Abstract
Background
Phylogenetic group D Escherichia coli clones (ST69, ST393, ST405) are increasingly reported as multidrug resistant strains causing extra-intestinal infections. We aim to characterize inter- and intraclonal diversity of a broad sample (isolates from different geographic locations and origins with variable antibiotic resistance profiles, 1980-2010) and their ability to adhere and form biofilm by both a modified quantitative biofilm producing assay and Field Emission Scanning Electron Microscopy (FESEM).
Results
High virulence scores were observed among ST69 (median 14/range 9–15) and ST393 (median 14/range 8–15) clones, particularly enriched in pap alleles, iha, kpsMTII-K5 and ompT, in contrast with ST405 (median 6/range 2–14) isolates, exhibiting frequently fyuA, malX and traT. All ST69 and ST393 and only two ST405 isolates were classified as ExPEC. Biofilm production was detected in two non-clinical ST69 and three ST393 isolates from different origins showing variable virulence profiles. Within each clonal group, and despite the high diversity of PFGE-types observed, isolates from different countries and recovered over large periods of time were clustered in a few groups sharing common virulence gene profiles among ST69 (n = 10 isolates) and ST393 (n = 9 isolates) (fimH-iha-iutA-kpsMTII-K5-(traT)-sat-(ompT)-papA-papEF-papGII-papC) or ST405 (n = 6 isolates) (fimH-traT-fyuA-malX).
Conclusions
This study highlights the circulation of highly transmissible ST69, ST393 and ST405 variants among different settings. Biofilm production seems not to be directly correlated with their epidemiological success.
Keywords: ExPEC, High-risk clones, ESBL, Virulence, Adhesion, Biofilm
Background
Multidrug resistant Escherichia coli clones of the phylogenetic group D causing extraintestinal human infections are increasingly reported all over the world [1-4]. Among them, E. coli clonal groups D-ST69 (also recognized as clonal group A or CGA) and D-ST393 (also known as O15:K52:H1 clonal group) are widely spread among different hosts, often causing urinary tract infections (UTI) and conferring resistance to antibiotics [5-10]. Isolates belonging to the D-ST405 have been involved in the spread of genes encoding extended spectrum β-lactamases (ESBLs) (mainly CTX-M enzymes), cephamycinases (AmpC), carbapenemases (NDM) or methylases (AmrA, RmtB) [2,11-15]. Recent studies have reported variants of ST69, ST393 and ST405 among ESBL and non-ESBL-producing strains from specific locations [4,5,8,9,13,16]. The presence of genes possibly involved in biofilm production detected in some of these surveys (fimH, papC, papG, fyuA or kpsMT II) suggests the ability of these clones to adhere and form biofilm, which could be favouring their persistence; however, such ability has not been specifically evaluated [5,8,13,17,18]. Biofilm growing ability of bacteria is commonly assessed by a quantitative measure of their adherence to microtiter plates, although electron microscopy analyses provide more accurate information on the biofilm structure and presence of matrix [17-19].
In this study, we aim to characterize the intraclonal diversity of extraintestinal pathogenic E. coli (ExPEC) isolates from phylogenetic group D (ST69, ST393, ST405) isolated from different geographic locations and sources, and to assess their ability to adhere and form biofilm on abiotic surfaces in order to evidence a possible contribution of biofilm formation to their persistence and epidemicity.
Methods
Bacterial isolates
We analysed thirty-five E. coli isolates belonging to ST69 (n = 13), ST393 complex (10 ST393, 1 ST2321) and ST405 complex (10 ST405, 1 ST964) isolated from multiple sources and countries. They include either isolates associated with nosocomial or community outbreaks in different countries [2,4,9,12,20] or isolates collected from non-clinical sources from distinct countries and showing variable antibiotic resistance profiles, selected from published papers by the end of 2010. They were recovered from nosocomial (66%) and community-acquired infections (17%), healthy volunteers (8%), food products (6%) or animals (3%) and produced diverse ESBL or AmpC enzymes. Their epidemiological features are shown in Table 1. Isolates were taken as part of standard patient care and no ethical approval was required for their use.
Table 1.
