Abstract
This chapter will describe the most common immunocytochemical method utilized in the stem cell field – using fluorescently tagged secondary antibodies to detect a primary antibody that is bound to an epitope on a molecule of interest. Secondary antibodies recognize the heavy chain of the primary antibody’s isotype. Generally, these methods employ an incubation period of the sample with the primary antibody, a series of washes to remove unbound primary antibody, a secondary incubation period of the sample with the fluorescently conjugated secondary antibody, followed by washes and preparation for microscopy.
Keywords: immunocytochemistry, antibodies, fluorescent tags, immunofluorescence
1. Introduction
Immunocytochemistry, using antigen-specific antibodies, is a fast and easy way to determine whether a population of cells is homogeneous or heterogeneous with regard to a particular molecular marker. Immunocytochemistry allows for the visualization of individual cells within a colony or culture and thus provides an overall assessment of expression of a particular marker throughout the culture under specific culture conditions (1–4). Antibodies, in combination with specific stains/dyes such as the commonly utilized nuclear stain DAPI, can also reveal the subcellular localization of the particular antigen in question. In addition, translocation of signaling factors from one cellular location to another following signal transduction may be easily examined following staining under alternate conditions. Primary antibodies are raised against an antigen, which may be a protein, glycolipid (such as the SSEA-4 epitope), carbohydrate, small molecule, or DNA.
Antibodies (also known as immunoglobulins, Igs), first described by Paul Ehrlich in 1891, have proven to be one of the most useful research tools available. They are typically made of basic structural units – each with two large heavy chains and two small light chains – to form, for example, monomers with one unit, dimers with two units or pentamers with five units. Antibodies are produced by white blood cells called plasma cells. There are several different types of antibody heavy chains, and several different kinds of antibodies, which are grouped into different isotypes based on which heavy chain they possess.
Though the general structure of all antibodies is very similar, a small region at the tip of the protein is extremely variable, allowing millions of antibodies with slightly different tip structures, or antigen-binding sites, to exist. This region is known as the hypervariable region. Each of these variants can bind to a different target, known as an antigen. The unique part of the antigen recognized by an antibody is called an epitope.
Primary antibodies vary widely in their binding affinities and specificities and must be tested to determine whether they recognize the antigen when the specimen is prepared for immunocytochemistry. Antibodies bind to specific epitopes on antigens. Epitopes may consist of short stretches of amino acids in a protein, conformational characteristics such as an exposed alpha helix, or structural elements of a small molecule. Polyclonal antibodies contain multiple antibodies that usually recognize several different epitopes on a single molecule. In contrast, monoclonal antibodies are of a single defined antibody type and recognize a single epitope on a single molecule (2, 5–8).
Specimens are often described as “weakly positive” or “strongly positive.” When using a new antibody or testing a new sample, it is usually a good idea to confirm the presence of the antigen using an alternate method, such as RT-PCR, if the antigen is a protein. In general, “weakly positive” samples must always be verified. If both mRNA and protein are present in your cells, then there is compelling evidence that the antigen you are examining is present. Other methods used for confirmation of antibody staining include the use of a second antibody that recognizes another epitope on the same molecule, and immunoblots (“Western blots”), in which predefined or predicted molecular weight determination adds confirmation of the identity of the antigen.
Immunocytochemistry for cultured cells uses an amplification technique to make submicroscopic molecules visible. Ideally, every experiment should include negative controls (such as no primary antibody) and positive controls (such as a cell type known to express the antigen) in order to assess the efficacy of staining.
2. Materials
2.1. Preparation of Samples
Chamber Culture slides, Lab-Tek II, (Thermo Fisher Nunc).
Extracellular Matrix Component such as Matrigel, laminin, or fibronectin.
Bromodeoxyuridine (BrdU, Sigma B-9285), 10 µM final concentration.
2.2. Fixation
Fume hood for working with paraformaldehyde.
0.2 M sodium phosphate buffer, pH 7.4.
4% Paraformaldehyde. In the fume hood: Add 40 g of paraformaldehyde to 500 mL of dH2O, heat to 60°C (do not exceed this temperature), and stir. Add a few drops of 1 N NaOH until solution is clear (the solution will not completely clear without the addition of NaOH as the basicity is needed to depolymerize the paraformaldehyde). Filter (0.2 or 0.45 µm) and add 500 mL of 0.2 M sodium phosphate buffer, pH to 7.4 (recheck pH after cooling and adjust if necessary with phosphoric acid or sodium hydroxide). Store at 4°C up to 1 week or alternatively store aliquots at −20°C up to 6 months.
2.3. Immunostaining
Dulbecco’s phosphate-buffered saline with Ca++ and Mg++ (DPBS).
Blocking buffer: DPBS, 0.3% (v/v) Triton X-100, 3% (v/v) serum from secondary antibody host species: rat, mouse, goat, donkey, etc.
Antibody dilution buffer: DPBS, 0.3% (v/v) Triton X-100, 1% (v/v) serum from secondary antibody host species: rat, mouse, goat, donkey, etc.
-
Antibodies, primary and secondary:
Primary antibodies can be purchased from various commercial vendors, such as BD Biosciences, Millipore, R&D Systems, Santa Cruz Biotechnology, Serotec, Sigma, or Developmental Studies Hybridoma Bank, or provided by colleagues.
Secondary antibodies (ex.: AMCA, Cy2, Cy5, RRX, AlexaFluors, DyLights) Jackson ImmunoResearch Laboratories, Invitrogen and other commercial sources.
ProLong Gold antifade reagent (Invitrogen, P-36934).
ProLong Gold antifade reagent with DAPI (Invitrogen, P-36934).
Cover slips, No. 1 thickness range for high magnification objectives (Thermo Fisher Scientific, 12-548-5P).
Nail polish “Clear” Top coat.
Sodium azide (Sigma-Aldrich, S8032).
Hoechst 33342 (Invitrogen, H3570).
