Summary
Pausing of RNA polymerase II (Pol II) 20-60 bp downstream of transcription start sites is a major checkpoint during transcription in animal cells. Mechanisms that control pausing are largely unknown. We developed permanganate-ChIP-seq to evaluate the state of Pol II at promoters throughout the Drosophila genome, and a biochemical system that reconstitutes promoter-proximal pausing to define pausing mechanisms. Stable open complexes of Pol II are largely absent from the transcription start sites of most mRNA genes, but are present at snRNA genes and the highly transcribed heat shock genes following their induction. The location of the pause is influenced by the timing between when NELF loads onto Pol II and how fast Pol II escapes the promoter region. Our biochemical analysis reveals that the sequence-specific transcription factor, GAF, orchestrates efficient pausing by recruiting NELF to promoters before transcription initiation and by assisting in loading NELF onto Pol II after initiation.
Keywords: promoter proximal pausing, RNA polymerase II, GAGA factor, NELF, permanganate-ChIP-seq
INTRODUCTION
Genome-wide analyses of Pol II in mammals and Drosophila reveals that Pol II is often concentrated at the 5′ end of genes irrespective of the level of gene expression (Adelman and Lis, 2012). Hence, transcriptional regulation after Pol II has associated with promoters is widespread, and may be a major form of gene regulation on par with transcription factor-mediated recruitment of the transcription machinery (Ptashne, 2005).
Based on studies of individual genes, the enrichment of Pol II at promoters has been linked to promoter-proximal pausing that occurs after Pol II initiates transcription and elongates downstream from the start site (Lis, 1998). However, much of our understanding of promoter-proximal pausing on a genomic scale has been defined by low resolution ChIP assays and transcript mapping, which either lack the resolution or do not permit the detection of events occurring between the site of preinitiation complex (PIC) assembly and the pause sites (Core et al., 2008; Nechaev et al., 2010). Even the recent high resolution transcript mapping technique, PRO-seq, cannot detect open complexes near the transcription start site (Kwak et al., 2013). Hence, possible open complexes formed over the transcription start or within the first 20 nucleotides cannot be distinguished or detected. Biochemical results argue for the existence of intermediates in the transcription cycle within the first 20 nucleotides (Nock et al., 2012; Pal et al., 2005). Whether these are major rate-limiting steps in vivo is not known.
The most widely accepted assay for monitoring the position of Pol II along DNA once it has melted the DNA (the Pol II “bubble”) is permanganate footprinting, which detects unpaired T residues in single-stranded open complexes (Adelman and Lis, 2012). While the permanganate assay is powerful in both its spatial resolution and definitive assessment of open complexes, it has thus far not been performed on a genomic scale. We developed permanganate ChIP-seq, which combines the single base pair spatial resolution of permanganate reactivity with the high signal-to-noise selection of Pol II ChIP and deep sequencing. For the first time, we can discern on a genomic scale whether Pol II resides in open complexes upstream of the pause site, or is only present in a kinetically trapped state at pause sites downstream from the transcription start site (TSS).
Promoter-proximal pausing depends on DSIF and NELF (Wu et al., 2003; Yamaguchi et al., 1999), two proteins that associate with the Pol II elongation complex. Reactivation of paused Pol II involves P-TEFb, a kinase that phosphorylates Pol II, DSIF, and NELF (Chiba et al., 2010; Price, 2008). Apart from the identity of these factors involved in pausing, there is almost no widely-accepted concept of the pausing mechanism. There could be a specifically positioned protein such as a nucleosome that physically blocks Pol II elongation (Brown et al., 1996; Mavrich et al., 2008). Alternatively, some feature of the promoter-proximal DNA sequence could inhibit Pol II elongation (Hendrix et al., 2008; Nechaev et al., 2010). A third possibility that we explore here is that sequence-specific transcription factors establish a pausing competency to Pol II that is available shortly after it initiates transcription. We find that promoters with the highest levels of paused Pol II tend to associate with the sequence-specific DNA binding protein, GAGA factor (GAF). Deletion of the GAGA element at the hsp70 promoter was previously shown to cause loss of paused Pol II (Lee et al., 1992), but it is unclear if GAF has a direct effect on the elongation complex or only functions in the initiation step that must precede pausing. To obtain a mechanistic understanding of how GAF affects pausing, we have developed a cell-free system that reconstitutes robust promoter proximal pausing. Together with genomic analyses, our results provide insight into how a sequence specific transcription factor can control the location and efficiency of promoter proximal pausing.
RESULTS
Paused genes do not accumulate open PICs
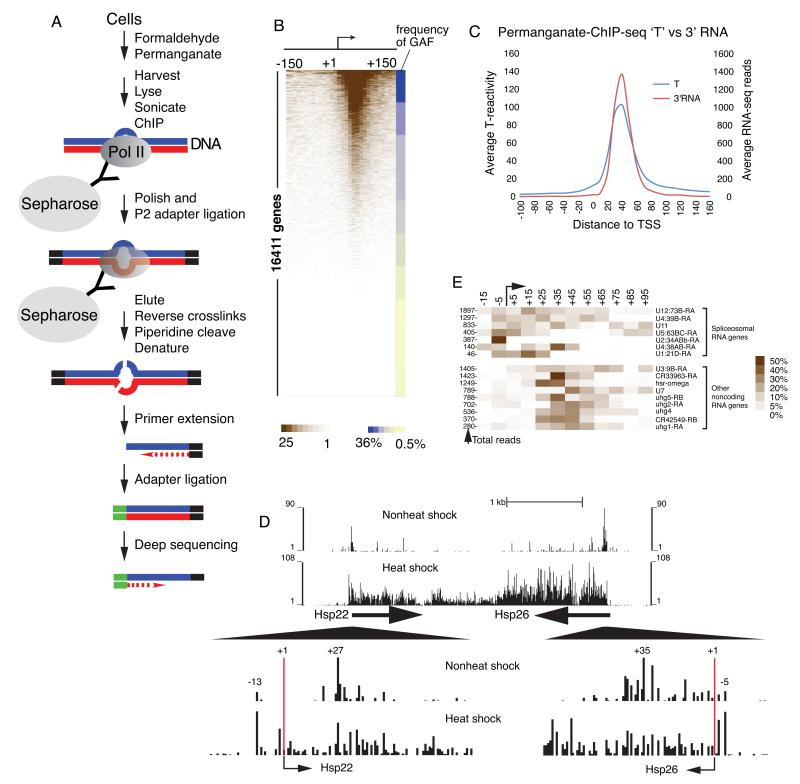
While it is clear that Pol II pauses on a genomic scale 20-60 bp after transcription initiation, it is not known on the same scale whether other polymerases might be engaged on the DNA just upstream. This is an important question because in yeast, Pol II is stably present within a open PIC at core promoters (Rhee and Pugh, 2012), and biochemical results provide evidence for open complexes at the TSS (Nock et al., 2012). To address this, we developed permanganate-ChIP-seq to detect open complexes on a genomic scale (Figure 1A). Cells were treated with formaldehyde to crosslink Pol II to DNA and then with permanganate to oxidize thymines in transcription bubbles. Sheared chromatin derived from these cells was immunoprecipitated with antibody against the Rpb3 subunit of Pol II. While the ChIP DNA was still associated with the resin, the first sequencing adaptor was ligated to both ends. The DNA was eluted from the immunoprecipitate, and the formaldehyde crosslinks were reversed. The DNA was treated with piperidine to cleave the DNA backbone at the oxidized thymines, and then denatured and subjected to second strand synthesis via priming off of the ligated adaptor. Finally, a second adapter was ligated to the newly polished end generated from piperidine cleavage, and the resulting library was subjected to deep sequencing. The 5′ end of each sequencing read corresponds to the nucleotide next to the piperidine cleavage site within a Pol II complex, thus identifying the location of hyper-reactive, unpaired thymines.