Epidemiological data and diversity among ST69, ST393 and ST405 clonal groups
| ST (N°) | Serotype | PFGE-type (Cluster) | Country (N° isolates) | Date | Origina | Source (N° isolates) | ESBL / AmpC | Antibiotic resistance profileb | Virulence gene profileb | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 69 |
O11, O73, O77 |
69_1 (I) |
US (4), NW (1) |
1999-2002 |
H |
Urine (3), blood (2) |
|
(Cm), Sm, Su, (Te), Tp, Ts |
fimH,iha, iutA, kpsMTII, K5, traT, sat, (ompT), (fyuA), papA, papEF, papGII, papC,(papGIII), (iroN), (iss), (ireA) |
[20,21] |
| 69 |
O77 |
69_2 (I) |
SP (1) |
- |
H |
Urine |
- |
Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, traT, sat, ompT, fyuA, papA, papEF, papGII, papC, papGIII |
[20] |
| 69 |
- |
69_3 (I) |
BR (1) |
- |
H, C |
- |
- |
(Ak), Cm, (Gm), (Km), (Nt), Sm, Su, (Tb), Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, traT, sat, ompT, fyuA, papA, papEF, papGII, papC |
[12,22] |
| NW (1) |
2006 |
Urine |
CMY-2 |
|||||||
| 69 |
- |
69_4 (I) |
PT (1) |
2007 |
H |
Urine |
- |
Cp, Na, Sm, Su, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, traT, sat, ompT, papEF, iroN, iss |
This study |
| 69 |
O17 |
69_5 (I) |
US (1) |
- |
H |
Blood |
- |
Cm, Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, traT, sat, ompT, fyuA, papA, papEF, papGII, papC, papGIII |
[20] |
| 69 |
- |
69_6 (II) |
PT (2) |
2010 |
S |
- |
- |
Km, Sm, Su, Te, Tp, Ts |
fimH, kpsMTII, K5, traT, ompT, papA, papEF, papC, papGIII, bmaE, gafD, iroN, iss |
[23] |
| 69 |
- |
69_7 (II) |
NW (1) |
2002 |
A |
Poultry meat |
- |
Sm, Su, Te, Ts |
fimH, iutA, traT, ompT, papC, iroN, iss, tsh, ireA |
[21] |
| 393 |
O15 |
NAc |
US (1) |
1980 |
H |
- |
- |
- |
fimH, iha, iutA, kpsMTII, K5, sat, ompT, fyuA, papA, papEF, papGII, papC, papGIII, tsh |
[4] |
| 393 |
O15 |
NAd |
US (1) |
1998 |
H |
- |
- |
Cm, Gm, Km, Nt, Sm, Su, Tb, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, sat, ompT, fyuA, papA, papEF, papGII, papC |
[4] |
| 393 |
O15 |
NAe |
US (1) |
1999 |
H |
- |
|
Cp, Km, Na, Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, sat, fyuA, papA, papEF, papGII, papC, tsh, papGI |
[4] |
| 393 |
O15 |
NAe |
KO (1) |
2006-7 |
C |
Urine |
- |
Cp, Gm, Km, Na, Nt, Sm, Su, Tb, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, K1, traT, sat, ompT, fyuA, papA, papEF, papGII, papC |
[9] |
| 393 |
O15 |
NAe |
FR (1) |
2006 |
F |
Feces |
- |
Cm, Cp, Gm, Km, Na, Nt, Sm, Su, Tb, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, traT, sat, ompT, papA, papEF, papGII, papC, iss, tsh |
[24] |
| 393 |
O25 |
NA |
FR (1) |
2006 |
F |
Feces |
- |
Cp, Na, Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, sat, ompT, fyuA, papA, papEF, papGII, papC, iss |
[24] |
| 393 |
O15 |
NAe |
FR (1) |
2006 |
F |
Feces |
- |
Cm, Cp, Gm, Km, Nt, Sm, Su, Tb, Te, Ts |
fimH, iha, iutA, kpsMTII, K5, K1, traT, sat, fyuA, papA, papEF, papGII, papC, iss, tsh |
[24] |
| 393 |
O15 |
NA |
SP (1) |
2002 |
C |
Urine |
CTX-M-14 |
Cp, Na, Sm, Su, Ts |
fimH, iha, iutA, kpsMTII, K5, sat, papA, papEF, papGII, papC, iss, tsh |
[25] |
| 393 |
O15 |
NAe |
KO (1) |
2006-7 |
C |
Urine |
- |
Cp, Km, Na, Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, papEF, papGII, papC |
[9] |
| 393 |
O15 |
NAe |
NW (1) |
2005 |
H |
Urine |
CMY-2 |
Cp, Km, Na, Nf, Sm, Su, Tp, Ts |
fimH, iutA, kpsMTII, K5, K1, ompT, fyuA, iss, tsh |
[12] |
| 2321 |
O25 |
NAe |
PT (1) |
2008 |
H |
Urine |
TEM-like |
Cp, Na, Sm, Su, Te, Tp, Ts |
fimH, iha, iutA, kpsMTII, K5, K1, sat, fyuA, papGII, papC, papGIII, iss, tsh, malX, iroN |
This study |
| 405 |
- |
405_1 (I) |
SP (1), KU (1)f NW (1) |
2002-2004 |
H |
Wound (1) Urine (1) Respiratory (1) |
CTX-M-15 (2), CTX-M-3 (1) |
(Cp), Cm, Gm, Km, Na, (Nt), Sm, Su, Tb, Te, Tp, Ts |
FimH, (iha), iutA, (kpsMTII), traT, sat, (malX), fyuA, (kpsMTIII), (iss), (tsh) |