2.4. Imaging
Fluorescence microscope.
Objectives: 10×, 20×, 40×, and perhaps 60× or 100×.
Filter cubes appropriate for secondary antibody fluorophores. It is important to make sure that the cubes will give maximal signal for one fluorophore but not allow bleed-through excitation of another fluorophore.
Digital Camera.
Image Pro 4.0 and AFA Plug-in (or other imaging software).
Adobe Photoshop.
3. Methods
The protocol described below, which has routinely produced high quality images for publication, is easy and can be performed by devoting only a short period of time each day. If rapid analysis is desired, the alternative protocol can be used, with timing indicated at the end of each section.
3.1. Preparation of Slides
3.1.1. Growth on Glass surface
Several days prior to staining, passage the cells to Lab-Tek glass chamber slides coated with extracellular matrix such as laminin or a feeder layer of cells, such that the cells will adhere strongly to the surface and not wash off during the staining process. Fluorescent antibody staining on plastic culture dishes is not advised. It is also advisable to incubate the slides in a large (165 mm) culture dish so that the slides do not need to be handled – handling increases the probability of breaking the seal between the wells. For a detailed description of pluripotent stem cell culture on glass slides, see Chapter 12.
3.1.2. Bromodeoxyuridine (BrdU) Labeling
BrdU (10 µM final concentration) should be incubated with the cells for 2–24 h prior to fixation (in some cases it will be desirable to remove the BrdU-containing medium and culture the cells in regular medium for a few days before fixation).
BrdU-labeled cells should be treated with HCl (1 N HCl for 20–30 min at 37°C) after fixation, but prior to blocking and antibody incubation.
Wash well with DPBS after HCl incubation.
3.2. Fixation
Carefully aspirate the growth medium and rinse cells one time with DPBS (see Note 1).
Fix cells for 10 min at room temperature with 4% paraformaldehyde in DPBS (see Note 2). Dispense the solution down the side of the well so that it slowly floods the well without disturbing the cell surface. Use this same gentle technique at all times while adding any solution to the wells.
Wash cells twice with DPBS, allowing the cells to incubate in the wash for approximately 5 min before aspirating the wash.
For best results, stain fixed cells within 24 h of fixation. Alternatively, store fixed cells at 4°C in DPBS, 0.05% (w/v) sodium azide.
3.3. Immunostaining
The method described is used for simultaneous staining with more than one antibody. Staining for more than one antigen involves use of multiple primary antibodies, each of a unique class or animal species, followed by use of multiple secondary antibodies, each specific for one of the primary antibodies and each carrying a unique fluorophore (see Note 3 for a summary of the entire procedure).
3.3.1. Day 1
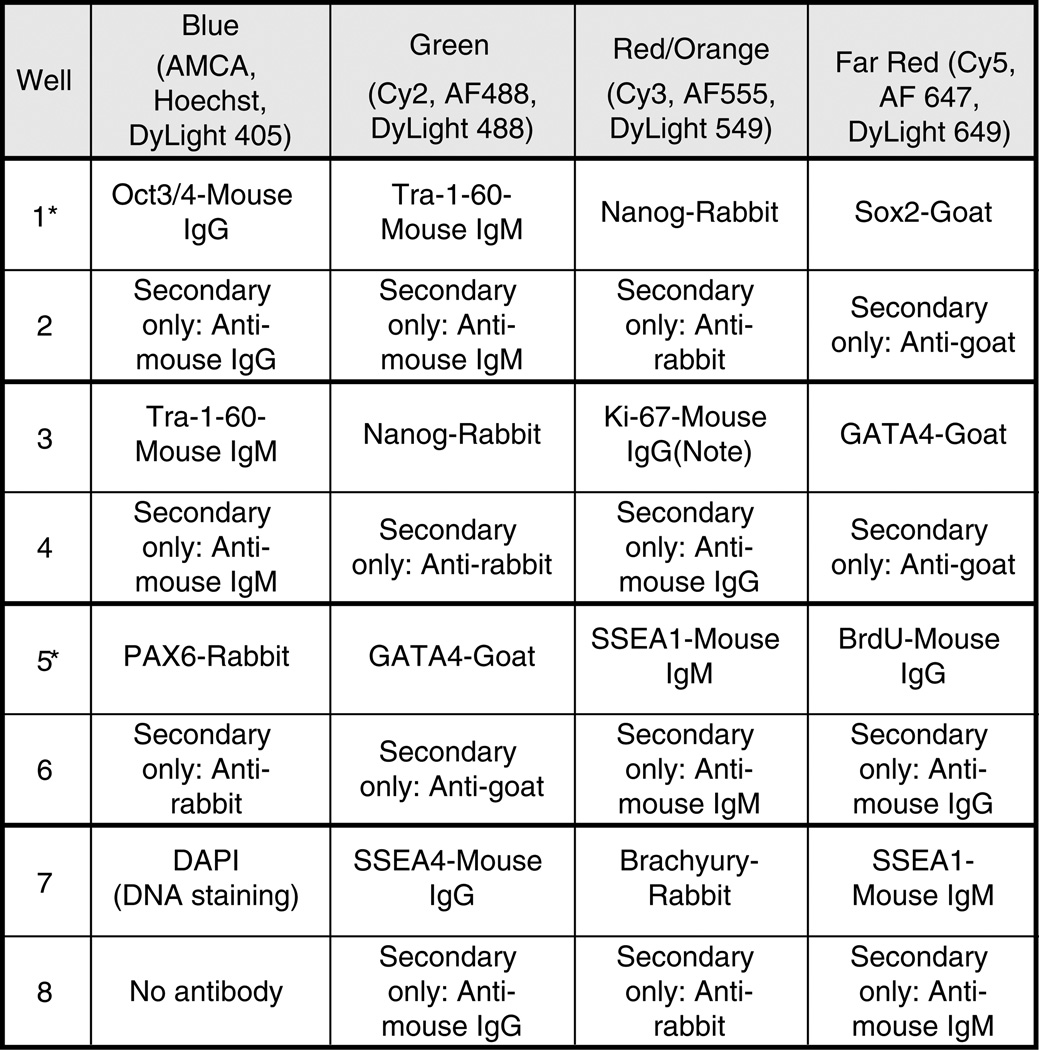
Design a plan for each sample well as in Fig. 1. Make certain antibody isotypes do not overlap within a given well (see Notes 4 and 5).