Figure 1. Detection of Pol II open complexes by permanganate-ChIP-seq.
(A) Schematic of permanganate-ChIP-seq technique.
(B) Heat map of T-reactivities by permanganate-ChIP-seq at all promoters of protein-coding genes. 16411 protein-coding genes are ranked by T-reactivity from −150 to +150 bp, mapped to 10 bp bins. Each row represents an individual gene. The frequency with which GAGA factor (GAF) associated with each decile of promoters is displayed on the right. The association of GAF with the promoters is defined by the appearance of GAF ChIP-chip peaks within −500 to +300 bp from the TSS (Lee et al., 2008).
(C) Positional overlap of T-reactivities (T) and 3′ ends of small capped RNAs at the promoters of 3725 active genes. Active genes were defined as those associated with 5′ small capped RNAs and the 3′ ends of these small capped RNAs were used in this analysis (Nechaev et al., 2010). Genes were further selected to have no neighboring TSS within 500 bp, and to have focused initiation (defined as genes having > 75% of their 5′ ends of small capped RNAs mapping to within 10 bp of a common TSS.)
(D) Permanganate-ChIP-seq analysis of heat shock genes before and after heat shock induction. The upper two tracks show the pattern of T-reactivity in a region spanning the heat shock genes, Hsp22 and Hsp26. The lower two tracks magnify the promoter regions of the two heat shock genes. Numbers above lines represent locations relative the TSS.
(E) Enrichment of open complexes at the TSS of spliceosomal snRNA genes. Rows correspond to genes, aligned by their TSS. T-reactivity is displayed as a heat map, where color intensity represents the percentage of total T-reactivity in the region from −20 to +100 for each gene, mapped to 10 bp bins. The numbers on the left are the total number of sequencing reads acquired for the region from −20 to +100 for each gene.
Our permanganate ChIP-seq revealed that hyper-reactive thymines were highly enriched between +20 and +60 at thousands of Drosophila promoters (Figure 1B and 1C), exactly where permanganate reactivity had been observed in single gene assays (Lee et al., 2008). The majority of cleavages were at thymines and the sequence composition did not bias the pattern of cuts (see methods and Figures S1A – C). Essentially identical results were obtained in two different Drosophila cell lines (S2R+ and BG3, Figure S1D, E). Clusters of T-reactivity corresponded to the location of 3′ ends of small, capped RNAs, thereby validating them as transcription bubbles (Figure 1C and S1F).
Little or no T-reactivity was detected upstream from +20 at the core promoter (the average T-reactivity from +20 to +60 is 25-fold higher than from −30 to +10). This could reflect the absence of stable open Pol II complexes at the transcription start site or an inability of permanganate ChIP-seq to detect them. To test the latter possibility, we did permanganate ChIP-seq on heat shocked cells and compared the T-reactivity on heat shock genes to that of control cells. Heat shock genes are highly induced by heat shock, and permanganate footprinting of individual genes detects open complexes at their transcription start sites (Giardina et al., 1992). In contrast to the pattern of T-reactivity in nonheat shocked cells, the T-reactivity at the TSS of heat shock-induced hsp22 and hsp26 was comparable to the T-reactivity in the +20 to +60 region indicating that permanganate-ChIP-seq can detect open complexes at the TSS (Figure 1D). Hence, the absence of T-reactivity upstream from +20 on the majority of mRNA genes in nonheat shocked cells indicates that when Pol II assembles into a pre-initiation complex, it rapidly moves into a transcriptionally engaged paused state.
Small, noncoding nuclear RNA genes encoding components of the splicing machinery have relatively more open complexes at the TSS than other Pol II genes
To identify genes that harbor open complexes at the transcription start site in nonheat shocked cells, we searched for genes that had higher or similar levels of T-reactivity at the TSS than the promoter proximal region. Remarkably, the only genes with this feature encoded snRNAs that are part of the spliceosome (Figure 1E and Figure S1G). This pattern of T-reactivity was not observed on other noncoding RNA genes (Figure 1E and Figure S1H) nor was it related to the overall density of Pol II engaged in the promoter region (Figure 1E, compare Total reads). Thus, a significant level of open complex at the transcription start is a special feature of these snRNA genes. Notably, these open complexes are distributed evenly into several of these genes so escape of the open complex at the TSS into the gene does not appear to be rate limiting.
Biochemical analysis of promoter proximal pausing in Drosophila nuclear extracts
We searched the sequences of promoters with a high level of T-reactivity for conserved DNA elements to obtain leads for understanding mechanisms of pausing. Two previously reported elements were found over-represented on promoters with paused Pol II. The GAGA element was enriched among genes with the highest level of T-reactivity while another previously described consensus called Motif 1 (Ohler et al., 2002) was enriched among genes with moderate T-reactivity (Figure S2). Motif 1 binds a novel DNA binding protein that will be described elsewhere (manuscript submitted). Here, we focus on the function of the GAGA element in promoter proximal pausing. The GAGA element associates with GAF, and both are implicated in promoter proximal pausing [Figure 1B and (Fay et al., 2011; Lee et al., 2008; Lee et al., 1992)]. How GAF contributes to pausing is not known.
To investigate the function of GAF, we developed a robust biochemical system that paused Pol II in a GAF-dependent manner. Previously, we showed that promoter proximal pausing could be reconstituted on the hsp70 promoter in nuclear extracts from Drosophila embryos (Li et al., 1996). However, this system was limited because the paused Pol II was detected by permanganate footprinting and it was not possible to ascertain when permanganate reactivity was lost whether this was due to transcriptional read-through, premature termination, or inhibition of initiation. To overcome these limitations, we developed an alternative means for monitoring the behavior of Pol II. A pulse-chase procedure was employed to restrict radiolabel to the 5′ region of transcripts so that the signals from transcripts on gels were proportional to the amount rather than the length of the transcripts. Also, a biotinylated oligonucleotide was used to isolate hsp70 transcripts produced during the transcription reaction in nuclear extracts so these transcripts were not obscured by other radiolabelled nucleic acids generated by the extract independently of Pol II.