[2,12] |
| 405 |
- |
405_2 (I) |
NW (1) |
2003 |
H |
Wound |
CTX-M-14 |
Cm, Km, Sm, Su, Te, Tp, Ts |
fimH, traT, malX, fyuA, kpsMTIII |
[12] |
| 405 |
- |
405_3 (I) |
NW (1) PT (1) f |
2003-2006 |
H |
Abcess (1) Urine (1) |
CMY-2 (1) CTX-M-15 (1) |
(Gm), (Km), Na, Sm, Su, (Tb), Te, Tp, Ts |
fimH, (iha), iutA, (kpsMTII), traT, malX, fyuA, (papEF), (papGIII), (iroN), (iss), (tsh), (ireA) |
[2,12] |
| 405 |
- |
405_4 (II) |
SW (1) SP (1) |
2000-2005 |
H, C |
Urine (1) Blood (1) |
CTX-M-15 TEM-24 |
(Cm), Cp, (Gm), Km, Na, (Nt), Sm, Su, Te, Tb, Tp, Ts |
(fimH), (iutA), traT, malX, fyuA, kpsMTIII, (iss), (tsh) |
[2,26] |
| 405 |
- |
405_5 (II) |
SP (1) |
2001 |
H |
Wound |
TEM-52 |
Cm, Cp, Gm, Km, Na, Sm, Su, Tb, Te, Ts |
traT, fyuA, kpsMTIII |
This study |
| 405 |
- |
NA |
SP (1) |
2008 |
C |
Urine |
CTX-M-15 |
Ak, Cp, Gm, Na, Tb, Ts |
fimH, fyuA |
[27] |
| 964 | - | 405_7 (III) | NW (1) | 2003 | H | Respiratory | CTX-M-15 | Cm, CpGm, Km, Na, Nf, Sm, Su, Tb, Te, Tp, Ts, | fimH, traT, sat, fyuA, papEF | [12] |
aH Hospitalized humans (obtained from representative outbreaks), C Community acquired infections (obtained from representative outbreaks), F Healthy humans (feces), A Animals, S Ready-to-eat salads. b Variability among isolates is represented in parenthesis. cIsolates identified as biotype A, dIsolates identified as biotype B; eIsolates identified as biotype C. f Isolate considered ExPEC. ND Not determined, NA, Not applicable, Ak Amikacin, Cm Chloramphenicol, Cp Ciprofloxacin, Gm Gentamicin, Km Kanamycin, Na Nalidixic acid, Nt Netilmicin, Nf Nitrofurantoin, Sm Streptomycin, Su Sulphonamides, Tb Tobramycin, Te Tetracyclin, Tp Trimethoprim, Ts Trimethoprim-Sulfamethoxazole, Definitions: fimH (type 1 fimbriae), papA (P fimbriae major subunit, pyelonephritis-associated), papC (P fimbriae assembly), papEF (P fimbriae minor tip pilins), papG allele I (papG variant), papG allele II (papG variant, pyelonephritis-associated), papG allele III (P fimbriae adhesion, cystitis-associated), sfa/focDE (S and F1C fimbriae), bmaE (Blood group M-specific adhesin), gafD (glucosamine-specific adhesin), iha (iron-regulated-gene-homologue adhesion), sat (secreted autotransporter toxin), tsh (serine protease autotransporter), fyuA (yersiniabactin receptor) iutA (ferric aerobactin receptor), iroN (catecholate siderophore receptor), ireA (Iron-regulated element ), kpsMTII (group II capsular polysaccharide), kpsMTII K1 (variant K1), kpsMTII K5 (variant K5), kpsMTIII (group III capsular polysaccharide), traT (serum survival associated), iss (increased serum survival), usp (uropathogenic-specific protein), ompT (outer membrane protease), malX (pathogenicity-associated island marker.
Clonal diversity
Relatedness among isolates was established by XbaI-pulsed-field gel electrophoresis (PFGE), multi-locus sequence typing (MLST, http://mlst.ucc.ie/mlst/dbs/Ecoli), and identification of E. coli phylogenetic groups and serogroups by PCR [28]. Isolates exhibiting ≥85% homology were considered to belong to the same PFGE-type. XbaI-profiles were compared using InfoQuest™ FP version 5.4 software (BioRad Laboratories), by applying the UPGMA algorithm based on the Dice coefficient (1.0% band tolerance; 1.0% optimization).
Virulence genes profile
Screening of 38 virulence factors (VFs) including adhesins, toxins, siderophores, polysaccharide coatings and others (malX, usp, ibeA, iss, tsh) presumptively associated with ExPEC isolates was performed by PCR as previously described [8,28]. The Fisher’s exact test was used for each comparison, a p value <0.05 being considered to reveal significant differences. A strain satisfied the criteria for being ExPEC if it carried two or more of the following genes: papA, papC, sfa/focDE, afa/draBC, iutA and kpsMII[8].