Aliquot antibody dilution buffer (ADB) into single 0.65 mL micro-centrifuge tubes for each well. If using eight-well culture slides, you will need a final volume of 250 µL per well. For four-well culture slides, use 400 µL per well (adjust volume per well accordingly for wells that are other sizes). Add appropriate volume of primary antibody (or antibodies) to each tube with ADB and gently mix. We typically dilute primary antibodies 1:100. Note that secondary-only control wells (see Fig. 1) should be incubated in ADB alone (no primary antibody) or with a control Ig diluted in ADB.
Remove protein precipitates from the primary antibody solution by spinning at 16,000 × g for 5 min in a microcentrifuge.
Gently remove primary antibodies to new tubes, leaving a small amount of liquid at the bottom where the sediment remains (if the hinge of the tube is placed toward the outside of the rotor, then the sediment, if any, will be directly under the hinge). Keep diluted antibodies on ice until added to cells.
Wash cells gently with DPBS. Note – incubate any BrdU-treated wells with HCl then rinse with DPBS (see notes on BrdU above in Subheading 3.1.2).
Remove DPBS and add approximately 250 µL of Blocking Buffer to each well. Incubate for 15 min at room temperature.
Wash cells gently with DPBS.
Remove DPBS and immediately add the diluted primary antibodies to the wells.
Remove the covers from the (eight-well) slides and place slides into a humidity controlled bin (i.e., covered Tupperware with damp Kimwipes). Condensation on the eight-well slide cover increases the probability of cross-contamination among the wells.
-
Recommended method: Incubate chamber slides overnight at 4°C.
Alternate method: incubate slides 1–2 h at room temperature.
Fig. 1.
An example staining plan for an eight-well slide. Note: This well must be treated with HCl prior to applying primary antibody. See Subheading 3.1.2 above.
3.3.2. Day 2
Dilute secondary antibody (or antibodies, see Table 1 for fluorophore selection criteria) in ADB using the concentration recommended by vendor or determined empirically to give the best results. We typically dilute secondary antibodies 1:250, Alexafluor 1:1,000.
Remove the primary antibody from each well (see Note 6).
Wash cells twice with DPBS. Replace aspirator tips after each use.
Spin secondary antibodies at 16,000 × g for 5 min, to remove any protein precipitates (see steps 3 and 4 of Day 2 above).
Carefully add secondary antibodies to aspirated wells immediately after aspiration.
-
Recommended: Incubate slides overnight at 4°C in a humidity controlled bin (i.e., covered Tupperware with damp Kimwipes).
Alternate method: Incubate 1 h at room temperature.
Table 1.
Common fluorophores and their peak excitation and emission spectra
| Fluorophore | Excitation peak (nm) | Emission peak (nm) |
|---|---|---|
| AMCA, Hoechst, DAPI | ~350 | ~450 |
| FITC, Cy2, Alexa488 | ~492 | ~520 |
| TRITC, Cy3, Alexa555 | ~550 | ~570 |
| Cy5, Alexa647 | ~650 | ~670 |
3.3.3. Day 3
Wash wells three times with DPBS, incubating for 5 min during each wash (cells in the chamber slide can be visualized under the fluorescence microscope during this procedure to ensure that enough washes have been performed to adequately reduce background signal).
If nuclear counterstaining is desired, cells can be incubated for a short period with the counterstaining reagent following by washing and mounting of the slide. Alternatively, the use of a mounting medium which already contains DAPI may eliminate the need for a separate staining step. In the first method, the counterstaining reagent (Hoechst 33342 Invitrogen) at 1 mg/mL in DMSO (stored at 4°C in the dark) is diluted 1:500 in DPBS and incubated with cells for 1–5 min at room temperature, followed by washing excess stain away with DPBS prior to mounting. If a mounting medium such as ProLong Gold Antifade reagent with DAPI (Invitrogen P36935) is used, excess moisture is removed from the slide by gently tapping the side of the slide or coverslip onto a clean Kimwipe prior to addition of the prewarmed-to-room-temperature reagent. It is often useful to have a cellular counterstain if it does not interfere with an antibody being detected by a fluorophore in the blue channel, such as 7-Amino-4-methylcoumarin-3-acetic acid (AMCA). A nuclear counterstain is also helpful when evaluating the nuclear localization of an antigen (particularly in stem cells that have a high nucleus-to-cytoplasm ratio).
Prepare mounting medium (used to minimize photobleaching of fluorescence) in accordance with manufacturer’s instructions. Examples of mounting media are as follows: Vectashield (Vector Labs), Slow Fade (Invitrogen), and Prolong Gold Antifade Reagent (Invitrogen). It should be noted that certain antifade reagent solutions contain glycerol and may be incompatiable with certain applications, such as specimens that contain lipophilic plasma membrane stains such as DiI.
Aspirate wells.
Snap off plastic wells according to the manufacture’s recommendations. Carefully use a razor on one of the short ends of the silicone gasket (if present; otherwise, skip this step). Using fine tweezers peel back the gasket slowly.
Pipette a bead of the mounting medium along the long end of the slide (approximately 300 µL). Be careful not to allow bubbles to form on the bead. Gently lower a rectangular cover slip at a 45° angle on the slide. Allow the mounting medium to spread.
Using two fingers very gently squeeze out the extra mounting medium and/or trapped air bubbles over a disposable paper towel. Pressing too hard will displace and/or damage cultures. Aspirate the extra medium off the slide.