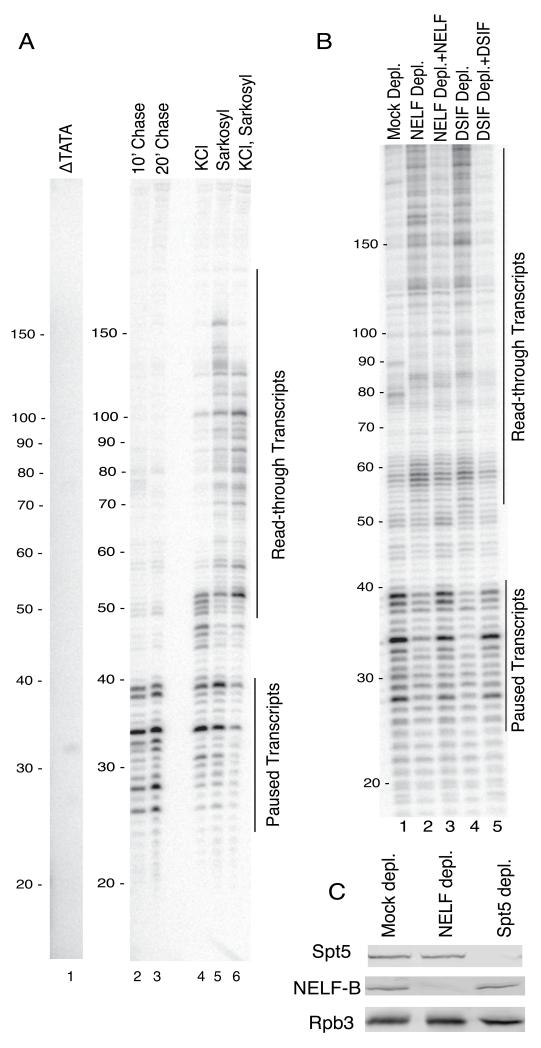
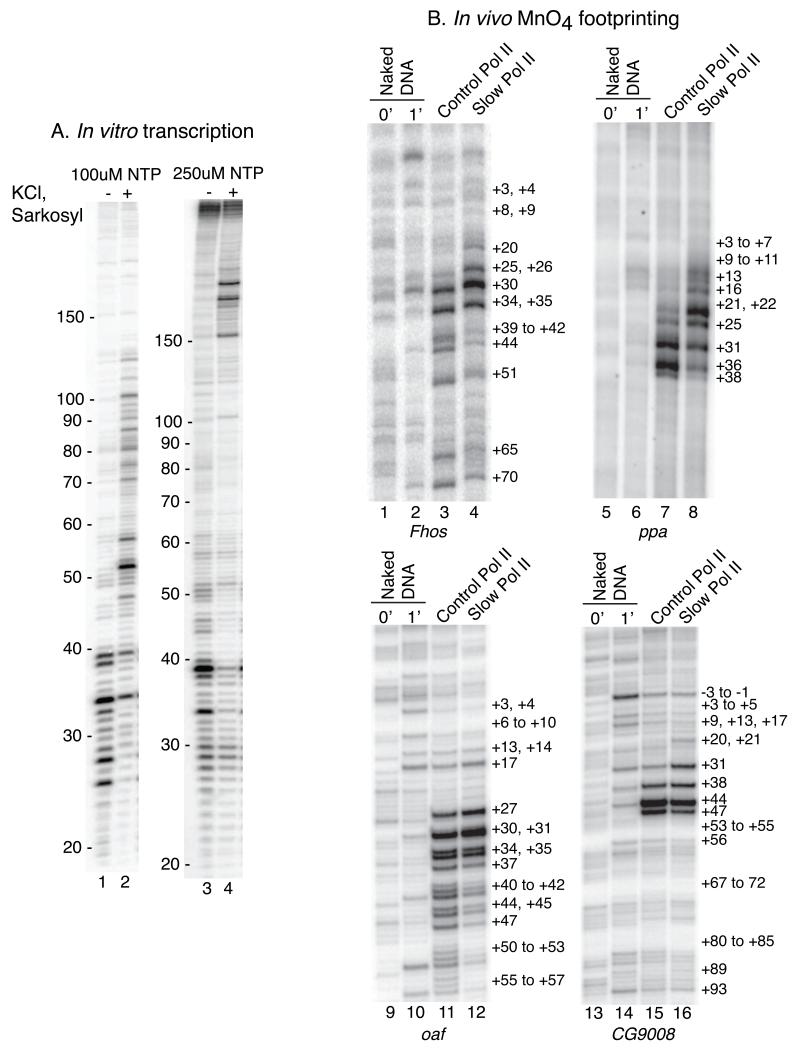
In vitro transcription of hsp70 produced short transcripts in a size range matching those observed in cells (Rasmussen and Lis, 1993) (Figure 2A, lanes 2 and 3). These transcripts were not produced from an hsp70 promoter lacking the TATA box (Figure 2A, lane 1), a mutation that inactivates the promoter (Gilmour et al., 1988). Sarkosyl or KCl reactivate the paused Pol II in isolated nuclei (Rougvie and Lis, 1988). Likewise, the addition of sarkosyl, KCl or both to our cell-free system resulted in lengthening of the transcripts (Figure 2A, lanes 4-6). Thus, the short transcripts detected in lanes 2 and 3 are associated with paused Pol II rather than being products of premature termination. In addition, the location of the paused Pol II inferred from transcript lengths corresponded to the location of paused Pol II detected by permanganate footprinting (Figure S3A).
Figure 2. Promoter-proximal pausing on hsp70 in nuclear extracts from Drosophila embryos.
(A) Promoter proximal pausing occurs during transcription in nuclear extracts. hsp70 transcripts produced during transcription were radiolabeled by a pulse-chase procedure and isolated as described in the methods. ΔTATA has the TATA box deleted and is transcriptionally inactive (lane 1). Normal hsp70 produced paused transcripts of 20 to 40 nucleotides that persist for 10 to 20 min (lanes 2 and 3). KCl and sarkosyl added 10 min after the chase causes the paused Pol II to resume elongation (lanes 4 – 6).
(B) Pausing depends on NELF and DSIF. Nuclear extracts were depleted with antibodies against NELF (lanes 2 and 3), DSIF (lane 4 and 5), or with control IgG (lane 1). Pausing was restored by the addition of purified DSIF or NELF to depleted extracts (lanes 3 and 5). These results are representative of 3 experiments.
(C) Western blotting analysis of nuclear extracts immunodepleted of NELF or DSIF. Spt5 is the largest subunit of DSIF. Rpb3 is a subunit of Pol II and NELF-B is a subunit of NELF.
To validate our biochemical system, we tested if pausing was dependent on NELF and DSIF. Depletion of either protein decreased paused transcripts and increased read-through transcripts (Figure 2B, compare lanes 1, 2, and 4). Promoter proximal pausing was rescued by adding back purified forms of the depleted proteins (Figure 2B, compare lanes 2 to 3 and 4 to 5). Together these results indicate that our cell-free system reconstitutes promoter proximal pausing that recapitulates key aspects of the pausing at hsp70 in vivo.
