Abstract
HIF-2alpha plays a critical role in renal tumorigenesis. HIF-2alpha is stabilized in Von Hippel-Lindau (VHL)-deficient renal cell carcinoma through mechanisms that require ongoing mRNA translation. Mammalian target of Rapamycin (mTOR) functions in two distinct complexes, Raptor-associated mTORC1 and Rictor-associated mTORC2. Rictor-associated mTORC2 complex has been linked to maintaining HIF-2alpha protein in the absence of VHL, however the mechanisms remain to be elucidated. Although Raptor-associated mTORC1 is a known key upstream regulator of mRNA translation, initiation and elongation, the role of mTORC2 in regulating mRNA translation, is not clear. Complex assembly of the mRNA cap protein, eIF4E, with activators (eIF4G) and inhibitors (4E-BP1) are rate-limiting determinants of mRNA translation. Our laboratory has previously demonstrated that reactive oxygen species, mediated by p22phox-based Nox oxidases, are enhanced in VHL-deficient cells and play a role in the activation of Akt on S473, a site phosphorylated by the mTORC2 complex. In this study, we examined the role of Rictor-dependent regulation of HIF-2alpha through eIF4E-dependent mRNA translation and examined the effects of p22phox-based Nox oxidases on TORC2 regulation. We demonstrate for the first time that mTORC2 complex stability and activation is redox sensitive and further defined a novel role for p22phox-based Nox oxidases in eIF4E-dependent mRNA translation through mTORC2. Furthermore, we provide the first evidence that silencing of p22phox reduces HIF-2alpha-dependent gene targeting in vitro and tumor formation in vivo. The clinical relevance of these studies is demonstrated.
Keywords: mTOR, Nox Oxidase, Rictor, p22phox, HIF-2alpha, von Hippel-Lindau, renal cancer, oxidative stress
Introduction
Hypoxia Inducible Factor-2alpha (HIF-2alpha) is stabilized in Von Hippel-Lindau (VHL)-deficient renal cell carcinoma (RCC) (1-3). Our laboratory has demonstrated that ongoing mRNA translation is necessary for maintaining HIF-2alpha protein expression in the absence of VHL through mechanisms involving p22phox-based Nox oxidases (4). mRNA translation is regulated at multiple levels and deregulation of mRNA translation is evident in many cancers (5-8). In part, initiation of cap-dependent mRNA translation requires activation of the Raptor-associated mammalian target of rapamycin (mTORC1) signaling pathway and formation of the eIF4F complex which include the cap binding protein, eIF4E, the ATPase/RNA helicase, eIF4A, and the scaffold protein, eIF4G. eIF4E has two key mutually exclusive binding partners; eukaryotic translation initiation factor 4 gamma (eIF4G) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), which lead to mRNA translation activation or repression respectively (9-11). Phosphorylation of 4E-BP1 by mTORC1, results in 4E-BP1/eIF4E dissociation allowing eIF4E to bind eIF4G and activate mRNA translation. mTOR functions in two distinct complexes, mTORC1 and mTORC2, which contain common (mLST8/GβL) and unique (Raptor and Rictor) protein partners (13-15). We have previously demonstrated that p22phox-based Nox oxidases mediate mRNA translation of HIF-2alpha, in part, through Akt-dependent inactivation and degradation of the mTORC1 inhibitor, Tuberin (16). Overexpression of a dominant-negative form of 4E-BP1, which harbors mutations in the mTORC1-dependent phosphorylation sites, reduces HIF-2alpha protein expression in VHL-deficient cells, suggesting that initiation of mRNA translation is necessary for maintaining HIF-2alpha in the absence of VHL (16). More recently it has been demonstrated that knock-down of Rictor, a subunit of the mTORC2 complex, but not Raptor, a subunit of the mTORC1 complex reduces HIF-2alpha subunit in VHL-deficient cells suggesting that mTORC2 complex play a role in the regulation of HIF-2alpha; although the mechanism have yet to be elucidated (17). In this study, we examined the role of p22phox-based Nox oxidases in Rictor-dependent regulation of HIF-2alpha protein expression and elucidated the mechanisms by which Rictor regulates HIF-2alpha in the absence of VHL. We demonstrate for the first time that mTORC2 complex stability and activation is redox sensitive and further defined a novel role for p22phox-based Nox oxidases in eIF4E-dependent mRNA translation through mTORC2. Furthermore, we provide the first evidence that silencing of p22phox reduces HIF-2alpha-dependent gene targeting in vitro and tumor formation in vivo. The clinical relevance of these studies is demonstrated.
