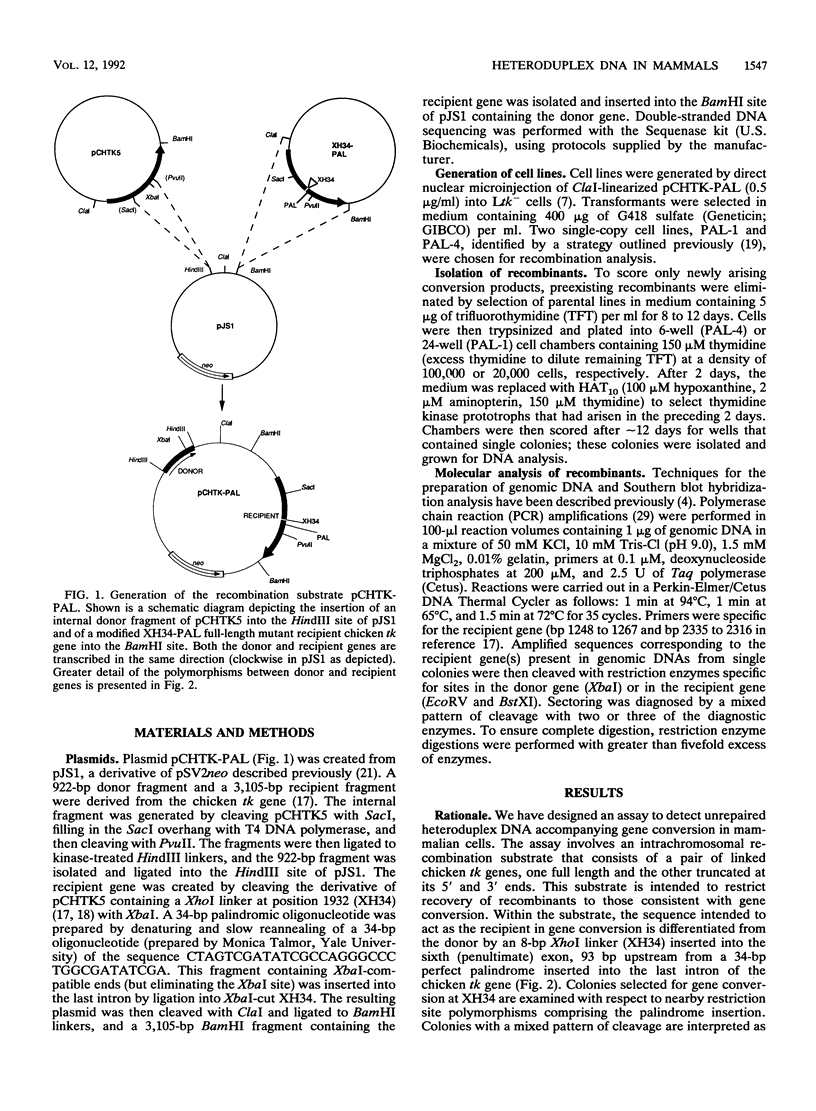
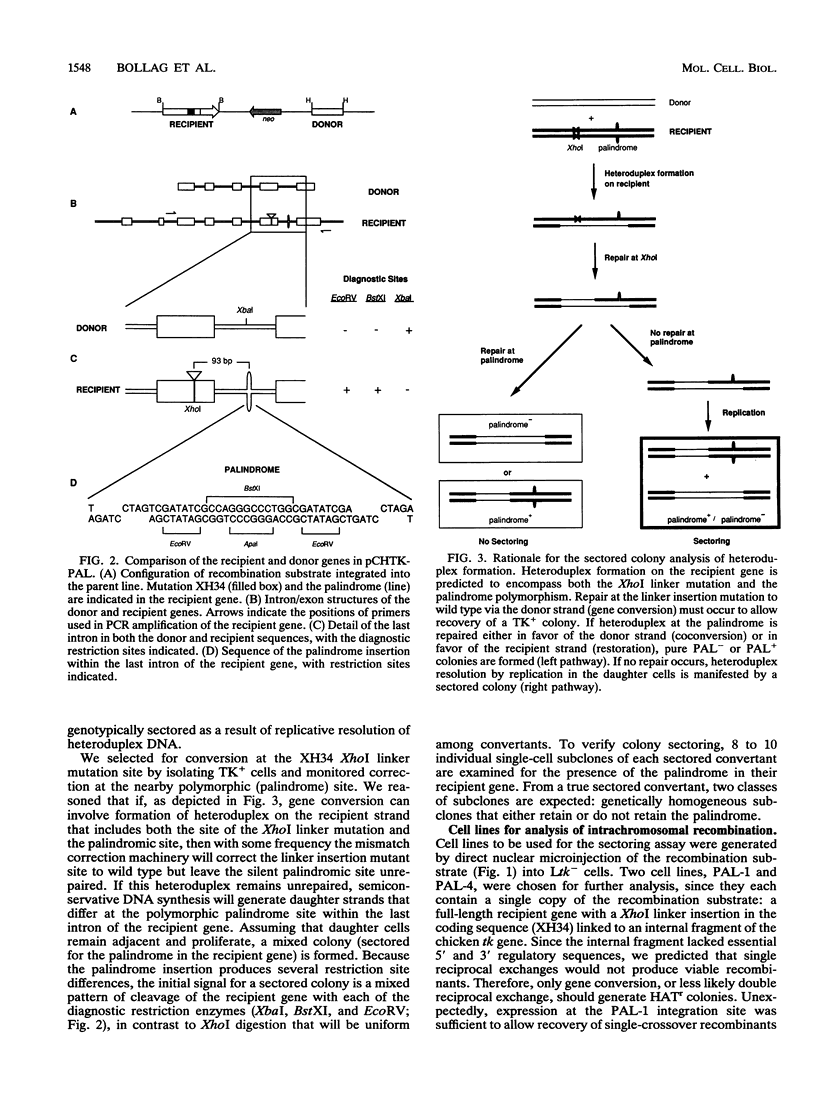
Abstract
We have studied intrachromosomal gene conversion in mouse Ltk- cells with a substrate designed to provide genetic evidence for heteroduplex DNA. Our recombination substrate consists of two defective chicken thymidine kinase genes arranged so as to favor the selection of gene conversion products. The gene intended to serve as the recipient in gene conversion differs from the donor sequence by virtue of a palindromic insertion that creates silent restriction site polymorphisms between the two genes. While selection for gene conversion at a XhoI linker insertion within the recipient gene results in coconversion of the nearby palindromic site in more than half of the convertants, 4% of convertant colonies show both parental and nonparental genotypes at the polymorphic site. We consider these mixed colonies to be the result of genotypic sectoring and interpret this sectoring to be a consequence of unrepaired heteroduplex DNA at the polymorphic palindromic site. DNA replication through the heteroduplex recombination intermediate generates genetically distinct daughter cells that comprise a single colony. We believe that the data provide the first compelling genetic evidence for the presence of heteroduplex DNA during chromosomal gene conversion in mammalian cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Cami B., Dinh T. H., Igolen J., Kourilsky P. Processing of complex heteroduplexes in Escherichia coli and Cos-1 monkey cells. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5792–5796. doi: 10.1073/pnas.81.18.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayares D., Ganea D., Chekuri L., Campbell C. R., Kucherlapati R. Repair of single-stranded DNA nicks, gaps, and loops in mammalian cells. Mol Cell Biol. 1987 May;7(5):1656–1662. doi: 10.1128/mcb.7.5.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987 Jul 23;328(6128):362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- Bollag R. J., Liskay R. M. Conservative intrachromosomal recombination between inverted repeats in mouse cells: association between reciprocal exchange and gene conversion. Genetics. 1988 May;119(1):161–169. doi: 10.1093/genetics/119.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Liskay R. M. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol Cell Biol. 1991 Oct;11(10):4839–4845. doi: 10.1128/mcb.11.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Capecchi M. R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980 Nov;22(2 Pt 2):479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- Esposito M. S. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Efficient correction of mismatched bases in plasmid heteroduplexes injected into cultured mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):70–74. doi: 10.1128/mcb.5.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer P. M., Sarkar S. N., Chisholm G. E., Summers W. C. DNA mismatch repair detected in human cell extracts. Mol Cell Biol. 1987 Jan;7(1):218–224. doi: 10.1128/mcb.7.