogeny trees; ALD, AP, MB and LD organized the logistics of the speleological expedition into the cave, collected the insect samples and recorded their in-situ behaviour, ASE provided the data of microbial colonization of the cave substrate moonmilk and discussed its similarity with the Cansiliella microbiota; AT and BB performed the fluorescent stereomicroscopy detection of bacteria on external appendages of the insect; GC performed the water chemical analysis of the cave environment; AS performed the bioinformatical analyses, the microbial ecology assessment and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Cluster analysis dendrogram obtained with the first 46 screened clones, Gram-negative portion.
Cluster analysis dendrogram obtained with the first 46 screened clones, Gram-positive portion.
Rarefaction curve for OTUs defined at 81% similarity.
Contributor Information
Maurizio G Paoletti, Email: paoletti@bio.unipd.it.
Luca Mazzon, Email: lmazzon@unipd.it.
Isabel Martinez-Sañudo, Email: isabel.martinez@unipd.it.
Mauro Simonato, Email: mauro.simonato@unipd.it.
Mattia Beggio, Email: mattia.beggio@gmail.com.
Angelo Leandro Dreon, Email: leandro.dreon@libero.it.
Alberto Pamio, Email: albertopamio@gmail.com.
Mauro Brilli, Email: mauro.brilli@igag.cnr.it.
Luca Dorigo, Email: luca.scorpio@libero.it.
Annette Summers Engel, Email: aengel1@utk.edu.
Alessandra Tondello, Email: alessandra.tondello@unipd.it.
Barbara Baldan, Email: barbara.baldan@unipd.it.
Giuseppe Concheri, Email: giuseppe.concheri@unipd.it.
Andrea Squartini, Email: squart@unipd.it.
Acknowledgements
The authors thank Enrico Ruzzier for his collaboration to the present study.
References
- Buchner P. Endosymbiosis of animals with plant microorganisms. New York: Interscience Publishers, Inc; 1965. [Google Scholar]
- Baumann P, Moran NA. Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie van Leeuwenhoek. 1997;72:39–48. doi: 10.1023/A:1000239108771. [DOI] [PubMed] [Google Scholar]
- Munson MA, Baumann P, Moran NA. Phylogenetic relationships of endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol Phylogen Evol. 1992;1:26–30. doi: 10.1016/1055-7903(92)90032-C. [DOI] [PubMed] [Google Scholar]
- Clark MA, Baumann L, Munson MA, Baumann P, Campbell BC, Duffus JE, Osborne LS, Moran NA. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodoidea) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr Microbiol. 1992;25:119–123. doi: 10.1007/BF01570970. [DOI] [Google Scholar]
- Campbell BC, Bragg TS, Turner CE. Phylogeny of symbiotic bacteria of four weevil species (Coleoptera: Curculionidae) based on analysis of 16S ribosomal DNA. Insect Biochem Mol Biol. 1992;22:415–421. doi: 10.1016/0965-1748(92)90136-3. [DOI] [Google Scholar]
- Aksoy S Molecular analysis of the endosymbionts of tsetse flies. 16S rDNA locus and over-expression of a chaperonin. Insect Mol Biol. 1994;4:23–29. doi: 10.1111/j.1365-2583.1995.tb00004.x. [DOI] [PubMed] [Google Scholar]
- Bandi C, Damiani G, Magrassi L, Gigolo A, Fani R, Sacchi L. Flavobacteria as intracellular symbionts in cockroaches. Proc R Soc Lond B. 1994;257:43–48. doi: 10.1098/rspb.1994.0092. [DOI] [PubMed] [Google Scholar]
- Baumann P, Lai C, Baumann L, Rouhbakhsh D, Moran NA, Clark MA. Mutalistic associations of aphid and prokaryotes: biology of the genus Buchnera. Appl Environ Microbiol. 1995;61:1–7. doi: 10.1128/aem.61.1.1-7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda H, Nakashima N, Koizumi M. Phylogenetic position of yeast-like symbiotes of rice planthoppers based on partial 18S rDNA sequences. Insect Biochem Mol Biol. 1995;25:639–646. doi: 10.1016/0965-1748(94)00107-S. [DOI] [PubMed] [Google Scholar]