Results
Blocking TORC2 reduces HIF-2alpha protein expression through inhibition of the mRNA initiation complex
To examine the role and elucidate the mechanisms of TORC1 and TORC2 in maintaining HIF-2alpha protein expression, we used the rapalog inhibitor of TORC1, temsirolimus (CCI-779) or pp242, a newly developed mTORC1/mTORC2 inhibitor (20). We find that treatment of VHL-deficient RCC 786-O cells with CCI-779 had no effect on HIF-2alpha protein expression; however, cells treated with pp242 efficiently reduced HIF-2alpha protein expression (Fig. 1A). Importantly, mRNA levels of HIF-2alpha did not significantly change (Fig. 1B). Due to the fact that pp242 reduces HIF-2alpha protein expression independent of mRNA levels, we next examined the effects of TORC inhibitors on mRNA translation. eIF4E is a eukaryotic translation initiation factor involved in directing ribosomes to the cap structure of mRNAs. eIF4E bound to 4E-BP1 indicates inhibition of mRNA translation. We treated RCC 786-O cells with CCI-779 or pp242 and affinity purified eIF4E using 7-methyl-GTP sepharose beads (7mGTP) that mimic the mRNA cap structure (21). 4E-BP1 was not detected in affinity purified eIF4E pull downs in RCC 786-O buffer or CCI-779 treated cells whereas 4E-BP1 was readily detected in affinity purified eIF4E pull downs in RCC 786-O cells treated with pp242 (Fig. 1C).
Figure 1.
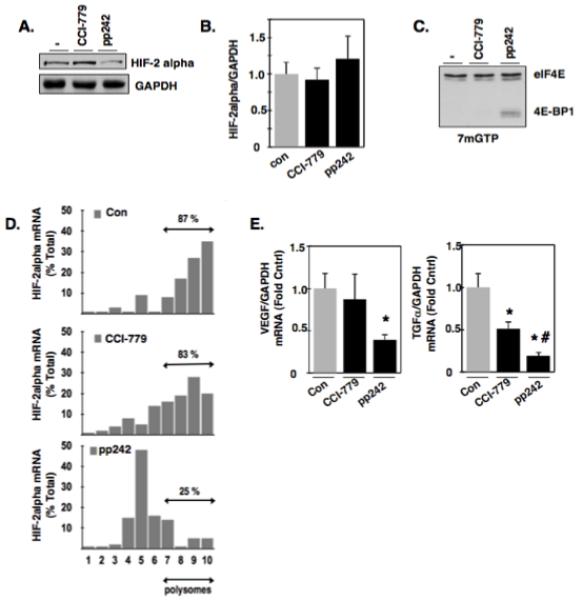
Effects of mTORC1 and mTORC2 inhibition on eIF4E binding to mRNA repressor, 4E-BP1. (a): VHL-deficient RCC 786-O cells were incubated with buffer alone (−), mTORC1 inhibitor (CCI-779), or the mTORC1/mTORC2 inhibitor (pp242) for 24 hours. Cellular lysates were analyzed by Western blot analysis for HIF-2alpha expression. GAPDH was used as a loading control. (b): In parallel to A, RNA was extracted and HIF-2alpha mRNA was examined by quantitative RT-PCR. (c): Cellular lysates were prepared as described in materials and methods from RCC 786-O cells treated with buffer alone (−), CCI-779, or pp242 for 24 hrs and eIF4E and its associated proteins were affinity purified using 7-methyl-GTP sepharose beads followed by Western blot analysis for total eIF4E and 4E-BP1. The data are representative of three independent experiments. (d): Polysomal fractions were prepared by passing cytosolic lysates from RCC 786-O cells treated with buffer treated (−), CCI-779, or pp242, over sucrose gradients as described in experimental methods. HIF-2alpha and GAPDH mRNAs were detected in each fraction by RT-PCR on RNA extracted from each fraction. Sedimentation analysis was examined in parallel of RCC 786-O cells by measuring optical density at 254 nm. (e): Quantitative RT-PCR was performed as outlined in materials and methods to examine the mRNA expression levels HIF-responsive genes (VEGF and TGF-alpha) in RCC 786-O cells treated with buffer alone (−), CCI-779, and pp242. The data is representative of three independent experiments and are expressed as fold control where the ratio of the buffer treated control cells was defined as 1. Values are the means +/− S.E, * p<0.05 and # p<0.05 compared to CCI-779 alone.