-
Allow the slide to dry at room temperature in a dark, dry place overnight.
Alternate method: Allow samples to dry briefly then proceed to the steps in Day 4 below. Note that the coverslips will still move around and should be handled with care.
3.3.4. Day 4
Remove excess mounting medium by gently wiping the slide with 70% ethanol (use Kimwipes or cotton swab).
Seal slide edges with nail polish.
Allow to dry.
View slides on fluorescence microscope. Afterward, store slides at −20°C (with desiccant for best preservation). Storage at −20°C can preserve the signal for months (depending on the sample, antibody, choice of antifade reagent, etc.).
3.4. Imaging
After immunostaining, cells are usually viewed on a fluorescent microscope and images of the stained cells captured with a digital camera. There are a variety of cameras and image capturing software packages available; therefore, we won’t go over the specific details of a particular program here (details about one program, ImagePro, can be found in the Appendix of this chapter). However, many scientists bring the captured images into Adobe Photoshop to create output for publications. Therefore, in the next section we will describe how to use several features in Photoshop and briefly introduce a program available for image quantization (NIH Image, also known as ImageJ).
3.4.1. Microscope Setup
Seat slide on microscope stage with the cover slip facing the objective lens.
Make sure the microscope shutter is closed. Turn on mercury lamp and incandescent lamp.
Using a phase-contrast 20× objective, bring the sample into the focal plane.
Turn off incandescent light and use mercury light (preferably through lower frequency filters). Bleaching of fluorochromes is accelerated during exposure to higher frequency light. We prefer an excitation of ~570 nm (Cy3 channel) to first evaluate staining. Open shutter and analyze cultures through the microscope’s binocular eyepieces.
Scan through areas of interest while cycling through the other channels. Remember to limit the exposure of the slide to mercury light. Close shutter when not analyzing samples.
3.4.2. Adobe Photoshop
Photoshop can open a wide variety of image files captured from a microscope-mounted camera, including “.tiff” and “.jpg” formats, and provides a variety of means to manipulate images; however, it may not be able to open 16-bit files (if this is the case, be sure to save your files as 8-bit files). Here, we will briefly describe how to set the color mode, alter the image size, create scale bars for an image, adjust the image brightness and contrast, and create color overlays of images.
Setting the Color Mode
Color digital images can either use RGB (red, green, blue) or CMYK (cyan, magenta, yellow, black) for color encoding. RGB images are more compatible for computer monitors or projectors, since they use an additive light system (printers rely on a subtractive light system such as CMYK). Bright greens, reds, and blues cannot be reproduced as readily in print as they can on a monitor, so prints of an RGB image may not convey the bright colors or fine detail visible on the computer monitor. For print purposes (and therefore for most journal submissions), it is best to convert an RGB image to CMYK. To convert to CMYK for printing, go to “Image” → “Mode” and select CMYK.
Adjusting the Image Size
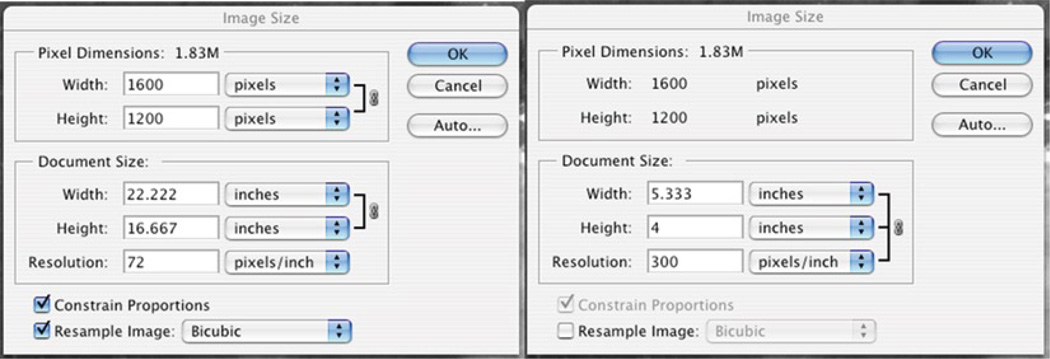
Images captured by image acquisition software programs can come in a variety of sizes and resolutions. To find the size of your image, go to “Image” → “Image Size.” Images often are captured at 72 pixels/in. and are of fairly large dimensions (in terms of inches). It is often desirable to set the resolution to 300 pixels/in. but not change the overall size of the file so that the dimensions (in inches) of the image are more suitable for printing or incorporating into a figure. To do this, make sure that the checkbox next to “Resample Image” is unchecked (as in Fig. 2) then adjust the resolution [see the figure and note that the overall pixel dimensions (1.83 M, 1,600 × 1,200 pixels) are the same for both while the document sizes (width, height, resolution) are different].
Fig. 2.
Adjusting the image size in Adobe Photoshop.
Scale Bars
One way to generate scale bars for your images and to make size/length determinations is to use a scale micrometer. These are slides that have lines etched a particular distance apart from each other. The micrometer can be placed on the microscope and an image taken using each of the microscope objectives. As the images are captured at the same width (in terms of pixels); you can determine a conversion factor that will allow you to measure real distances on your images. As an example, if an image taken with a camera on a particular microscope using a 20× objective has a total width of 580 µm (from the scale micrometer) and 1,600 pixels; this means that 100 µm would equal ~276 pixels on that image. Note that these measurements will be specific to the objective, microscope, and camera used, so attention must be paid to the conditions under which a particular image was captured in order to appropriately determine the scale. You can draw a line of a particular length (in pixels) in Photoshop by using the line tool (on the tool bar, which also contains the move tool, text tool, etc.) and watching the pixel location in the Navigator window (“Window” → “Navigator”; click on the “Info” tab in the Navigator window). The X and Y coordinates of the cursor location will be in pixels as long as the rulers for the image are set to “pixels” (“Preferences” → “Units and rulers”). Your image capture software, in most cases, also has the capacity for generating scale bars on your images.