Adhesion and biofilm-producing assays
The ability of D-E.coli strains to in vitro adhere was investigated by a modified quantitative biofilm production assay, as previously described [28]. The E. coli strain CFT073 and the culture medium supplemented with 1% (v/v) glucose were used as positive and negative controls, respectively. Assays were performed in quintuplicate and repeated at least 4 times. The cut-off optical density (ODc) was defined as three standard deviations above the mean OD of the negative control (culture medium), and strains were classified as non-adherent (OD ≤ ODc), weakly adherent (ODc < OD ≤ 2 × ODc), moderately adherent (2 × ODc < OD ≤ 4 × ODc), or strongly adherent (OD > 4 × ODc). The ultrastructural analysis of biofilm was performed by a Field Emission Scanning Electron Microscope (FESEM) (Zeiss, Germany). Briefly, adjusted inocula (200 μl, 0.5 McF) of each strain diluted with 1.8 ml of fresh LB supplemented with 1% (v/v) glucose were added to 24-well plates with round glass coverslips (1 cm diameter) put into each well and incubated at 37°C for 24 h. The content of each well was removed and the round coverslips were washed with PBS (1%) twice. Biofilms grown on coverslips were fixed with 2,5% glutaraldehyde in Na-cacodylate 0,1 M (pH 7.4) buffer solution (AppliChem, Germany) for 2 h at room temperature. Following three washing steps with the same buffer solution, samples were dehydrated through graded ethanol (30°, 50°, 70°, 85°, 95°, 100°) and dried with hexamethyldisilazane (Alfa Aesar, USA) for 1 h30'. Samples were air dried overnight and coated by sputtering with a gold target [19].
Results and discussion
Diversity among clonal groups of E. coli phylogroup D
Isolates belonging to the three analysed STs exhibited inter and intraclonal variability regarding the VF profile and the ability to form biofilm. On the basis of their virulence scores, all ST69 (n = 13/13; median = 14/range = 9-15) and all ST393 (n = 11/11; median = 14/range = 8-15), and only sporadic ST405 (n = 2/11; median = 6/range = 2-14) isolates were classified as ExPEC (Table 2). While most ST69 and ST393 carried pap alleles (papA, papC, papEF, papG II), iha, kpsMTII-K5 and ompT, ST405 isolates frequently contained fyuA, malX and traT, suggesting the presence of different genomic islands among E. coli phylogroup D isolates.
Table 2.
Virulence gene profiles of phylogenetic group D E. coli clonal groups
| Virulence genesa |
N° of isolates (%) |
P valuea |
|||||
|---|---|---|---|---|---|---|---|
| ST69 (n = 13) | ST393 (n = 11) | ST405 (n = 11) | ST69 vs ST393 | ST69 vs ST405 | ST393 vs ST405 | ||
|
Adhesins |
|
|
|
|
|
|
|
| |
fimH |
13 (100%) |
11 (92%) |
9 (82%) |
0.480 |
0.199 |
0.590 |
| |
papA |
11 (85%) |
8 (67%) |
0 (0%) |
0.378 |
0.000 |
0.001 |
| |
papC |
12 (92%) |
10 (83%) |
0 (0%) |
0.593 |
0.000 |
0.000 |
| |
papEF |
12 (92%) |
9 (75%) |
2(18%) |
0.322 |
0.001 |
0.012 |
| |
papG allele I |
0 (0%) |
1 (8%) |
0 (0%) |
0.480 |
- |
1.000 |
| |
papG allele II |
9 (69%) |
10 (83%) |
0 (0%) |
0.645 |
0.001 |
0.000 |
| |
papG allele III |
9 (69%) |
2 (17%) |
1 (9%) |
0.015 |
0.005 |
1.000 |
| |
bmaE |
2 (15%) |
0 (0%) |
0 (0%) |