eIF4E association with 4E-BP1 inhibits initiation of mRNA translation, therefore, we examined the polysomal profile of HIF-2alpha mRNA as a readout of mRNA translation. The polysomal distribution of HIF-2alpha mRNA was examined in the presence or absence of the aforementioned mTORC inhibitors. Polysomal distribution of HIF-2alpha mRNA was assessed by ultracentrifugation of postnuclear fractions in 15-40% sucrose gradients. After centrifugation, gradients were separated in 10 fractions, RNA was extracted from each fraction, and HIF-2alpha and GAPDH mRNA abundance was measured by RT-qPCR. Heavy polysomal fractions (6-10) are indicated as assembled ribosomes as determined by the basal polysomal profile (data not shown) and suggestive of active translation where as lighter polysomal fractions (1-5) are indicated as unassembled ribosomes and is suggestive of inhibition of translation. Figure 1D shows that HIF-2alpha mRNA is most abundant in heavy polysomal fractions 6-10 in RCC 786-O cells treated with buffer alone or CCI-779 (87% and 83% respectively) whereas RCC 786-O cells treated with pp242 resulted in a reduction of HIF-2alpha mRNA in heavy polysomal fractions (~60%) concomitant with an increase of HIF-2alpha mRNA in lighter polysomal fractions, suggesting an inhibition of mRNA translation. GAPDH mRNA distribution was not affected by the treatments (data not shown). In parallel, HIF-2alpha target genes were examined as a read-out of HIF-2alpha activity in RCC 786-O cells treated with buffer alone, CCI-779, or pp242. We find that RCC 786-O cells treated with pp242 significantly reduced HIF-alpha target genes, VEGF and TGF-alpha compared to buffer treated cells, whereas CCI-779 partially reduces TGF-alpha (Fig. 1E).
Rictor regulates HIF-2alpha through eIF4E-dependent mRNA translational mechanisms
To further characterize and define the role of TORC2 in the regulation of 4E-BP1-eIF4E complex assembly involved in mRNA translation and to determine if this is a mechanism by which TORC2 regulates HIF-2alpha, we transiently knocked down an important subunit of the TORC2 complex, Rictor, using small interfering RNAs (siRictor) or scrambled control (Scr). As expected, downregulation of Rictor efficiently reduced phosphorylation of Akt (pAkt473), a known downstream target of TORC2, and reduced HIF-2alpha expression in VHL-deficient cells with no effect on total Akt levels (Fig. 2A, 17). Successful Rictor knockdown was confirmed by Western blot analysis and GAPDH was used as a loading control. Moreover, mRNA expression of HIF-2alpha was not altered in siRictor transfected cells compared to scr control (Fig. 2B). Affinity purification of eIF4E using 7-methyl-GTP sepharose beads in RCC 786-O cells silenced of Rictor (siRictor) resulted in enhanced eIF4E binding to 4E-BP1 in 4 independent transfections compared to RCC 786-O cells transfected with 3 independent scrambled controls (Scr) (Fig. 2C). Importantly, polysomal profiling of HIF-2alpha mRNA in RCC 786-O cells transiently silenced of Rictor demonstrates an approximate 50% reduction of the HIF-2alpha mRNA in heavy polysomal fractions compared to scrambled transfected controls (Fig. 2D). Similarly, transient silencing of Rictor using small inhibitory RNA in an independent VHL-deficient cell line which only expresses HIF-2alpha, A498, resulted in reduced HIF-2alpha expression (Fig. S1A) and a reduction of HIF-2alpha mRNA in heavy polysomal fractions of approximately 45% compared to scrambled transfected control (Fig. S1B). In support of these data, examination of HIF-2alpha target genes (VEGF and TGF-alpha) in RCC 786-O cells silenced of Rictor demonstrated a significant reduction of VEGF and TGF-alpha mRNA compared to scrambled control (Fig. 2E). Taken together, this suggests Rictor-associated mTORC2 regulates HIF-2alpha and HIF-2alpha target genes through regulation of 4E-BP1-eIF4E complex assembly.