Adjusting the Brightness/Contrast of an Image
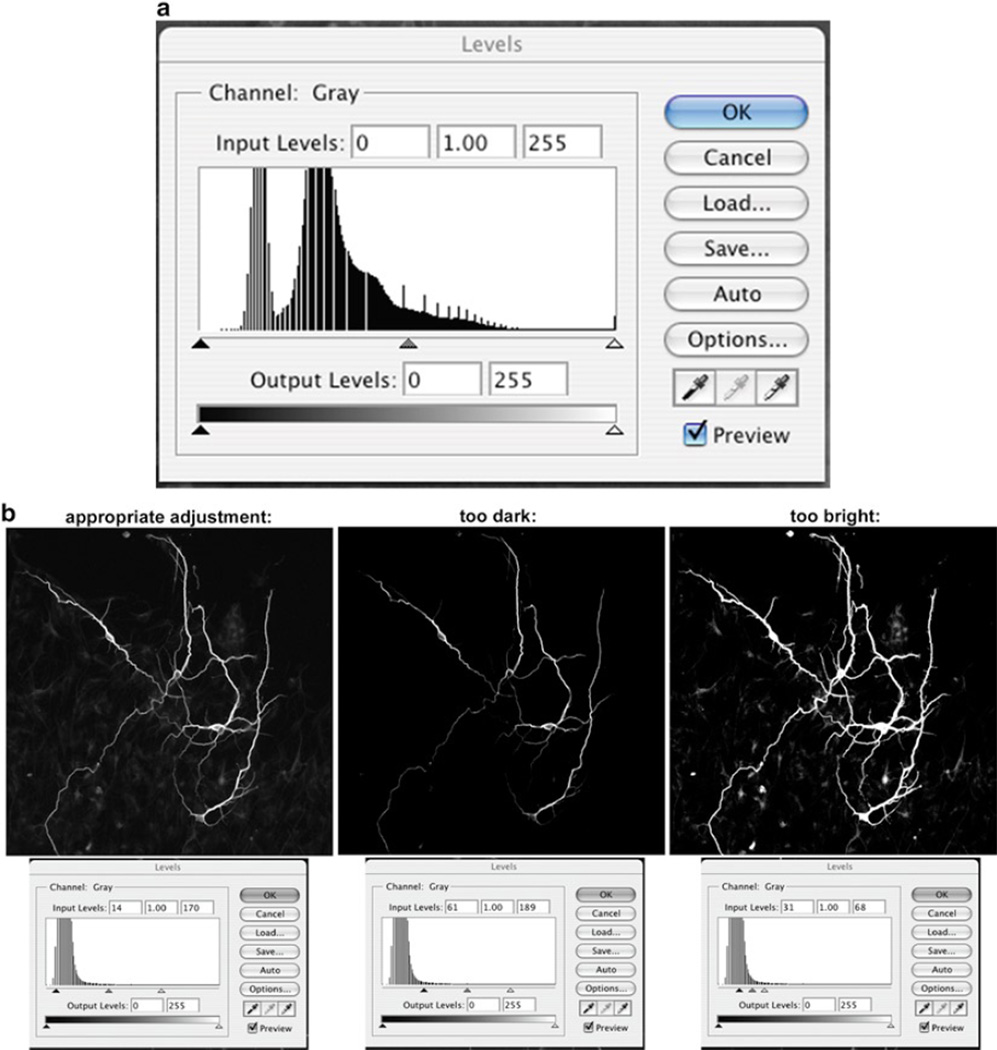
There are multiple ways to adjust images in Photoshop, and most are found under “Image” → “Adjustments.” One straightforward way to adjust the brightness/contrast is to use the “Levels” option (“Image” → “Adjustments” → “Levels”) and adjust the sliders under the histogram (see Fig. 3). The advantage of this option is that by viewing the histogram, you can more accurately adjust the intensity of the image without altering the data. It is IMPERATIVE when using any image adjustment for data images to be extremely careful not to alter the data with the adjustment. For example, decreasing the brightness should not remove signal and increasing the brightness should not create signal or expand the signals zone. See examples below for images that have been appropriately and inappropriately adjusted.
Fig. 3.
Adjusting the brightness/contrast of an image. (a) By using the histogram to adjust brightness/contrast levels, you can more accurately adjust the intensity of the image without altering the data. (b) Examples for images that have been appropriately and inappropriately adjusted.
Changing Grayscale Images to Color and Overlaying Color Images
Cells or tissues are often double- or triple-labeled with different fluorescent molecules to allow visualization of multiple signals. Photoshop can be used to convert captured grayscale images to color and overlay the color images so that all fluorescent signals can be visualized simultaneously. In order to create a color overlay, the images of the different fluorescent channels are brought together into a single file. The separate images are maintained on individual layers and then assigned a different color. To begin, select the entire image (“Select” → “All”) and copy (“Edit” → “Copy”). Make a new file (“File” → “New”) and the size, resolution, etc., will be identical to what you just copied. In the window that opens and describes the new file, switch from “Grayscale” to “RGB” (or “CMYK” if the image is solely for print media). Once the new file is created, paste in the copied image (“Edit” → “Paste”). Select all and copy the other images to be overlaid then paste them into the new file. Each image will automatically be pasted into a different layer (“Window” → “Layers”). To change the color of an image in a layer, open the “Levels” option (“Image” → “Adjustments” → “Levels”) and use the tab marked “RGB” to select either the Red, Green, or Blue channel. Use the “Output Levels” to alter the color: for a Red image, make the Green and Blue output levels 0 (change the number in the box on the right from 255 to 0), for a Green image, make the Red and Blue output levels 0, and for a Blue image, make the Red and Green output levels 0. These steps can be repeated for different layers within the same document to create layers that are of different colors. To overlay differently colored layers, position one colored layer directly above the other colored layer (in the “Layers” window) and then change the button under the “Layers” tab from “Normal” to “Screen”. You should now see both layers overlaid. Be sure, however, to keep the original, unchanged B&W files (raw data) of all images for future reference or alternative image production.