GAF regulates pausing at steps before and after transcription initiation
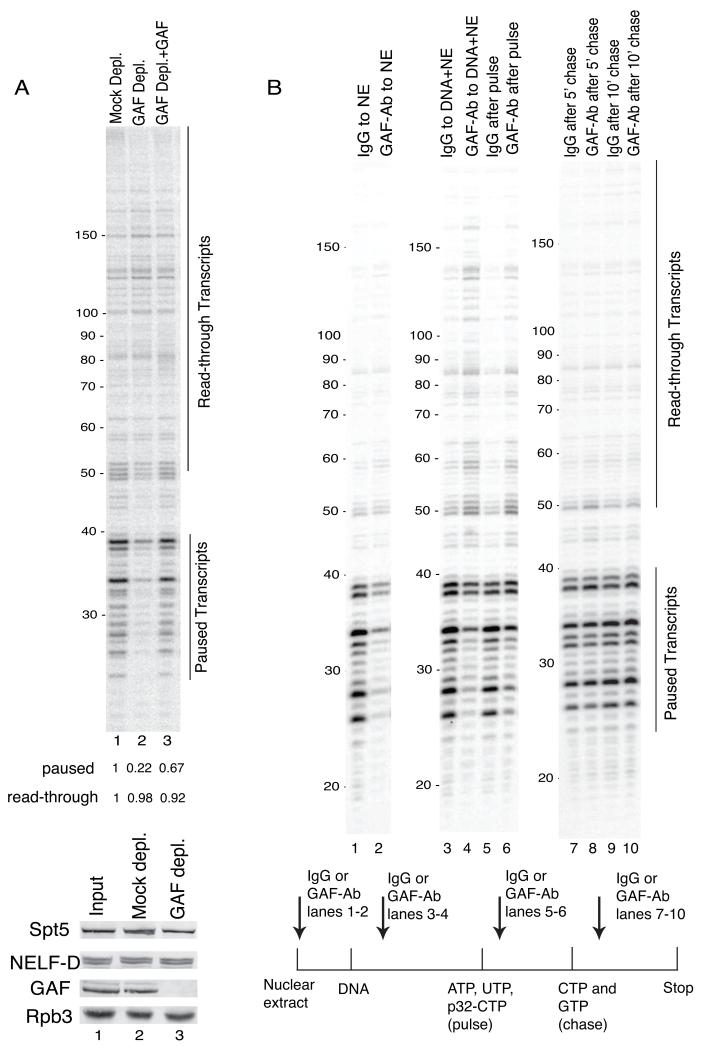
Immunodepletion of GAF from our nuclear extract caused a marked decrease in paused transcripts, and a significant portion of these transcripts were restored by adding back purified GAF (Figure 3A). Notably, the level of read-through transcripts was unchanged in the GAF-depleted sample suggesting that the efficiency of pausing in these reactions was diminished (Figure 3A, compare lanes 1 and 2).
Figure 3. GAF regulates pausing during the elongation phase of transcription.
(A) Upper panel: Immunodepletion of GAF from extracts impairs pausing. Nuclear extracts were depleted with control (lane 1) or GAF (lane 2) antibody. Addition of FLAG-tagged GAF purified from Drosophila embryos restores pausing (lane 3). The signal intensities of bands in the region of the paused or read-through transcripts relative to the Mock depleted sample (lane 1) are presented below the lanes. Lower panel: Western blot of extract before antibody depletion (lane 1) or after depletion with control IgG (lane 2) or GAF antibody (lane 3).
(B) GAF functions during initiation and pausing. GAF activity was disrupted at different stages of the transcription reaction with GAF antibody. The schematic indicates when GAF or control IgG was added. Lanes 1 and 2: antibody added to the extract before adding DNA to inhibit GAF function prior to the assembly of preinitiation complexes. Lanes 3 and 4: antibody added after incubating extract and DNA without nucleotides to inhibit GAF function after preinitiation complex formation. Lanes 5 and 6: antibody added after allowing transcription to start with limiting nucleotides to inhibit GAF function after the start of transcription. Lanes 7 and 8: antibody added after allowing transcription to the pause for 5 min to inhibit GAF function after Pol II has paused. Lanes 9 and 10: antibody added after allowing transcription to the pause for 10 min. The results are representative of 3 experiments.
To determine when GAF contributed to promoter proximal pausing, we examined the effects of disrupting GAF’s function before, during or after transcription initiation by adding GAF antibody at different stages of the transcription reaction. GAF clearly contributed to initiation since incubation of the nuclear extract with GAF antibody before adding hsp70 DNA substantially diminished the overall level of transcripts (Figure 3B, lanes 1 and 2). Addition of GAF antibody after formation of preinitiation complexes, but before initiation with NTPs, diminished paused transcripts and increased read-through transcripts (Figure 3B, compare lanes 3 and 4). The same effect was observed when the GAF antibody was added after the pulse but before the chase (Figure 3B, compare lanes 5 and 6). Importantly, GAF antibody had no effect after the Pol II had paused (Figure 3B, lanes 7 to 10). Corroborating results were obtained by permanganate footprinting (Figure S3B - D). GAF antibody had similar effects on the GAF-associated Mrp4 promoter but had no effect on the GAF-less promoter, oaf (Figure S4). Thus, GAF specifically regulates transcription initiation and the establishment of the pause before and after Pol II initiates transcription.
GAF can recruit NELF to the hsp70 promoter
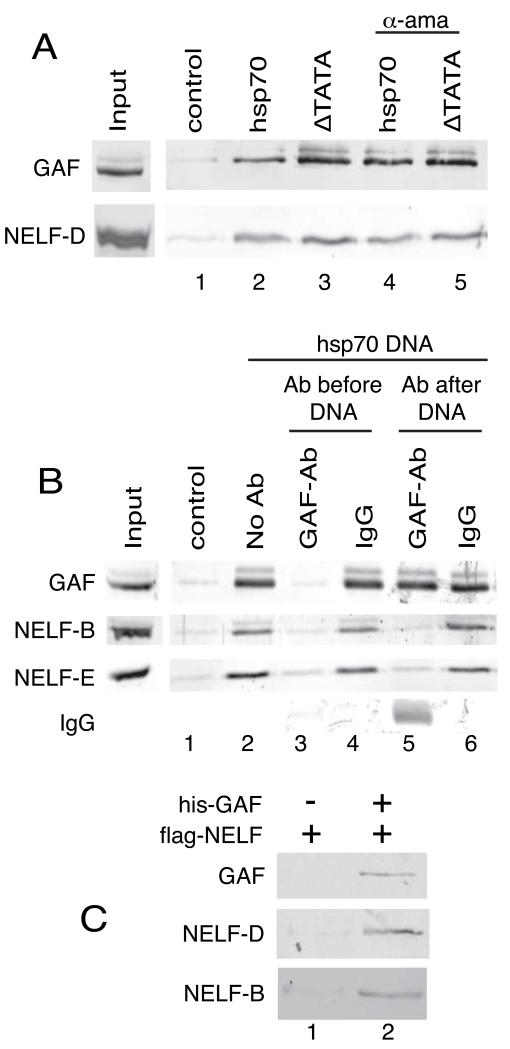
To investigate the mechanism by which GAF regulates pausing during elongation, we monitored the association of GAF and NELF with immobilized DNA in nuclear extracts. Substantially more GAF and NELF associated with the hsp70 DNA than with a control DNA (Figure 4A, compare lanes 1 and 2). This association occurred in the absence of transcription since neither deletion of the TATA box nor the presence alpha-amanitin affected the binding of GAF and NELF (Figure 4A, lanes 3 – 5).
Figure 4. GAF recruits NELF to hsp70 promoter DNA.
(A) NELF associates with the hsp70 promoter in the absence of transcription. DNA immobilized on magnetic beads was incubated with nuclear extract. Proteins associating with the immobilized DNA were isolated and detected by western blotting. 10 μg/ml alpha-amanitin (α-ama) was used to inhibit low levels of transcription that might occur due to nucleotides in the extract (lanes 4 and 5). Control DNA (lane 1) lacks the hsp70 promoter.
(B) GAF antibody disrupts the association of NELF with the immobilized hsp70 DNA. GAF or control antibody was added before (lanes 3 and 4) or after (lanes 5 and 6) combining nuclear extract with immobilized hsp70 DNA. Proteins bound to the immobilized DNA were detected by western blotting.