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S. Coincident gene conversion during mitosis in saccharomyces. Genetics. 1984 Jul;107(3):355–365. doi: 10.1093/genetics/107.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S. Mitotic recombination: mismatch correction and replicational resolution of Holliday structures formed at the two strand stage in Saccharomyces. Mol Gen Genet. 1981;183(2):252–263. doi: 10.1007/BF00270626. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwoh T. J., Engler J. A. The nucleotide sequence of the chicken thymidine kinase gene and the relationship of its predicted polypeptide to that of the vaccinia virus thymidine kinase. Nucleic Acids Res. 1984 May 11;12(9):3959–3971. doi: 10.1093/nar/12.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwoh T. J., Zipser D., Wigler M. Mutational analysis of the cloned chicken thymidine kinase gene. J Mol Appl Genet. 1983;2(2):191–200. [PubMed] [Google Scholar]
- Letsou A., Liskay R. M. Effect of the molecular nature of mutation on the efficiency of intrachromosomal gene conversion in mouse cells. Genetics. 1987 Dec;117(4):759–769. doi: 10.1093/genetics/117.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L., Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. D., Holliday R. Evidence for the formation of hybrid DNA during mitotic recombination in Chinese hamster cells. Cell. 1976 Aug;8(4):573–579. doi: 10.1016/0092-8674(76)90225-7. [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Genetic evidence for preferential strand transfer during meiotic recombination in yeast. Genetics. 1990 Aug;125(4):753–761. doi: 10.1093/genetics/125.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A., Petes T. D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989 Jul 27;340(6231):318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., Moore P. D. Molecular recombination and the repair of DNA double-strand breaks in CHO cells. Nucleic Acids Res. 1979 Jul 11;6(9):3145–3160. doi: 10.1093/nar/6.9.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H., Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2696–2700. doi: 10.1073/pnas.85.8.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]
- Weiss U., Wilson J. H. Heteroduplex-induced mutagenesis in mammalian cells. Nucleic Acids Res. 1988 Mar 25;16(5):2313–2322. doi: 10.1093/nar/16.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss U., Wilson J. H. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., DiMartino J. F., Anderson R. W., Lusnak K., Hilbert D., Fogel S. A DNA sequence conferring high postmeiotic segregation frequency to heterozygous deletions in Saccharomyces cerevisiae is related to sequences associated with eucaryotic recombination hotspots. Mol Cell Biol. 1988 Mar;8(3):1253–1258. doi: 10.1128/mcb.8.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985 May 23;315(6017):350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- Wildenberg J. The relation of mitotic recombination to DNA replication in yeast pedigrees. Genetics. 1970 Oct;66(2):291–304. doi: 10.1093/genetics/66.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]