Figure 2.
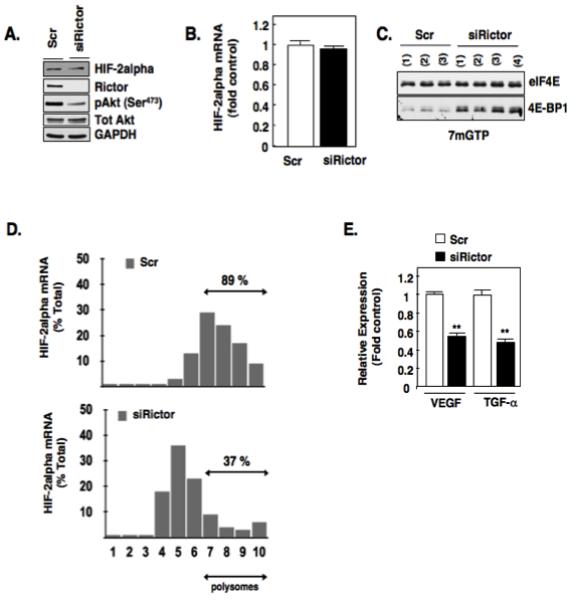
Rictor mediates eIF4E binding to mRNA translational repressor, 4E-BP1. (a): Equivalent amounts of cell lysates were analyzed for known downstream targets of Rictor, HIF-2alpha, pAkt (S473) by Western blot analysis in RCC 786-O cells silenced of Rictor using small inhibitory RNA (siRNA) or scrambled control (scr) as described in materials and methods. Total Akt and GAPDH were used as loading controls. (b): Steady state levels of HIF-2alpha mRNA levels were examined in RCC 786-O cells silenced of Rictor (siRictor) or scrambled control (scr). (c): eIF4E association with 4E-BP1 was examined as outlined in Fig. 1C using 7-methyl-GTP sepharose beads from RCC 786-O cells silenced for Rictor using siRNA (siRictor) or scrambled control (scr). The data are representative of at least three independent experiments. (d): Polysomal fractions were prepared by passing cytosolic lysates from RCC 786-O cells silenced of Rictor (siRictor) or scrambled control (scr), over sucrose gradients as described in experimental methods. HIF-2alpha and GAPDH mRNAs were detected in each fraction by RT-PCR on RNA extracted from each fraction. (e): Quantitative RT-PCR was performed as outlined in materials and methods to examine the mRNA expression levels HIF-responsive genes (VEGF and TGF-alpha) after siRNA down-regulation of Rictor or scrambled control. The data is representative of three independent experiments and are expressed as fold control where the ratio of the scrambled control was defined as 1. Values are the means +/− S.E, ** p<0.01.