- Schröder D, Deppisch H, Obermayer M, Krohne G, Stackebrandt E, Hölldobler B, Goebel W, Gross R. Intracellular endosymbiotic bacteria of Camponotus species (carpenter ants): systematics, evolution and ultrastructural analysis. Mol Microbiol. 1996;21:479–489. doi: 10.1111/j.1365-2958.1996.tb02557.x. [DOI] [PubMed] [Google Scholar]
- Capuzzo C, Firrao G, Mazzon L, Squartini A, Girolami V. ‘Candidatus Erwinia dacicola’, a coevolved symbiotic bacterium of the olive fly Bactrocera oleae (Gmelin) Int J Syst Evol Microbiol. 2005;55:1641–1647. doi: 10.1099/ijs.0.63653-0. [DOI] [PubMed] [Google Scholar]
- Savio C, Mazzon L, Martinez-Sañudo I, Simonato M, Squartini A, Girolami V. Evidence of two lineages of the symbiont "Candidatus Erwinia dacicola" in Italian populations of Bactrocera oleae (Rossi) based on 16S rRNA gene sequence. Int J Syst Evol Microbiol. 2011;72:179–187. doi: 10.1099/ijs.0.030668-0. [DOI] [PubMed] [Google Scholar]
- Mazzon L, Piscedda A, Simonato M, Martinez-Sañudo I, Squartini A, Girolami V. Presence of specific symbiotic bacteria in flies of the subfamily Tephritinae (Diptera Tephritidae) and their phylogenetic relationships: proposal of ‘Candidatus Stammerula tephritidis’. Int J Syst Evol Microbiol. 2008;58:1277–1287. doi: 10.1099/ijs.0.65287-0. [DOI] [PubMed] [Google Scholar]
- Mazzon L, Martinez-Sañudo I, Simonato M, Squartini A, Savio C, Girolami V. Phylogenetic relationships between flies of the Tephritinae subfamily (Diptera, Tephritidae) and their symbiotic bacteria. Molecular Phylogenetics and Evolution. 2010;56:312–326. doi: 10.1016/j.ympev.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Mazzon L, Martinez-Sañudo I, Savio C, Simonato M, Squartini A, In: Manipulative Tenants: Bacteria Associated with Arthropods. Stammerula and other symbiotic bacteria within the fruit flies inhabiting Asteraceae flowerheads. CRC Press: Edited by Zchori-Fein E, Bourtzis K; 2011. pp. 89–111. [Google Scholar]
- Rouhbaksh D, Lai C-Y, von Dohlen CD, Baumann L, Baumann P, Moran NA, Voegtlin DJ. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated trpEG within the Aphididae. J Mol Evol. 1996;42:414–421. doi: 10.1007/BF02498635. [DOI] [PubMed] [Google Scholar]
- Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Nat Acad Sci. 2009;106:21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Borm S, Buschinger A, Boomsma JJ, Billen J. Tetraponera ants have gut-symbionts related to nitrogen-fixing symbionts. Proc R Soc Lond B. 2002;269:2023–2027. doi: 10.1098/rspb.2002.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. Specific Developmental Window for Establishment of an Insect-Microbe Gut Symbiosis. Appl Environ Microbiol. 2011;77:4075–4081. doi: 10.1128/AEM.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado SS, Almeida RPP. Phylogenetic Placement of Pentatomid Stink Bug Gut Symbionts. Curr Microbiol. 2009;58:64–69. doi: 10.1007/s00284-008-9267-9. [DOI] [PubMed] [Google Scholar]
- Worthen PL, Gode CJ, Graf J. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol. 2006;72:4775–4781. doi: 10.1128/AEM.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J. Symbiosis of Aeromonas veronii Biovar sobria and Hirudo medicinalis, the Medicinal Leech: a Novel Model for Digestive Tract Associations. Infection and Immunity. 1999;67:1–7. doi: 10.1128/iai.67.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sârbu SM, Kane TC, Kinkle BK. A chemoautotrophically based cave ecosystem. Science. 1996;272:1953–1955. doi: 10.1126/science.272.5270.1953. [DOI] [PubMed] [Google Scholar]
- Engel AS, Meisinger DB, Porter ML, Payn RA, Schmid M, Stern LA, Schleifer K-H, Lee NM. Linking phylogenetic and functional diversity to nutrient spiraling in microbial mats from Lower Kane Cave (USA) ISME J. 2010;4:98–110. doi: 10.1038/ismej.2009.91. [DOI] [PubMed] [Google Scholar]
- Paoletti MG. Un nuovo Catopide pholeuonoide del Cansiglio (Prealpi Carniche) (Col. Bathysciinae) Boll Mus Civ St Nat Venezia. 1972. pp. 119––131.