3.4.3. NIH Image (ImageJ)
NIH Image (or ImageJ) is a free program available for download that can be used to quantify a wide variety of parameters in an image. In addition to the basic features of ImageJ, there are Macros that others have created (or you can write yourself) that expand the functionality of the program. For details and downloads see: http://rsb.info.nih.gov/nih-image/Default.html.
Measurements in ImageJ
In order to do measurements in ImageJ, you must first know the scale of your picture in real dimensions (see “Scale Bar” section above). An easy way to convert this information to a scale in ImageJ is to draw a line across the entire width of your image (use the straight line tool on the toolbar). Then, go to “Analyze” → “Set Scale” and set the known distance to the numerical value and unit of length for your image width (for example, the width of the image described above would be 580 µm). Keep the Pixel Aspect Ratio as 1 and use “um” for “µm”. If you are analyzing multiple images that were taken under the same conditions and thus have the same scale, you can check “Global” in the “Set Scale” window and the scale will be automatically applied to all the images. After setting the scale, the length of any line drawn and measured will be given in the desired units. To measure an element in your image, you can draw a line (straight, segmented, or freehand) and then click on “Analyze” → “Measure”. “Analyze” → “Set measurements” allows you to decide what parameters will be measured. Note that you can also choose other types of shapes (other than a line) and measure parameters such as area.
3.4.4. Image Pro
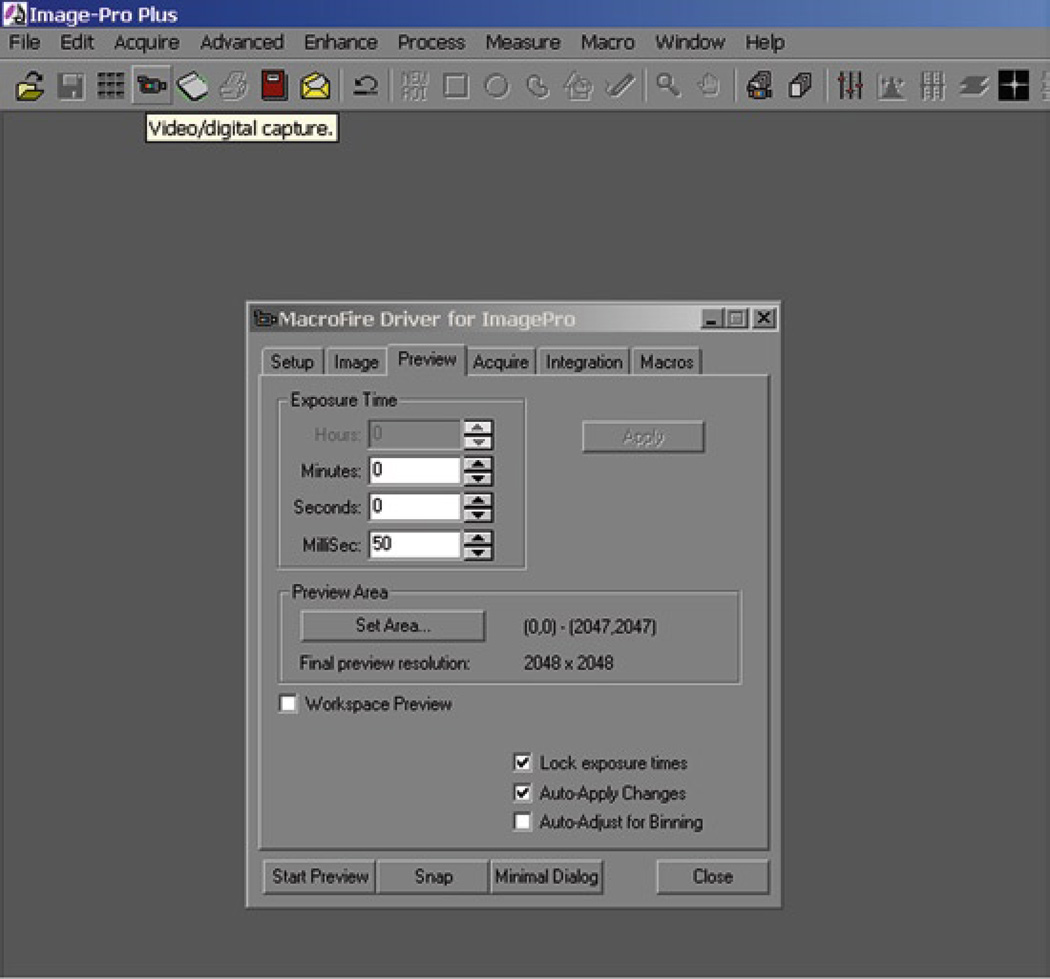
The following section will describe using Media Cybernetics’ Image-Pro plus (www.mediacy.com) to photograph snapshots of a field of interest using a digital camera.
Turn on digital camera.
Open Image Pro.
Under Acquire select “Video/Digital Capture.”
The following window will allow you to preview and snap pictures directly from the camera.
Click Start Preview.
Adjust the exposure time to brighten image without over-saturating digital feed (most digital camera drivers have a configuration setting to provide live saturation warnings).
Snap the image when you are satisfied with the previewed image.
Save image (a “.tiff” file format is recommended for preserving image detail).