0.480 |
0.482 |
- |
| |
gafD |
2 (15%) |
0 (0%) |
0 (0%) |
0.480 |
0.482 |
- |
| |
iha |
10 (77%) |
10 (83%) |
2 (18%) |
1.000 |
0.012 |
0.003 |
|
Toxins |
|
|
|
|
|
|
|
| |
sat |
10 (77%) |
9 (75%) |
6 (55%) |
1.000 |
0.390 |
0.400 |
| |
tsh |
1 (8%) |
7 (58%) |
3 (27%) |
0.011 |
0.300 |
0.214 |
|
Siderophores |
|
|
|
|
|
|
|
| |
fyuA |
8 (62%) |
8 (67%) |
11 (100%) |
1.000 |
0.041 |
0.093 |
| |
iutA |
11 (85%) |
11 (92%) |
6 (55%) |
1.000 |
0.182 |
0.069 |
| |
iroN |
5 (39%) |
1 (8%) |
1 (9%) |
0.160 |
0.166 |
1.000 |
| |
ireA |
2 (15%) |
0 (0%) |
1 (9%) |
0.480 |
1.000 |
1.000 |
|
Capsule |
|
|
|
|
|
|
|
| |
kspMT II |
12 (92%) |
11 (100%) |
2 (18%) |
1.000 |
0.001 |
0.000 |
| |
kpsMT III |
0 (0%) |
0 (0%) |
5 (46%) |
- |
0.011 |
0.014 |
| |
K1 |
0 (0%) |
4 (33%) |
0 (0%) |
0.039 |
- |
0.093 |
| |
K5 |
12 (92%) |
11 (100%) |
0 (0%) |
1.000 |
0.000 |
0.000 |
|
Protectins |
|
|
|
|
|
|
|
| |
traT |
13 (100%) |
3 (25%) |
10 (91%) |
0.000 |
0.458 |
0.003 |
| |
iss |
5 (39%) |
6 (50%) |
3 (27%) |
0.695 |
0.679 |
0.400 |
|
Miscellaneous |
|
|
|
|
|
|
|
| |
usp |
1 (8%) |
0 (0%) |
0 (0%) |
1.000 |
1.000 |
- |
| |
ompT |
12 (92%) |
6 (50%) |
0 (0%) |
0.030 |
0.000 |
0.014 |
| |
malX (PAI) |
0 (0%) |
1 (8%) |
7 (64%) |
0.480 |
0.001 |
0.009 |
| ExPEC statusb |
12 (100%) |
11 (100%) |
2 (18%) |
- |
0.000 |
0.000 |
|
| Virulence score |
13.23 (± 1.641) |
11.67 (± 3.576) |
6.27 (± 3.197) |
1.000 |
0.007 |
0.053 |
|
| Range | 9 – 15 | 8 – 15 | 2 – 14 | - | - | - | |
ap values (Fisher’s exact test) are shown in bold when p < 0.05. b ExPEC status defined by the presence of two or more among papA, papC, sfa/foc, afa/draBC, iutA and kpsMTII, as suggested [8].
Most of the isolates exhibited a weak adherence ability to abiotic surfaces (9 ST69, 8 ST393, 9 ST405; 0.13 < OD < 0.27) while a few strains were classified as moderately adherent (3 ST393, 2 ST69 and 1 ST405; 0.29 < OD < 0.47) or strongly adherent (2 ST69, 1 ST405; 0.49 < O.D < 0.71) (Figure 1), and were considered as presumptive biofilm producers. Among all the strains resulting to be moderately or strongly adherent, FESEM observations revealed the presence of aggregates and EPS matrix, both compatible with a biofilm development, only in two ST69 (69PT1S, 69PT2S) and three ST393 (393FR3F, 393N1H, 2321PT1H) isolates (Figure 2). These isolates corresponded to diverse clonal variants exhibiting variable virulence gene profiles, preventing from establishing a link between this phenotype and a given virulence gene or virulence gene profile.
Figure 1.
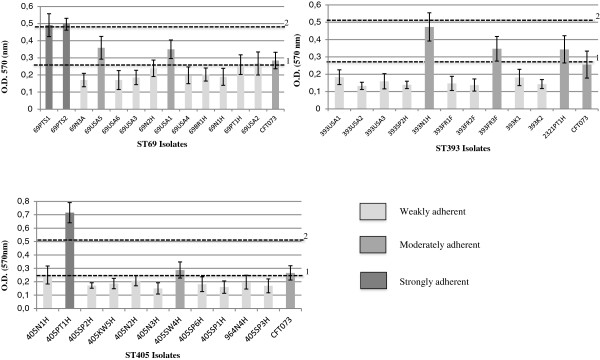
Quantitative biofilm-producing assay. The vertical axis represents the median optical density (OD) of at least 15 replicas of each isolate, determined at 570 nm. E. coli CFT073 was used as a positive control. Horizontal dotted lines represent the cut-off value between weakly adherent (light gray) and moderately adherent (gray) (1) and strongly adherent strains (dark grey) (2).
Figure 2.
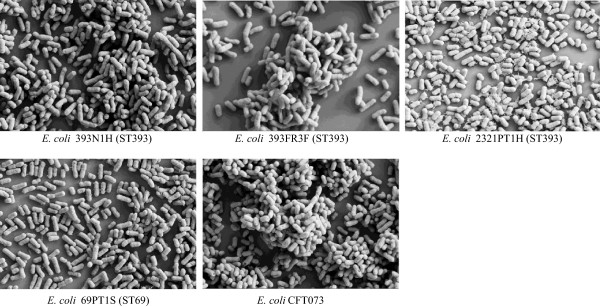
Biofilms of strongly and moderately adherent E. coli strains. FESEM micrographs of biofilm-growing E. coli strains were obtained at a magnification of 10.000 x using an EHT = 5.00 kV.