p22phox-based Nox oxidases play a novel role in the regulation of eIF4E-dependent mRNA translation
We have previously demonstrated that p22phox-based Nox oxidases maintain HIF-2alpha in the absence of VHL through an mRNA translational mechanism (4,16). We next explored a role for p22phox-based Nox oxidases in the regulation HIF-2alpha expression through 4E-BP1-eIF4E complex assembly. To maximize p22phox downregulation, we generated stable shp22phox knockdown clones and vector controls using lentivirus. RCC 786-O single cell clones stably reduced of p22phox were identified and characterized. shp22phox (1) and (2) are indicated as 2 independent single cell clones and demonstrate reduced p22phox protein expression and mRNA compared to RCC 786-O cells stably infected with independent vector control (shVector) (Fig. 3A left and right panels respectively). p22phox is a pivotal Nox subunit important for Nox-dependent superoxide and hydrogen peroxide production (4,16). We demonstrate successful reduction of NADPH-dependent superoxide generation (Nox activity) in all individual p22phox stable knockdown clones compared to vector controls (Fig. 3B). We next affinity purified eIF4E in the stable p22phox knockdown or vector cells and find that stable reduction of p22phox in 3 independent clones leads to a greater abundance of eIF4E in association with 4E-BP1 compared to 2 independent stable shVector clones (Fig. 3C). Chemical inhibition of Nox catalytic isoforms using the flavoprotein inhibitor, diphenyleneoidonium, DPI (4) also resulted in more eIF4E binding to 4E-BP1 in RCC 786-O cells (Fig. S2A). On the other hand, reverse experiments demonstrate that overexpression of p22phox in normal renal epithelial cells (HK2) led to a reduction of eIF4E in association with 4E-BP1 as examined by affinity purification using 7mGTP sepharose beads (Fig. S2B).
Figure 3.
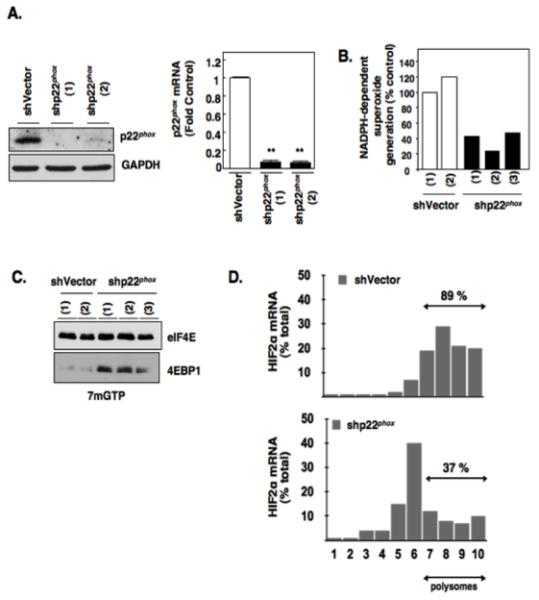
Effects of p22phox inhibition on eIF4E-dependent mRNA translation. (a): p22phox was stably knocked down using lentiviral short hairpin loop RNA (shp22phox) or vector control (shVector) as described in materials and methods. Western blot analysis (left panel) was carried out to confirm p22phox downregulation in indicated independent single cell shp22phox clones. GAPDH was used as loading control and quantitative RT-PCR (right panel) was carried out to confirm p22phox mRNA down regulation (b): NADPH oxidase activity was measured in parallel. (c): eIF4E association with 4E-BP1 was examined as outlined in Fig. 1C using 7-methyl-GTP sepharose beads from RCC 786-O cells stably silenced for p22phox (shp22phox) or Vector control (shVector). The data are representative of at least three independent experiments. (d): Polysomal analysis was performed from RCC 786-O cells stably silenced for p22phox (shp22phox) or Vector control (shVector) as outlined in 2D and as described in experimental methods.
Polysomal profiling of HIF-2alpha mRNA in shp22phox RCC 786-O cells demonstrated a reduction of the HIF-2alpha mRNA in heavy polysomal fractions compared to shVector RCC 786-O cells (Fig. 3D). HIF-2alpha expression and polysomal profiling of HIF-2alpha mRNA was also examined in an independent VHL-deficient cell line, A498, silenced of p22phox. Similar to VHL-deficient 786-O cells, a reduction of HIF-2alpha mRNA in heavy polysomal fractions was observed concomitant with reduced HIF-2alpha expression in A498 cells transiently silenced of p22phox using shRNA compared to shVector transfected A498 cells (Fig. S3A, S3B). Taken together, our data demonstrate a novel role for p22phox-based Nox oxidases in the regulation of eIF4E-dependent mRNA translation of HIF-2alpha and suggests a putative role for p22phox-dependent redox regulation of mTORC2 complex formation/stability.