3.4.5. Using Image-Pro AFA Plug-In
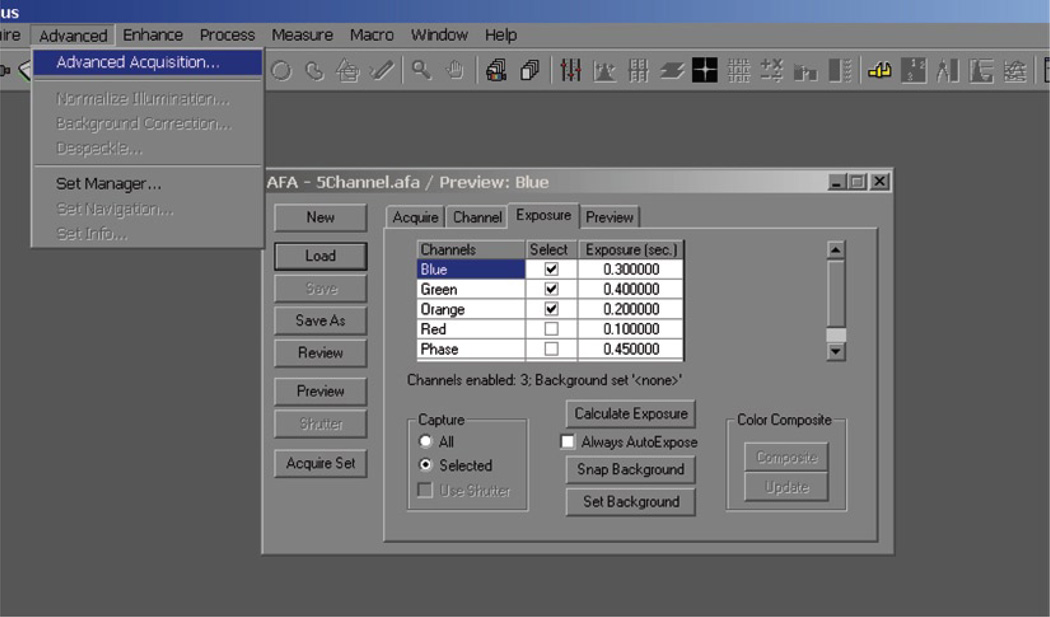
The Image-Pro AFA Plug-in is a useful tool for organizing and managing multiple channels from a field of interest. Exposures may be optimized for each channel before imaging the field of interest as a set. After a set of images is obtained, the color composite tool may be used to pseudo-color and merge channels.
Open the “Advanced Acquisition window.”
Click on preview (note: the exposure times for the preview are set for the first channel).
Adjust exposure times for each channel.
Check the boxes for the channels you want to photograph.
Click Acquire Set.
If the microscope used is fully automated it will automatically rotate the filter cubes and photograph the samples. If it is not, a prompt will ask you to manually turn the wheel between pictures.
Once the set has been acquired, you can save it using the “Set Manager.”
Fig. 4.
Acquiring an image in Image Pro.
Fig. 5.
The Image-Pro AFA Plug-in allows the user to optimize multiple channels from a field of interest.
Acknowledgments
We gratefully acknowledge intellectual contributions from Lisa A. Flanagan, Richard Pepple, Boback Ziaeian, and Theo Palmer.
Footnotes
General caution for antibody staining:
Importantly, the cells should never be allowed to dry out, so you should not completely aspirate all the liquid from the well and you should always have the next solution at hand to add immediately after aspiration.
Fixative preparation and storage:
Paraformaldehyde: With heat and basic pH, paraformaldehyde will depolymerize to a very pure form of formaldehyde. Solubilization of paraformaldehyde powder is often accomplished with heat and strong base but take care not to heat the solution above 55–60°C and add just enough base to depolymerize the paraformaldehyde (pH ≤ 10). If the solution goes over 65°C during preparation the formaldehyde degrades rapidly to formic acid and water. Therefore, do not use it as it will produce a strong autofluorescence in cells or tissues.
37% Formaldehyde: Storage of 37% formaldehyde over several months also results in degradation to formic acid and water. Old formaldehyde stocks should be disposed of every 12–24 months in accordance with your institution’s chemical policies.
10% Buffered formalin: 10% buffered formalin will pH drift due to degradation of formaldehyde to formic acid. Do not use if below pH 6.5 and rotate stocks regularly.
Summary of immunostaining procedure:
Remove medium from cells, wash with DPBS +/+.
Add Fixative, 10 min, RT.
Wash with DPBS +/+, 2 × 5 min.
Add HCl if BrdU-treated cells, 20–30 min, 37°C, wash DPBS +/+, 2 × 5 min
Add Blocking Buffer, 15 min, RT, remove
Add diluted primary antibodies, overnight, 4°C
Wash DPBS +/+, 2 × 5 min.
Add diluted secondary antibodies, 1 h, RT.
Wash with DPBS +/+, 2 × 5 min.
Add Hoechst (1:500 in DPBS +/+), 1–5 min, RT.
Wash with DPBS +/+, 1 × 5 min.
Mount and coverslip. Seal with nail polish.
View on microscope.
Choosing the Right Antibodies:
Most fluorescence microscopes have the ability to discern several unique fluorochromes using various optical filter arrangements. In designing a plan for co-staining for more than one antigen, it is important first to select primary antibodies of unique species or subtypes (i.e., Mouse IgG, Mouse IgM, Rabbit IgG, Goat IgG, Chicken IgG, Guinea Pig IgG, Rat IgG). If the primary antibodies for different antigens are from the same species and subtype, secondary antibodies will indiscriminately bind to both markers. For multiple antibody staining, care should be taken to use secondary antibodies that are highly specific for the class and species of primary antibody that needs to be detected. Some vendors provide secondary antibody reagents that are validated to have minimal cross reactivity to a wide spectrum of antibody classes and species (Jackson Immuno Research Laboratories is a reliable source). In addition, the fluorophores chosen for the secondary antibodies must match the particular filter sets present on your microscope to prevent optical overlap between the fluorophores (Table 1).
Antibody concentration:
Most manufacturers provide recommendations for antibody concentrations for specific applications. When using an antibody for the first time, it’s a good idea to try a range of concentrations around that recommended by the manufacturer. For example, if the recommended concentration is 1:100, try a range from 1:10 to 1:1,000. If no recommended concentration is given, start with 1 µg/100 µL antibody dilution buffer.