The presence of a characteristic virulence gene profile for isolates of different E. coli clonal groups confirms results obtained in previous studies [5,8]. However, small differences in the virulence profile observed among closely related isolates might be explained by the plasticity of the genomic islands where these genes are commonly clustered [29]. Most of the isolates were associated with extraintestinal infections (n = 25/35; 71%), including the non-ExPEC ST405 isolates. The lack of correspondence between ExPEC status and the ability to cause extraintestinal disease further suggests that other non-explored virulence factors might influence their pathogenicity [30]. Our results indicate that biofilm production seems not to be directly related with their epidemiological success, as already observed for the pandemic ST131 E. coli clone [28]. Moreover, when observed in particular strains, this feature could not be linked to a specific virulence gene or virulence profile.
Intraclonal diversity of ST69 isolates
Thirteen isolates corresponding to 7 PFGE types were classified in different serogroups (O11, O17, O73, O77), and clustered in two groups on the basis of the similarity of the XbaI restriction profiles. Cluster I comprised closely related isolates (n = 10, 73.8% homology) causing hospital or community acquired infections that exhibited a common virulence gene profile (80%, fimH-iha-iutA-kpsMTII-K5-traT-sat-ompT-papA-papEF-papGII-papC). Cluster II (n = 3, 71.8% homology) included two indistinguishable isolates recovered from different samples of ready-to-eat salads in Portugal and from poultry meat in Norway. They differ in the presence of iroN, iss, bmaE (n = 2/3) and gafD (n = 2/3), and the lack of iha, sat and papGII, observed for isolates of cluster I. All ST69 isolates exhibited resistance to streptomycin and trimethoprim-sulfamethoxazole, and they were frequently resistant to tetracycline (85%), and to chloramphenicol (46%). None of the isolates produced ESBL, but one encoded CMY-2.
Isolates belonging to cluster I seem to have been circulating among different continents since at least 1999, as reflects this and other studies [31-33]. Despite of the small sample analysed, differences among ST69 isolates from human and non-human origins suggest independent evolution of particular E. coli variants in different hosts.
Intraclonal diversity of ST393 isolates
These isolates corresponded to serogroups O15 (n = 9) or O25 (n = 2, one of them corresponding to ST2321, a single locus variant of ST393), and they mainly were biotype C (non-lactose fermenters and maltose fermenters; n = 7, 58.3%), which seem to be more commonly observed than those of biotype A (lactose and maltose fermenters) [4,6,34]. Most isolates analysed (n = 9/75%) were recovered from patients and healthy individuals in France, Spain, Korea and the USA and shared a pool of ten virulence genes (fimH-iha-iutA-kpsMTII-K5-sat-papA-papEF-papGII-papC) (Table 1). The ST2321 isolate belonged to O25 serotype and shared eight out of the ten frequent VFs, suggesting a common origin. Most isolates were resistant to trimethoprim-sulfamethoxazole (91%), streptomycin (91%), ciprofloxacin (82%), tetracycline (73%) and nalidixic acid (73%). Resistance against kanamycin (64%), gentamicin (36%), tobramycin (36%), netilmicin (36%) or chloramphenicol (27%) was also observed. ESBL or AmpC production was sporadically detected (1 CTX-M-14, 1 TEM-like and 1 CMY-2).
The study highlights the spread of ST393 isolates of biotype C with highly similar virulence gene profile in different continents over almost three decades, supporting previous observations in specific countries [5,8]. Unfortunately, clonal relatedness among different strains could not be analysed due to the spontaneous lysis of DNA, also reported by other groups [6,34].
Intraclonal diversity of ST405 isolates
Isolates of this clonal complex (n = 11, 6 PFGE types) were recovered from human infections (82% hospital, 18% community), and exhibited a common virulence profile (fimH-traT-fyuA-malX, n = 6, 55%) (Table 1). Most isolates belonging to cluster I (n = 6, 2 ExPEC; 77% homology) identified in hospitalized patients from Portugal, Spain, Norway and Kuwait contained additionally iutA and sat (n = 5/6, 83%) whereas cluster II (n = 3 from Spain and Switzerland; 80% homology) showed consistently kpsMTIII but not iutA and sat. Cluster III comprised only one isolate from Norway corresponding to a single locus variant of ST405 (ST964). ST405 isolates were commonly resistant to streptomycin, sulphonamides, trimethoprim (91% each), kanamycin, tetracycline, nalidixic acid (82% each), gentamicin (73%), tobramycin (64%), ciprofloxacin (45%) and chloramphenicol (45%) (Table 1).
These results suggest that several ST405 variants seem to be circulating in distinct countries. In contrast with ST69 and ST393, isolates frequently produced either ESBLs (mostly CTX-M-15, but also CTX-M-3, CTX-M-14, TEM-24 or TEM-52) or AmpC (CMY-2) enzymes, which might have facilitated the selection and successful spread of diverse ST405 variants [2,13,14,35].