mTORC2 but not mTORC1 complex is redox sensitive
Down regulation of p22phox or pharmacological inhibition of Nox oxidases results in reduction of Akt phosphorylation at the mTORC2 site, 473 (pAkt473) suggesting p22phox functions upstream of mTORC2 (4,16). Therefore, we next determined if the mechanism by which p22phox-based Nox oxidases regulate phosphorylation of Akt 473 and mRNA translation of HIF-2alpha may be mediated through redox regulation mTORC complexes. mTORC1 and mTORC2 complexes were examined in RCC 786-O cells stably reduced of p22phox (shp22phox) and vector controls. Rictor-associated mTOR complex and Raptor-associated mTOR complex were examined by immunoprecipitating mTOR using mTOR antibodies. Western blot analysis using the mTOR immunoprecipitates demonstrated significant reduction of Rictor in association with mTOR (60%) whereas there was no reduction of Raptor in association with mTOR in shp22phox stable RCC 786-O cells compared to vector control (shVec) (Fig. 4A, upper and lower panels). In support of this finding, pharmacological inhibition of the Nox catalytic subunits using DPI also reduced Rictor-associated mTOR complexes with no effect on Raptor associated mTOR complexes (Fig S4A). Successful immunoprecipitation of mTOR was examined by Western blot analysis and Rabbit IgG was used as a negative control for the immunoprecipitations.
Figure 4.
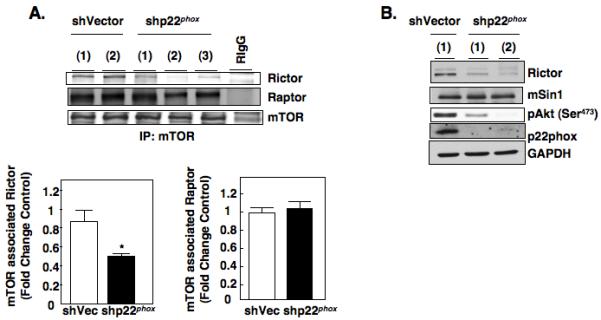
Redox regulation of mTORC complexes. (a): Association of Rictor and Raptor with mTORC complexes were examined in stable shVector or shp22phox RCC 786-O independent single cell clones by immunoprecipitating mTOR with anti-mTOR antibodies or equivalent amounts of rabbit IgG for control from cell lysates prepared in mTOR lysis buffer as described in materials and methods followed by Western blot analysis for Rictor, Raptor, and mTOR. Histogram (lower panel) represents the ratio of the intensity of the Rictor or Raptor bands to the mTOR band in shp22phox knock down clones compared to vector controls (Fold change control) quantified by densitometry. Values are the means ± S.E. (n =3), * p<0.05. (b): Cellular lysates were prepared from stable shVector or shp22phox RCC 786-O independent single cell clones and analyzed by Western blot analysis for Rictor, mSin1, pAkt473, and p22phox expression. GAPDH was used as a loading control.
Changes in protein expression can result in stabilization/destabilization of protein complexes. For example, down regulation of Rictor or mSin1 destabilizes the mTORC2 complex and thus inhibits mTORC2 activity (22,23). Therefore, we examined Rictor protein expression in p22phox knockdown or DPI treated RCC 786-O cells to determine if this is a mechanism by which Rictor-associated mTOR complex is reduced. When RCC 786-O cellular lysates from p22phox-stable knockdown clones and vector controls were probed for Rictor expression, we found a reduction in Rictor abundance and phosphorylation of Akt at the mTORC2 site (pAkt473), no significant changes of mSin1, another subunit of the mTORC2 complex, were observed (Fig. 4B). Similarly, we also find an ~30% reduction of Rictor abundance in A498 cells transiently silenced of p22phox (Fig. S3B) and a marked reduction of Rictor abundance in RCC 786-O cells treated with DPI (Fig. S4B) compared to vector transfected or buffer treated controls respectively. On the other hand, when p22phox is overexpressed in normal proximal tubular epithelial cells, HK2, Rictor expression is slightly but significantly increased compared to vector transfected or RCC 786-O cells alone (Fig. S4C left and right panels).