- Paoletti MG. Notizie sistematiche ed ecologiche su di un nuovo interessante genere del Cansiglio Cansiliella. Suppl Boll Mus Civ S Na. Venezia. 1973;24:81–88. [Google Scholar]
- Paoletti MG. Dati aggiuntivi alla conoscenza del genere Cansiliella Paoletti (Col. Bathysciinae) Redia Firenze. 1980;63:67–80. [Google Scholar]
- Paoletti MG, Beggio M, Pamio A, Gomiero T, Brilli M, Dreon AL, Toniello V, Engel AS. In: Proc 15th Int Cong Speleol. 1. White WB, editor. Kerrville, Texas; 2009. Comparison of three moonmilk cave habitats associated with troglobitic beetles; pp. 400–403. [Google Scholar]
- Paoletti MG, Beggio M, Dreon AL, Pamio A, Gomiero T, Brilli M, Dorigo L, Concheri G, Squartini A, Engel AS. A new foodweb based on microbes in calcitic caves: The Cansiliella (Beetles) case in northern Italy. Int J Speleol. 2011;40:45–52. doi: 10.5038/1827-806X.40.1.6. [DOI] [Google Scholar]
- Hill CA, Forti P. Cave Minerals of the World. Huntsville, Alabama: National Speleological Society; 1997. p. 446. [Google Scholar]
- Sket B. The cave hygropetric - a little known habitat and its inhabitants. Arch Hydrobiol. 2004;160:413–425. doi: 10.1127/0003-9136/2004/0160-0413. [DOI] [Google Scholar]
- Borsato A, Frisia S, Jones B, van der Borg K. Calcite moonmilk: crystal morphology and environment of formation in caves in the Italian Alps. J Sediment Res. 2000;70:1179–1190. [Google Scholar]
- Northup DE, Dahm CN, Melim LA, Crossey LJ, Lavoie KH, Mallory L, Boston PJ, Cunningham KI, Barn SM. Evidence for geomicrobiological interactions in Guadalupe caves. J Cave Karst Stud. 2000;62:80–90. [Google Scholar]
- Northup DE, Lavoie K. Geomicrobiology of caves: a review. Geomicrobiol J. 2001;18:199–222. doi: 10.1080/01490450152467750. [DOI] [Google Scholar]
- Mulec J, Zalar P, Zupan–Hajna N, Rupnik M. Screening for culturable microorganisms from cave environments (Slovenia) Acta Carsologica. 2002;31:177–187. [Google Scholar]
- Cañaveras JC, Cuezva S, Sanchez-Moral S, Lario J, Laiz L, Gonzalez JM, Saiz-Jimenez C. On the origin of fiber calcite crystals in moonmilk deposits. Naturwissenschaften. 2006;93:27–32. doi: 10.1007/s00114-005-0052-3. [DOI] [PubMed] [Google Scholar]
- Blyth AJ, Frisia S. Molecular evidence for bacterial mediation of calcite formation in cold high-altitude caves. Geomicrobiol J. 2008;25:101–111. doi: 10.1080/01490450801934938. [DOI] [Google Scholar]
- Engel AS, Paoletti MG, Beggio M, Dorigo L, Pamio A, Gomiero T, Furlan C, Brilli M, Dreon AL, Bertoni R, Squartini A. Comparative microbial community composition from secondary 1 carbonate (moonmilk) deposits: implications for the Cansiliella servadeii cave hygropetric food web. International Journal of Speleology. 2013. in press.
- Moldovan OT, Jalzic B, Erichsen E. Adaptation of the mouthparts in some subterranean Cholevinae (Coleoptera, Leiodidae) Nat Coroat. 2004;13:1–18. [Google Scholar]
- Jeannel R. Monographie des Bathsyciinae. Arch Zool Exp Gén. 1924;63:1–436. [Google Scholar]
- Remy P. Sur le mode de vie des Hadesia dans la grotte Vjetrenica. Rev France Entomol. 1940;7:1–8. [Google Scholar]
- Giachino PM, Vailati D. Kircheria beroni, a new genus and new species of subterranean hygropetricolous Leptodirinae from Albania. Subterranean Biol. 2006;4:103–116. [Google Scholar]
- Gasparo F. La grotta della Foos presso Campone (Prealpi Carniche) Mondo Sotterraneo. 1971;1:37–52. [Google Scholar]
- Palmano S, Firrao G, Locci R. Sequence analysis of domains III and IV of the 23S rRNA gene of verticillate streptomycetes. Int J Syst Evol Microbiol. 2000;50:1187–1191. doi: 10.1099/00207713-50-3-1187. [DOI] [PubMed] [Google Scholar]
- Osborn AM, Moore ERB, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Non-parametric estimation of the classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- Magurran AE. Measuring biological diversity. Oxford, UK: Blackwell Publishing; 2004. p. 256. [Google Scholar]
- Andert J, Marten A, Brandl R, Brune A. Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.) FEMS Microbiol. 2010;74:439–449. doi: 10.1111/j.1574-6941.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- Schmitt-Wagner D, Friedrich MW, Wagner B, Brune A. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.) Appl Environ Microbiol. 2003;69:6007–6017. doi: 10.1128/AEM.69.10.6007-6017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, Stingl U, Dyhrberg Bruun L, Pommerenke B, Brune A, Friedrich MW. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae) Appl Environ Microbiol. 2005;71:4556–4566. doi: 10.1128/AEM.71.8.4556-4566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, Wagner B, Lemke T, Brune A, Friedrich MW. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae) Appl Environ Microbiol. 2003;69:6659–6668. doi: 10.1128/AEM.69.11.6659-6668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MD. Breznak JA Effect of host diet on production of organic acids and methane by cockroach gut bacteria Appl Environ Microbiol. 1991. pp. 2628–2634. [DOI] [PMC free article] [PubMed]
- Santo-Domingo JW, Kaufman MG, Klug MJ, Holben WE, Haris D, Tiedje JM. Influence of diet on the structure and function of the bacterial hindgut community of crickets. Mol Ecol. 1998;7:761–767. doi: 10.1046/j.1365-294x.1998.00390.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cluster analysis dendrogram obtained with the first 46 screened clones, Gram-negative portion.
Cluster analysis dendrogram obtained with the first 46 screened clones, Gram-positive portion.
Rarefaction curve for OTUs defined at 81% similarity.