Conclusion
Factors responsible for the increased ability of particular E. coli clones to successfully spread and persist are poorly understood, and our work represents one of the few studies exploring the phenotypic traits involved in the increased epidemicity of emerging antibiotic resistant E. coli clonal groups [28,36]. The results highlight the inter and intraclonal diversity of E. coli clones of phylogroup D and further suggest the circulation of highly transmissible ST69, ST393 and ST405 variants, some of them being particularly widespread in different geographic areas and settings. The lack of association between the ability to produce biofilm exhibited by a few strains and specific virulence gene or virulence gene profiles points out the need to further explore factors involved in the selection of particular epidemic variants with enhanced ability to colonize and persist for extended periods of time.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
AN was responsible for study conception and design, data acquisition and analysis and drafted the manuscript. LP participated in the conception and design, analysis of data and preparation of the manuscript. CV, JP and CM contributed with data acquisition and analysis. TC and GD were implicated in data analysis and preparation of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ângela Novais, Email: angelasilvanovais@gmail.com.
Claudia Vuotto, Email: c.vuotto@hsantalucia.it.
João Pires, Email: jp.dcpires@gmail.com.
Carolina Montenegro, Email: carolina8.montenegro@gmail.com.
Gianfranco Donelli, Email: g.donelli@hsantalucia.it.
Teresa M Coque, Email: teresacoque@gmail.com.
Luísa Peixe, Email: lpeixe@ff.up.pt.
Acknowledgements
We thank (in alphabetical order) Anette Hammerum (Statens Serum Institut, Denmark), So Hyun Kim (Asian Bacterial Bank of the Asia Pacific Foundation for Infectious Diseases), Marie-Hélène Nicolas-Chanoine (Hôpital Beaujon, France), Lee W. Riley (School of Public Health, Berkeley, Canada), Flemming Scheutz (WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella, Copenhagen, Denmark), Arnfinn Sundsfjord (Department of Medical Biology, University of Tromsø, Norway), for the strains’ gift. We are also grateful to Dr. Martinotti for the gift of E. coli CFT073 and Carla Rodrigues for statistical analysis support. This work was supported by Fundação para a Ciência e Tecnologia (grants no. PEst-C/EQB/LA0006/2011, PTDC/AAC-AMB/103386/2008, EXPL/DTP-EPI/0196/2012 and FCOMP-01-0124-FEDER-027745) and Universidade do Porto/Santander TOTTA (grant no. PP-IJUP2011-277). AN was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (PIEF-GA-2009-255512) and an ESCMID research grant 2012. Work in Teresa M. Coque´s lab is funded by grants from the European Union (EVOTAR-LSHM-2011-282004), the Ministry of Economy and Competitiveness-ISCIII of Spain (PI12/01581) and the regional government of Madrid (S2010/BMD2414_PROMPT-CM).
References
- Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008;14(2):195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Menard ME, Lauderdale TL, Kosmidis C, Gordon D, Collignon P, Maslow JN, Andrasevic AT, Kuskowski MA. Global distribution and epidemiologic associations of Escherichia coli clonal group A, 1998–2007. Emerg Infect Dis. 2011;17(11):2001–2009. doi: 10.3201/eid1711.110488. [DOI] [PubMed] [Google Scholar]
- Olesen B, Scheutz F, Menard M, Skov MN, Kolmos HJ, Kuskowski MA, Johnson JR. Three-decade epidemiological analysis of Escherichia coli O15:K52:H1. J Clin Microbiol. 2009;47(6):1857–1862. doi: 10.1128/JCM.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, Garcia-Garrote F. et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother. 2011;66(9):2011–2021. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol. 2008;46(8):2605–2612. doi: 10.1128/JCM.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother. 2012;67(2):346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother. 2009;53(7):2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Choi HJ, Choi JY, Song M, Song Y, Kim SW, Chang HH, Jung SI, Kim YS, Ki HK. et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infect. 2010;60(2):146–153. doi: 10.1016/j.jinf.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Hammerum AM, Frimodt-Moller N. Detection of clonal group A Escherichia coli isolates from broiler chickens, broiler chicken meat, community-dwelling humans, and urinary tract infection (UTI) patients and their virulence in a mouse UTI model. Appl Environ Microbiol. 2010;76(24):8281–8284. doi: 10.1128/AEM.01874-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother. 2011;66(6):1263–1268. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, Simonsen GS, Sundsfjord A. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2009;117(7):526–536. doi: 10.1111/j.1600-0463.2009.02465.x. [DOI] [PubMed] [Google Scholar]