Effect of p22phox knockdown on HIF-2alpha transcriptional activity in vitro and tumor growth in vivo
We next elucidated the biological significance of p22phox-dependent reduction of HIF-2alpha expression. HIF-2alpha is a master transcription factor that is known to up-regulate genes involved in the pathogenesis of clear cell RCC. We examined the effect of p22phox knockdown on HIF-dependent gene regulation in vitro. p22phox was transiently silenced in RCC 786-O cells using small inhibitory RNA for p22phox (sip22phox) or scrambled control (scr). RCC 786-O renal carcinoma cell lines containing wild-type VHL (VHL) were used as a negative control. HIF transcriptional activity was measured using a hypoxia responsive element (HRE) reporter plasmid. Co-transfection of a mixture of scr or sip22phox and HRE reporter plasmid was performed to ensure that luciferase activity derived from HRE reporter plasmid was measured. These experiments revealed a significant decrease in the hypoxia-dependent reporter gene activation in p22phox knockdown cells and in 786-O + VHL (VHL) cells compared to the 786-O scr control (Fig. 5A, left panel). In parallel, cell lysates were prepared and analyzed for HIF-2alpha and p22phox protein expression. RCC 786-O cells transiently transfected with sip22phox demonstrated a decrease in p22phox protein expression (Fig. 5A, right panel). As previously demonstrated, 786-O + VHL cells also reduces p22phox protein levels compared to 786-O cells transfected with scr control, Fig. 5A, right panel and (4). As expected, HIF-2alpha was decreased in the 786-O + VHL cells and the siRNA p22phox knockdown cells (Fig 5A, right panel (2,4)).
Figure 5.
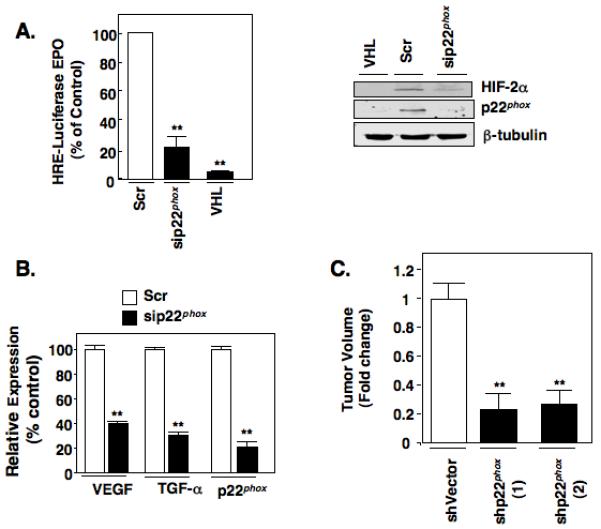
HIF2α dependent transcriptional activity and gene regulation is regulated by p22phox. (a): Left panel. Transcriptional activity of a HIF-responsive gene promoter (EPO) was measured in RCC 786-O cells transiently transfected with scrambled control (scr) or siRNA against p22phox (sip22phox) as described in materials and methods. RCC 786-O renal carcinoma cell lines containing wild-type VHL (786-O + VHL) were used as a negative control for HIF-transcriptional activity. Right panel. In parallel to 5A, cell lysates were prepared and protein expression of HIF-2α and p22phox were analyzed by Western blot analysis. β-tubulin was used as a loading control. (b): Quantitative RT-PCR was performed to examine the mRNA expression levels of HIF-responsive genes (VEGF, TGFα) after siRNA down-regulation of the p22phox gene. The data is representative of three independent experiments and are expressed as percent control where the ratio of the scrambled control was defined as 100%. Values are the means +/− S.E, ** p<0.01. (c): Nude mice were injected with stable shVector or shp22phox RCC 786-O independent single cell clones as outlined in materials and methods. Animals were sacrificed at 7 weeks and tumor volumes measured using calipers. There were 10 animals in each individual group and are expressed as fold control (shVector) where the ratio of the shVector control was defined as 1. Values are the means +/− S.E, ** p<0.01.