(C) Recombinant GAF recruits purified NELF to the hsp70 promoter. hsp70 promoterDNA was immobilized on magnetic beads and incubated with NELF in the absence (lane 1) or presence (lane 2) of GAF. Bound proteins were detected by western blotting.
To understand how GAF contributed to pausing, we investigated how the GAF antibody affected the associations of GAF and NELF with hsp70 in the nuclear extract. Incubation of the nuclear extract with GAF antibody prior to addition of the immobilized hsp70 DNA prevented both GAF and NELF from associating with the DNA (Figure 4B, compare lanes 3 and 4). In contrast, addition of GAF antibody after incubating the immobilized DNA with nuclear extract caused release of NELF but not GAF from the DNA (Figure 4B, compare lanes 5 and 6). Instead, GAF antibody associated with the promoter presumably by binding GAF (Figure 4B, lane 5).
A simple explanation for the effects of the GAF antibody on the promoter binding of GAF and NELF is that GAF recruits NELF to the DNA template, and that the GAF antibody displaces NELF from GAF. To directly test if GAF can recruit NELF to the promoter, we incubated purified NELF with immobilized DNA in the presence and absence of GAF (Figure 4C). NELF associated with the template in the presence of GAF but not in its absence indicating that GAF recruits NELF to the promoter.
Inhibition of GAF function shifts the location of the pause downstream
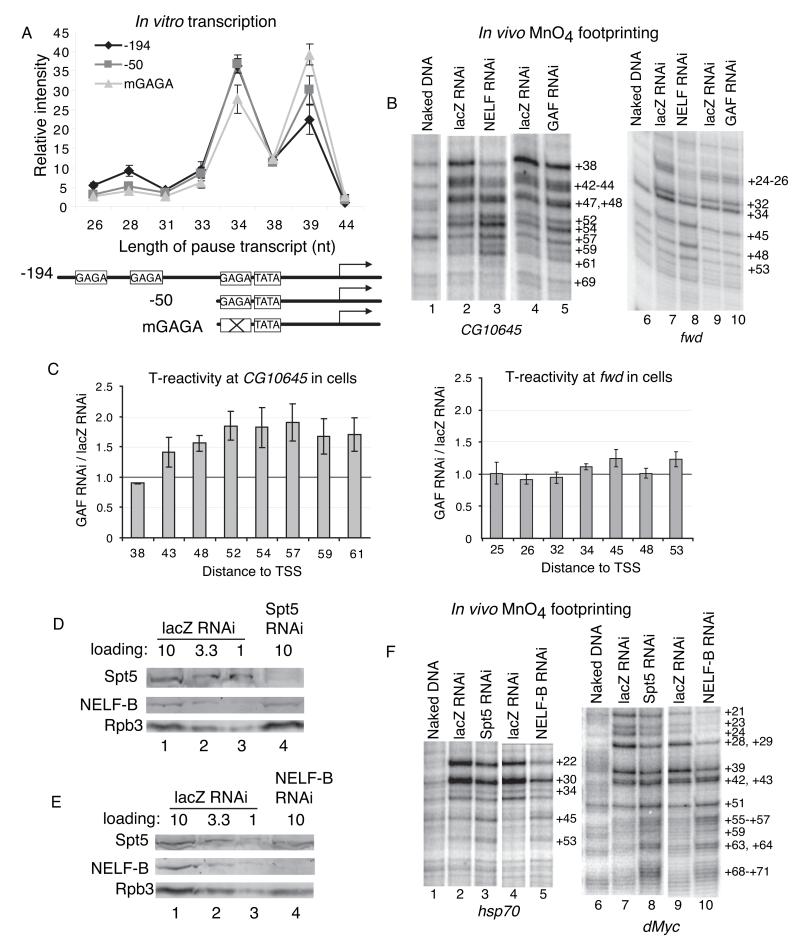
If GAF recruits NELF to the promoter, how might this recruitment contribute to pausing? There are multiple GAGA elements (GAF binding sites) in the hsp70 promoter (Gilmour et al., 1989; Weber et al., 1997) and we reasoned that deleting subsets of elements might provide information about the effect of GAF on pausing in the cell-free reaction. Deleting the two distal elements (−50 construct) caused the proportion of pauses to shift downstream and destroying an additional GAGA element caused an even greater proportion of the pauses to shift downstream (Figures 5A and S5A). Thus, GAF is influencing where the Pol II pauses.
Figure 5. Inhibition of GAF, NELF or DSIF shifts the location of pauses in the downstream direction.
(A) Distribution of paused transcripts transcribed from the wild-type (−194) and mutant hsp70 promoters in nuclear extracts. The −50 promoter deletes GAGA elements located between −194 and −50, and mGAGA mutates a GAGA element between −50 and −40. Band intensities corresponding to transcripts ending at the nucleotide positions shown on the abscissa were quantified and normalized to the total band intensities occurring from +26 to +44 (this compensated for reduced initiation occurring on promoters lacking GAGA elements, Figure S5A). Error bars are the SEM of three reactions done with different nuclear extract preparations. See Figure S5A for a representative gel.
(B) RNAi-mediated depletion of GAF from Drosophila S2 cells causes the permanganate reactivity on the GAF-associated gene, CG10645, to shift downstream from its normal position, but has no effect on the GAF-less gene, fwd. Cells were treated with GAF RNAi, NELF RNAi or the control lacZ RNAi prior to permanganate footprinting. The regimen of RNAi treatment for NELF and GAF differed so the corresponding regimens of lacZ RNAi treatment differed accordingly.
(C) Quantification of T-reactivity at the CG10645 and fwd promoters in GAF or lacZ RNAi-treated cells. The histograms show the ratio of the intensity of each major band from the GAF RNAi and lacZ RNAi samples. Error bars are the SEM of three independent RNAi experiments. Numbers above the red line correspond to increases in band intensity upon depletion of GAF.
(D) Western blot analysis of Drosophila S2R+ cells showing that Spt5 RNAi-treatment specifically depleted Spt5, the largest subunit of DSIF. Lanes 1 to 3 contain 3-fold dilutions of whole cell lysate from lacZ RNAi-treated cells. Lane 4 contains an amount of lysate from Spt5 RNAi-treated cells comparable to lane 1.
(E) Western blot analysis of Drosophila S2R+ cells showing that NELF-B RNAi-treatment specifically depleted NELF-B.
(F) Permanganate footprinting analyses of Drosophila S2 cells depleted of NELF (NELF-B RNAi) or DSIF (Spt5 RNAi). LacZ RNAi served as the negative control.
We performed a complementary experiment in vivo by depleting GAF from cells with RNAi. Based on a genomic analysis of GAF in Drosophila cells that had been depleted of GAF (J.T. Lis and M. Guertin, personal communication), we selected promoters that retained approximately 25% of the normal level of GAF following RNAi treatment so that the promoters would retain some transcriptional activity. Figures 5B and 5C show permanganate footprinting results in cells for a GAF gene, CG10645 and a GAF-less gene, fwd. Only CG10645 exhibited a downstream shift in the permanganate reactivity upon depletion of GAF. In contrast, both exhibited downstream shifts upon depletion of NELF (Figure 5B). GAF depletion caused a similar downstream shift of paused Pol II on a second GAF gene, CG11798 (Figure S5B). Importantly, the depletion of GAF did not alter the level of NELF or DSIF in cells (Figure S5C).