- Shin J, Kim DH, Ko KS. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J Infect. 2011;63(1):39–47. doi: 10.1016/j.jinf.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother. 2011;66(9):2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- Tian GB, Rivera JI, Park YS, Johnson LE, Hingwe A, Adams-Haduch JM, Doi Y. Sequence type ST405 Escherichia coli isolate producing QepA1, CTX-M-15, and RmtB from Detroit, Michigan. Antimicrob Agents Chemother. 2011;55(8):3966–3967. doi: 10.1128/AAC.00652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Blanco M, Lopez C, Mamani R, Blanco JE, Alonso MP, Garcia-Garrote F, Dahbi G, Herrera A, Fernandez A. et al. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents. 2011;37(1):16–21. doi: 10.1016/j.ijantimicag.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Hancock V, Ferrieres L, Klemm P. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology. 2008;154(Pt 1):167–175. doi: 10.1099/mic.0.2007/011981-0. [DOI] [PubMed] [Google Scholar]
- Naves P, Del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte Mdel C, Soriano F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb Pathog. 2008;45(2):86–91. doi: 10.1016/j.micpath.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol Med Microbiol. 2012;65(2):318–325. doi: 10.1111/j.1574-695X.2012.00962.x. [DOI] [PubMed] [Google Scholar]
- Tartof SY, Solberg OD, Manges AR, Riley LW. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol. 2005;43(12):5860–5864. doi: 10.1128/JCM.43.12.5860-5864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobos M, Christensen H, Sunde M, Nordentoft S, Agerso Y, Simonsen GS, Hammerum AM, Olsen JE. Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology. 2009;155(Pt 3):831–836. doi: 10.1099/mic.0.024190-0. [DOI] [PubMed] [Google Scholar]
- Queiroz ML, Antunes P, Mourao J, Merquior VL, Machado E, Peixe LV. Characterization of extended-spectrum beta-lactamases, antimicrobial resistance genes, and plasmid content in Escherichia coli isolates from different sources in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2012;74(1):91–94. doi: 10.1016/j.diagmicrobio.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Campos J, Peixe L, Mourão J, Pires J, Silva A, Costa C, Nunes H, Pestana N, Novais C, Antunes P. Are ready-to-eat salads an important vehicle of pathogenic and commensal bacteria resistant to antibiotics? Clin Microbiol Infect. 2011;17(4):S703. [Google Scholar]
- Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine MH. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol. 2008;46(12):3900–3905. doi: 10.1128/JCM.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53(12):5204–5212. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A, Baquero F, Machado E, Cantón R, Peixe L, Coque TM. International spread and persistence of TEM-24 is caused by the confluence of highly penetrating enterobacteriaceae clones and an IncA/C2 plasmid containing Tn1696::Tn1 and IS5075-Tn21. Antimicrob Agents Chemother. 2010;54(2):825–834. doi: 10.1128/AAC.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais Â, Viana D, Baquero F, Martínez-Botas J, Cantón R, Coque TM. Contribution of IncFII and broad-host IncA/C and IncN plasmids to the local expansion and diversification of phylogroup B2 Escherichia coli ST131 clones carrying blaCTX-M-15 and qnrS1 genes. Antimicrob Agents Chemother. 2012;56(5):2763–2766. doi: 10.1128/AAC.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L. Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010) Antimicrob Agents Chemother. 2012;56(7):3973–3976. doi: 10.1128/AAC.00475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33(2):376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao BH, Chang YF, Scaria J, Chang CC, Chou LW, Tien N, Wu JJ, Tseng CC, Wang MC, Hsu YM. et al. Identification of Escherichia coli genes associated with urinary tract infections. J Clin Microbiol. 2012;50(2):449–456. doi: 10.1128/JCM.00640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias RC, Marangoni DV, Riley LW, Moreira BM. Identification of uropathogenic Escherichia coli clonal group A (CgA) in hospitalised patients. Memorias do Instituto Oswaldo Cruz. 2009;104(5):787–789. doi: 10.1590/S0074-02762009000500021. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Murray AC, Kuskowski MA, Schubert S, Prere MF, Picard B, Colodner R, Raz R. Distribution and characteristics of Escherichia coli clonal group A. Emerg Infect Dis. 2005;11(1):141–145. doi: 10.3201/eid1101.040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Eng J Med. 2001;345(14):1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- Prats G, Navarro F, Mirelis B, Dalmau D, Margall N, Coll P, Stell A, Johnson JR. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J Clin Microbiol. 2000;38(1):201–209. doi: 10.1128/jcm.38.1.201-209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaila L, Wyplosz B, Clermont O, Garry L, Hipeaux MC, Vittecoq D, Dussaix E, Denamur E, Branger C. Probable intrafamily transmission of a highly virulent CTX-M-3-producing Escherichia coli belonging to the emerging phylogenetic subgroup D2 O102-ST405 clone. J Antimicrob Chemother. 2010;65(7):1537–1539. doi: 10.1093/jac/dkq155. [DOI] [PubMed] [Google Scholar]
- Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61(5):1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]