Antibody aspiration and washing:
To save time, set up your aspirator accordingly: Attach a Pasteur pipet to the end of your aspirator. Now, place a disposable P100 or smaller pipette tip on the end of the attached Pasteur pipette and replace only the P100 tips for each aspiration. A used aspirator tip greatly increases the likelihood of cross-contaminating adjacent wells.
Background staining:
- Spin the antibodies to remove precipitates before adding the antibody to the sample (see steps 3 and 4 of Day 2 above).
- Use fresh antibodies. Over time antibodies will degrade and increase the amount of background and nonspecific staining. To avoid multiple freeze–thaw cycles, aliquot the antibody upon receipt into smaller working volumes.
- Primary antibody and/or secondary antibody concentrations are too high.
- Increase DPBS rinsing time or number of washes.
- Incorrect blocking serum or insufficient blocking time. One can also try blocking with IgG-free BSA rather than animal serum (use 5% w/v in DPBS for blocking buffer and 1% w/v in DPBS for antibody dilution buffer).
- Cell cultures were stressed during growth. Refine growth conditions.
- Attempt to use a different antibody for the antigen (try to choose an antibody that recognizes a different epitope on the same molecule).
Species mismatch:
Problem: Same-species antibodies yield high background. For example, when mouse primary antibodies are used on mouse tissues, detection with anti-mouse secondary antibodies will detect all mouse immunoglobulins that are native to the mouse tissue.
Solution: Use species-mismatched primary antibodies or block the endogenous antibodies by preincubating with an unconjugated secondary antibody. When blocking, it is necessary to use Fab fragments and important to use a Fab preparation that matches the conjugated secondary antibody that will be used for detection. Vendors often sell unconjugated Fab preparations that match the detecting secondary antibody for this purpose.
Note: Why use Fab fragments for blocking endogenous Ig? Whole Ig is multivalent and a block with a multivalent antibody will leave many Fab ends unbound. Subsequent treatment with the primary antibody will simply bind these exposed ends and aggravate the background problems.
Fc Receptors in sample:
Problem: Fc receptors expressed by cells nonspecifically bind primary and secondary antibodies. Particularly problematic for tissues that have been damaged and contain activated immune cells.
Solution: Use Fab preparations for detection rather than whole antibodies or block using unconjugated Fc fractions that match both primary and secondary antibody preparations.
Note: When using Fab fragments for detection, the secondary antibody must be one that recognizes a Fab fragment. Typically, the secondary antibody used will recognize light-chain rather than heavy chain and one must take care to determine the class of light chain present in the Fab fragment (i.e., either kappa or lambda light chain).
Generalized Background
Problem: Very high overall background.
Solution: Titrate antibodies (both primary and secondary) for optimum signal to noise. Primary or secondary antibody may recognize nonspecific antigens. To determine if the problem is with the primary or secondary antibody, prepare one sample that is treated with secondary antibody alone. If background is low, then problem is with primary antibody. If background is present in samples treated with secondary antibody alone, then the problem is with secondary antibody. In both cases, an alternate antibody should be tried (if available) or more aggressive means to improve specificity should be explored.
Note: Secondary antibody background can be reduced if the vendor provides unconjugated preimmune serum from the same species (ideally collected from the same animal prior to immunization). This is used in the initial blocking step to bind all non-specific sites prior to the final detection using the conjugated secondary antibody preps.
- Test the antibodies on known positive and negative controls.
- Try another antibody to the same antigen.
- Fixation and/or Permeabilzation – Follow the manufacturer’s specific instructions for methods of fixation and permeabilization to use with the antibody in question. In addition, check the literature for papers that have used the antibody (and have nice images of immunostained cells) and follow the protocol verbatim (call or email the authors, if necessary, to get details). Most antibodies are sensitive to the type of fixation and or permeabilization used. In addition, the concentration of chemical used and the time of exposure can also be critical (it is possible to over-fix).
- Increase the concentration of primary and/or secondary antibody.
- Increase the time of the primary antibody incubation. If positively staining slides have faded over time, be certain the nail polish sealant on slides is intact and that the slides are being stored in a desiccated, cold environment.
- Reduce primary antibody concentration.
- Reduce primary or secondary antibody incubation period.
- Attempt to use a different clone of antibody for the same antigen.
- See notes on blocking in “Background staining” section above.
References
- 1.Li F, Lu S, Vida L, Thomson JA, Honig GR. Bone morphogenetic protein 4 induces efficient hematopoietic differentiation of rhesus monkey embryonic stem cells in vitro. Blood. 2001;98:335–342. doi: 10.1182/blood.v98.2.335. [DOI] [PubMed] [Google Scholar]
- 2.Pera MF, Filipczyk AA, Hawes SM, Laslett AL. Isolation, characterization, and differentiation of human embryonic stem cells. Methods Enzymol. 2003;365:429–446. doi: 10.1016/s0076-6879(03)65030-5. [DOI] [PubMed] [Google Scholar]
- 3.Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Fenderson BA, De Miguel MP, Pyle AD, Donovan PJ. Staining embryonic stem cells using monoclonal antibodies to stage-specific embryonic antigens. Methods Mol. Biol. 2006;325:207–224. doi: 10.1385/1-59745-005-7:207. [DOI] [PubMed] [Google Scholar]
- 5.Draper JS, Moore HD, Ruban LN, Gokhale PJ, Andrews PW. Culture and characterization of human embryonic stem cells. Stem Cells Dev. 2004;13:325–336. doi: 10.1089/scd.2004.13.325. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 7.Loring JF, Rao MS. Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells. 2006;24:145–150. doi: 10.1634/stemcells.2005-0432. [DOI] [PubMed] [Google Scholar]
- 8.Ohnuki M, Takahashi K, Yamanaka S. Generation and characterization of human induced pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2009 doi: 10.1002/9780470151808.sc04a02s9. Chapter 4, Unit 4A.2. [DOI] [PubMed] [Google Scholar]