Decreasing NELF or DSIF levels in cells shifts the location of the pause downstream
The downstream shift in the location of the pause caused by perturbing GAF prompted us to posit that the location of the pause might be dependent on the rate at which pausing factors such as NELF associate with Pol II as Pol II transcribes the promoter proximal region. By recruiting NELF to the promoter, GAF could make the association of NELF with Pol II kinetically favorable.
If the rate at which NELF associates with the elongation complex affects the location of the pause, then reducing the amount of NELF in vivo should cause Pol II to pause downstream from its normal position. Moreover since NELF and DSIF bind cooperatively to the elongation complex (Missra and Gilmour, 2010), decreasing DSIF should also shift the location of the pause. In accordance with these predictions, RNAi-mediated depletion of NELF-B or the DSIF subunit Spt5 caused a downstream shift in the pattern of permanganate reactivity on hsp70 and dMyc, indicating the Pol II was pausing farther from the transcription start than normal (Figure 5F).
Slowing the elongation rate of Pol II in vitro or in vivo shifts the pause upstream
For the association rate between a pausing factor and the elongation complex to affect the location of the pause, this rate would need to be in competition with other processes that antagonize the action of the pausing factor. A possible candidate is the rate of Pol II elongation. To determine if the rate of elongation affects the location of the pause, we first determined what affect lowering the nucleotide concentration in our in vitro transcription reaction had on the location of the pause. Lowering the nucleotide concentration slows the elongation rate of Pol II, and this caused Pol II to pause closer to the TSS of hsp70, since the most abundant transcript at 100 uM NTP ends at +34 whereas the most abundant transcript at 250 uM NTP ends at +39 (Figure 6A, see lanes 1 and 3).
Figure 6. Slowing the rate of elongation shifts the location of pauses upstream.
(A) Altering the NTP concentration changes the location of paused Pol II in vitro. In vitro transcription was done with 100 μM NTP (lanes 1 and 2) or 250 μM NTP (lanes 3 and 4). High salt and sarkosyl were added to samples in lanes 2 and 4 to show that the short transcripts were engaged with paused Pol II.
(B) Permanganate footprinting analyses of salivary glands expressing normal Pol II (Control) or a slow mutant of Pol II (Slow). The first two lanes in each panel are purified DNA treated with permanganate for 0 or 1 min. The last 2 lanes in each panel show T-reactivity detected in salivary glands expressing normal (Control) or slow mutant Pol II.
To test if the elongation rate affected the location of the pause in vivo, we examined where Pol II paused on genes in Drosophila salivary glands expressing a mutant form of Pol II that slows the rate of elongation (Chen et al., 1996). The human counterpart of this slow mutant provided evidence that mRNA splice site selection is linked to the rate of elongation (de la Mata et al., 2003). Four promoters were selected based upon well-defined permanganate footprints between +30 and +50. Strikingly, T-reactivity for the slow Pol II was shifted closer to the TSS than for the wild-type Pol II (Figure 6B). These results indicate that where Pol II pauses in vivo is also impacted by the rate of transcription elongation.
Genomic analysis of the permanganate footprints indicates that GAF orchestrates efficient pausing
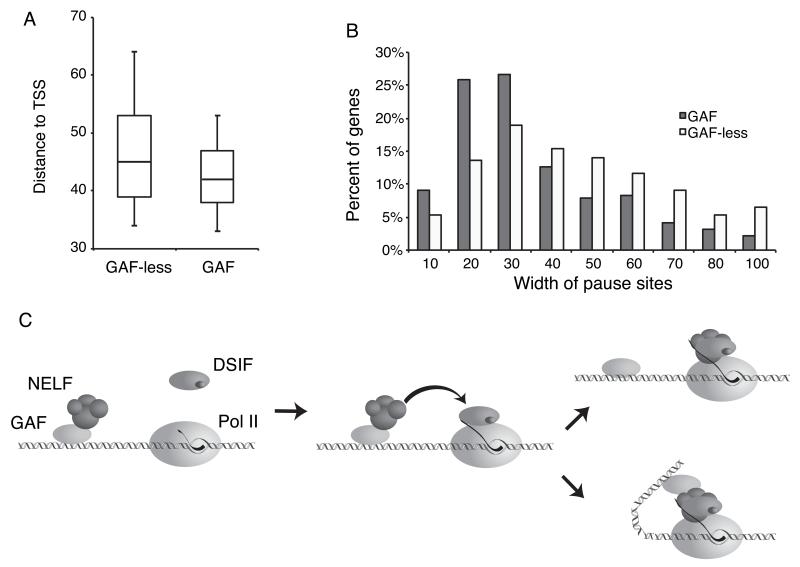
Finally, we sought to determine if GAF tips the kinetic competition in favor of pausing on a genomic scale. The high resolution of our permanganate-ChIP-seq allowed us to compare locations of the paused Pol II on GAF and GAF-less genes. GAF genes tend to have Pol II paused closer to the TSS than GAF-less genes (Figure 7A, P<0.0001 Mann-Whitney test). Moreover, the width of the intervals where pausing occurs is narrower on GAF genes than GAF-less genes (Figure 7B, P<0.0001 Mann-Whitney test). Hence, on a genomic scale, GAF corresponds with kinetically efficient pausing at promoters.
Figure 7. Genomic comparison of pausing on GAF and GAF-less genes provides evidence of efficient GAF-dependent pausing.
(A) Box plots of the distance from the center of transcription bubbles to the TSS of GAF-associated and GAF-less genes. Boxes depict 25th through 75th percentiles, and whiskers show 10th through 90th percentiles.
(B) Histograms of the width of pause sites for GAF and GAF-less genes.
(C) Models for how GAF could be involved in pausing after transcription initiation.
DISCUSSION
A rate-limiting step in the transcription of many genes in mammals and Drosophila is the pausing of Pol II in a region downstream from the transcription start site (Adelman and Lis, 2012). We have developed a comprehensive approach for investigating mechanisms that cause this pause. Our approach capitalizes on a new technique described here called permanganate-ChIP-seq that maps the distribution of transcriptionally engaged Pol II throughout the genome at near base-pair resolution. To investigate mechanistic aspects of promoter proximal pausing, we developed a biochemical system that recapitulates the promoter proximal pausing observed in vivo. This allowed us to investigate how GAF, a sequence specific transcription factor that binds upstream from the transcription start of over 1500 genes with paused Pol II, controls pausing.
We draw several significant conclusions. First, core promoters of protein-coding genes throughout the Drosophila genome contain little or no open Pol II promoter complexes. Thus immediately after assembly of the transcription complex at the core promoter, Pol II rapidly initiates and elongates into a paused state that precludes assembly of a second polymerase into an initiation complex. snRNA genes encoding components of the spliceosome and the induced heat shock genes are exceptional in that open Pol II promoter complexes are evident on these genes. Second, the location of the pause can be altered by perturbing the kinetics of elongation or the levels of pausing factors suggesting that the location of the pause depends on a kinetic competition between elongation and the binding of these factors. Third, GAF tips this kinetic competition in favor of pausing by recruiting NELF to the promoter.
Permanganate-ChIP-seq reveals that paused Pol II occludes assembly of a second polymerase at the core promoter of protein coding genes
The results of our permanganate-ChIP-seq shows that Pol II associates with the downstream promoter region of protein-coding genes primarily in a transcriptionally engaged state, 20 to 60 nucleotides downstream from the start site. In contrast to other available genomic methods, we can rule-out that there are Pol II molecules in open complexes at the transcription start sites of these genes. This situation differs dramatically from budding yeast, where a Pol II stably resides at the core promoter just upstream of the transcription start site (Rhee and Pugh, 2012), and indicates that open complexes detected at core promoters in vitro (Nock et al., 2012) are short-lived. Only on the induced heat shock genes, where the initiation frequency is estimated to be once every few seconds, is a substantial level of open complex evident at the transcription start site. For other protein coding genes, the paused Pol II becomes the dominant state at the promoter as it likely occludes other Pol II from assembling and initiating transcription until the paused Pol II resumes elongation. Thus, the pausing checkpoint might serve two purposes: ensuring that Pol II is properly licensed for elongation and ensuring that a second polymerase does not assemble until the first is well on its way.
Unlike protein-coding genes, snRNA genes encoding parts of the spliceosome show clear evidence of open Pol II complexes at the transcription start site. The promoters of these genes function differently from other Pol II-targeted promoters. snRNA genes use a complex called SNAP(c) instead of TFIID as a foundation for assembling preinitiation complexes (Hernandez, 2001). These genes are also regulated by an alternate elongation complex that lacks P-TEFb, the kinase that functions to reactivate paused Pol II at protein-coding genes (Smith et al., 2011). The distinction between snRNA genes and protein-coding genes revealed by our permanganate-ChIP-seq raises the possibility that TFIID itself could have some role in dictating the pause. Consistent with this possibility, the efficiency and location of the pause on hsp70 has been linked to sequence elements in the core promoter that are recognized by TFIID (Kwak et al., 2013). Alternatively, the SNAP(c) complex might antagonize Pol II pausing at the promoter.
Kinetic Competition between Pol II elongation and NELF dictates the location and efficiency of promoter proximal pausing on GAF genes
Permanganate-ChIP-seq shows that the location and efficiency of promoter proximal pausing varies among promoters. Our results indicate that the location of the pause can be controlled by a kinetic competition between the rate of Pol II elongation and the rate at which NELF associates with the elongation complex. This conclusion is based on the observations that perturbations in factors involved in pausing shift the location of the paused Pol II in the promoter proximal region. These are the first instances where such shifts have been observed.
The most compelling evidence that the Pol II elongation rate affects pausing is our finding that a slow Pol II mutant pauses closer to the TSS than wild-type Pol II in living cells. In accordance with this, lowering the concentration of nucleotides in the in vitro transcription reaction shifted the pause closer to the TSS. Several lines of evidence indicate that the association rate of NELF impacts pausing. First, we show that depleting NELF causes Pol II to pause farther away from the TSS. Second, GAF recruits NELF to the hsp70 promoter and depleting GAF from cells or deleting binding sites for GAF from the hsp70 promoter causes the location of the paused Pol II to shift downstream from the TSS. Finally, on a genomic level, we observe that GAF-associated promoters have higher levels of paused Pol II and the location of the pause is closer to the TSS and distributed over a narrower region than for other promoters.
DNA Binding Factors Recruit NELF to Control Pausing
By using an antibody to inhibit GAF function during specific stages of the transcription cycle, we show that GAF not only functions in initiation but also directly impacts the pause after Pol II has initiated transcription. Furthermore, we find that GAF can recruit NELF to the promoter in the absence of transcription initiation. Previously, we showed that the elongation complex must transcribe greater than 18 nucleotides to stably associate with DSIF, similar to what was observed for human Pol II elongation complexes (Cheng and Price, 2008; Missra and Gilmour, 2010). We also showed that DSIF and NELF bind cooperatively to the elongation complex (Missra and Gilmour, 2010). We propose that GAF recruits NELF to the promoter before transcription initiates and poises NELF to bind the elongation complex once the elongation complex has transcribed far enough to associate with DSIF (Figure 7C). GAF binds DNA as a large oligomeric complex (Katsani et al., 1999) so multiple points of NELF binding could be provided by GAF at the promoter. GAF might function simply to increase the local concentration of NELF at a promoter (Figure 7C upper panel), or it might act as an allosteric regulator of NELF to increase NELF’s affinity for the DSIF-Pol II elongation complex (Figure 7C lower panel).
GAF is probably not alone in being able to recruit NELF to a gene’s promoter. NELF was previously shown to associate with the estrogen receptor, c-fos, and c-jun (Aiyar et al., 2004; Zhong et al., 2004). In each of these cases, the DNA binding protein is an activator that associates with its target gene during induction yet NELF functions to attenuate the level of expression. These opposing activities could serve to fine-tune the level and duration of expression.
Unraveling control mechanisms for promoter proximal pausing
Our combination of in vitro and in vivo analyses of promoter proximal pausing has lead us to conclude that the timing of the association of pausing factors and Pol II elongation have a significant role in dictating the location and efficiency of pausing. While GAF facilitates pausing by recruiting NELF, other factors undoubtedly contribute to pausing. Pausing on the oaf promoter in the nuclear extract occurred independently of GAF and presumably nucleosomes, so other sequence specific transcription factors might exist that function analogously to GAF.
Previous studies have linked several other features to promoter proximal pausing. Chromatin structure has been implicated in pausing Pol II on the hsp70 gene in mammals (Brown et al., 1996). As demonstrated here and previously (Benjamin and Gilmour, 1998), chromatin is not essential to reconstitute paused Pol II on Drosophila hsp70 in vitro. Moreover, our analysis of the nucleosome organization on GAF and GAF-less genes reveals striking differences that suggest chromatin is unlikely to cause pausing on GAF genes (Figure S6). We speculate that by recruiting NELF, GAF circumvents the contribution that chromatin structure makes to these genes.
The stability of the DNA-RNA heteroduplex was found to correlate with the location of short transcripts (Nechaev et al., 2010). However, our analysis of the relationship of the energetic landscape of the nucleic acid framework in the elongation complex and the location of the transcription bubble indicates that much of the Pol II is not pausing at sites where the stability of the heteroduplex or the transcription bubble as a whole achieves a local minimum (Figure S7); such local minima are predicted to be places for Pol II to pause if pausing is dictated solely by the thermodynamic stability of the nucleic acid framework (Bai et al., 2004). Furthermore, our finding that the location of the pause can be shifted without changing the nucleic acid sequence indicates that the stability of the transcription bubble is not a major determinant of the location of the pause on the genes that were analyzed.
A computational analysis of approximately 1500 paused genes revealed enrichment of the GAGA element, initiator, DPE and a motif related to the DPE called the pause button (Hendrix et al., 2008). Paused genes in the Hendrix et al. study were defined by the ratio of the Pol II density in the promoter and the body of the gene. Our search for conserved sequences among the larger number of paused genes defined by permanganate-ChIP-seq only identified the GAGA element and another sequence element known as Motif 1 (Figure S2). We have identified a novel protein that recognizes Motif 1, and our results indicate that GAF and this novel protein are likely to orchestrate distinct mechanisms of pausing (manuscript submitted). The initiator, DPE and the pause button are also enriched among those promoters with the highest level of paused Pol II (Figure S2). These elements are likely to be recognized by TFIID, and therefore might be the complex recently implicated in contributing to pausing on hsp70 in Drosophila (Kwak et al., 2013).
Finally, recent biochemical analyses have identified proteins in addition to NELF and DSIF that could function in pausing Pol II in the promoter proximal region (Cheng et al., 2012). These include two proteins that associate with Pol II, Gdown1 and TFIIS, and an additional activity whose identity is currently unknown. Future biochemical studies with the Drosophila system could aid in identifying new pausing factors and assessing the function of current candidates. By using antibodies against factors that associate with the Pol II elongation complex, permanganate-ChIP-seq could provide detailed information about when and where factors associate with elongation complexes, thus, providing new insights into the function of these factors in vivo.
EXPERIMENTAL PROCEDURES
Permanganate-ChIP-seq
Permanganate-ChIP-seq was performed on Drosophila tissue culture cells and combined: Pol II ChIP (Petesch and Lis, 2008), permanganate genomic footprinting (Lee et al., 2008), and steps from ChIP-exo (Rhee and Pugh, 2011). A detailed description of the procedure is provided in the Supplemental Information.
The majority of cleavage sites throughout the genome mapped to thymine, and the second most frequent cleavage site mapped to cytosine (Figure S1A). This matched the permanganate sensitivity of nucleotides measured in vitro (Bui et al., 2003), indicating the specificity of the permanganate reaction. Unless specified, we used reads that mapped to thymine (T-reactivity) to avoid ‘background’ cleavage by piperidine at non-oxidized nucleotides that are not necessarily within transcription bubbles. Since we mapped the cleavage sites on both strands, permanganate reactivity is displayed at both A and T in the genomic sequence of one strand. These results were not biased by the nucleotide composition, because T-reactivity had no correlation with the AT content in the local genomic DNA sequence (Figure S1B, C).
Promoter proximal pausing in Drosophila nuclear extracts
In vitro transcription reactions were done in nuclear extracts derived from Drosophila embryos (Biggin and Tjian, 1988) using a pulse-chase labeling strategy to restrict labeling in newly synthesized transcripts to the 5′ region (Marshall et al., 1996), and oligonucleotide-directed purification of the transcripts (Rasmussen and Lis, 1993). Nuclear extracts were dialyzed to a conductivity equal to buffer with 150 mM KCl rather than 100 mM because this retained more DSIF and NELF in a soluble state. The oligonucleotide-directed purification of the transcripts separates the transcripts from radiolabelled products that were not produced by Pol II. The shortest transcripts that we have detected with this technique are 18 nucleotides so we can not rule out the possibility that some population of Pol II molecules are pausing closer to the transcription start site in the cell-free reactions. A detailed description of the protocol is provided in the Supplemental Information.
Immunodepletion of NELF, DSIF, and GAF, and sources of recombinant DSIF, NELF and GAF
NELF-, DSIF-, and GAF-depleted nuclear extracts were prepared as previously described (Wu et al., 2005). DSIF used in Figure 2B was purified from a baculovirus expression system (Wu et al., 2003). NELF complex used in Figures 2B and 4C was purified from a transgenic fly line expressing flag-NELF D (Missra and Gilmour, 2010). Flag-GAF used in Figure 3A was purified from embryo nuclear extracts of a transgenic fly line (Shimojima et al., 2003). The full-length GAF (isoform 519) used in Figure 4C was cloned into the pET28 vector with a His-tag at the C-terminus. His-GAF was expressed in BL21 DE3 and purified with TALON cobalt beads.
Pull-down assays with immobilized templates
A detailed description for the generation of biotinylated DNA templates and their immobilization on magnetic beads is provided in the Supplemental Information. For experiments in Figures 4A and 4B, immobilized templates were incubated with nuclear extract and magnetically isolated. Following washes, proteins were eluted and analyzed by western blotting. Similar pull-down experiments were done with recombinant GAF and purified NELF (Figure 4C). Further information is provided in the Supplemental Information.
Potassium permanganate analysis of pausing in vivo
Fly lines expressing a mutant Pol II (C4) or wild type Pol II were grown at 24 °C. The C4 mutant, stock number 3663, was obtained from the Bloomington Drosophila Stock Center. Salivary glands from both lines were dissected, and genomic footprinting with potassium permanganate was done as described (Gilmour and Fan, 2009). Permanganate genomic footprinting of tissue culture cells and the primers used in ligation-mediated PCR were as previously described (Lee et al., 2008).
Bioinformatic analyses
Supplementary Material
Highlights.
Genome-wide mapping of transcription bubbles reveals locations of paused Pol II
Open Pol II complexes rarely exist at the start sites
Where Pol II pauses depends on rates of NELF binding and transcription elongation
GAF facilitates loading NELF onto Pol II after transcriptional initiation
ACKNOWLEDGMENTS
We thank Mike Guertin and John Lis for providing unpublished results from their RNAi-mediated, GAF depletion analysis. We also thank Dr. Susumu Hirose at National Institute of Genetics in Japan for providing the flag-GAF fly line. This work was supported by grant GM47477 from the NIH to D.S.G, and by grant ES013768 from NIH to B.F.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Raw sequencing data are available at the NCBI Sequence Read Archive (accession number: GSE46620).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and seven figures.
REFERENCES
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- Benjamin LR, Gilmour DS. Nucleosomes are not necessary for promoter-proximal pausing in vitro on the Drosophila hsp70 promoter. Nucleic Acids Res. 1998;26:1051–1055. doi: 10.1093/nar/26.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Bui CT, Rees K, Cotton RG. Permanganate oxidation reactions of DNA: perspective in biological studies. Nucleosides Nucleotides Nucleic Acids. 2003;22:1835–1855. doi: 10.1081/NCN-120023276. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chafin D, Price DH, Greenleaf AL. Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem. 1996;271:5993–5999. [PubMed] [Google Scholar]
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Experimental cell research. 2010;316:2723–2730. doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol. 2011;21:1624–1634. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Dietz TJ, Elgin SC. TATA box-dependent protein-DNA interactions are detected on heat shock and histone gene promoters in nuclear extracts derived from Drosophila melanogaster embryos. Mol Cell Biol. 1988;8:3204–3214. doi: 10.1128/mcb.8.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Fan R. Detecting transcriptionally engaged RNA polymerase in eukaryotic cells with permanganate genomic footprinting. Methods. 2009;48:368–374. doi: 10.1016/j.ymeth.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Thomas GH, Elgin SC. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989;245:1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. Embo J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Li B, Weber JA, Chen Y, Greenleaf AL, Gilmour DS. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock A, Ascano JM, Barrero MJ, Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol Cell. 2012;48:837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends in biochemical sciences. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al. The little elongation complex regulates small nuclear RNA transcription. Mol Cell. 2011;44:954–965. doi: 10.1016/j.molcel.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Taxman DJ, Lu Q, Gilmour DS. Molecular architecture of the hsp70 promoter after deletion of the TATA box or the upstream regulation region. Mol Cell Biol. 1997;17:3799–3808. doi: 10.1128/mcb.17.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Lee C, Fan R, Smith MJ, Yamaguchi Y, Handa H, Gilmour DS. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhong H, Zhu J, Zhang H, Ding L, Sun Y, Huang C, Ye Q. COBRA1 inhibits AP-1 transcriptional activity in transfected cells. Biochem Biophys Res Commun. 2004;325:568–573. doi: 10.1016/j.bbrc.2004.10.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.