We next examined the effect of p22phox knockdown on VEGF and TGF-α gene expression; known targets of HIF-2alpha using real time PCR. Transient knockdown of p22phox in RCC 786-O cells using siRNA resulted in an 80% reduction of p22phox mRNA, a 60% reduction of VEGF, and a 70% reduction of TGF-alpha compared to scrambled transfected 786-O control (Fig. 5B). These results indicate that p22phox regulate HIF-2alpha dependent expression of genes that contribute to angiogenic and oncogenic potential of renal cell carcinoma. To test this, we injected nude mice with two independent RCC 786-O cells stably knockdown of p22phox, shp22phox (1) and shp22phox (2), or vector controls and monitored tumor growth for 7 weeks (Fig. 5C). 10 mice were included in each group and for each animal, stable vector cells were injected into the left flank whereas the RCC 786-O shp22phox stable cells were injected into the right flank of the same animal. Tumors started to form in vector controls at 1 week with no visible growth in the flanks of animals injected with RCC 786-O shp22phox cells until small tumors were observed in a minority of animals at 5 weeks with most of the animals demonstrating some growth at 7 weeks. At the end point, we noted a significant reduction of tumor volume in shp22phox compared to vector alone, suggesting a delay in tumor growth.
pp242 blocks eIF4E complex assembly in ex vivo RCC cells
VHL is mutated or silenced in ~80% of human RCC tumors which stabilizes HIF-2alpha expression. Our data presented here and elsewhere suggest that HIF-2alpha is actively translated in vitro VHL-deficient RCC cultured cells, which is necessary and sufficient for maintaining its expression (4,16). To determine if HIF-2alpha mRNA is actively translated in human RCC tumors, we examined HIF-2alpha mRNA expression and translational profile of HIF-2alpha mRNA using 2-independent tumors or normal adjacent control tissue from the same patient. We find increased HIF-2alpha mRNA expression (Figs. 6A,B left panel) and active translation of HIF-2alpha compared to normal adjacent control tissue, which showed no active translational profile (Figs 6A,B right panel). Downregulation of HIF-2alpha remains a therapeutic target; however, clinically, the treatment of renal cell carcinoma with the mTORC1 inhibitor, CCI-779 has largely been ineffective. Our in vitro data using established cultured VHL-deficient cell lines suggests that inhibition of mTORC1/mTORC2 using pp242 can effectively inhibit mRNA translation of HIF-2alpha. To determine if mTORC1/mTORC2 inhibitors demonstrate the same effect in human RCC cells, we isolated and cultured ex vivo RCC tumor cells and incubated the cells for 24 hours with buffer control (−), the mTORC1 inhibitor (CCI-779), or the mTORC1/mTORC2 inhibitor, pp242. We examined the effects of the aforementioned inhibitors on eIF4E-4E-BP1 association by affinity purification using 7-methyl GTP sepharose beads. Importantly, we find that dual inhibition of mTORC1 and mTORC2, but not mTORC1 alone, resulted in enhanced binding of eIF4E to 4E-BP1 compared to buffer treated cells (Fig. 6C). Together, this suggests that translation of HIF-2alpha is enhanced in RCC tumors and that Rapalog therapy alone may not be sufficient to reduce RCC tumorigenic phenotypes due to lack of mRNA translation inhibition.
Figure 6.
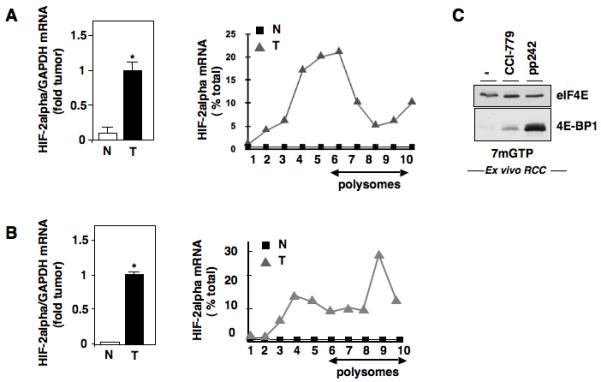
Effects of mTORC1/mTORC2 inhibitor on eIF4E complex formation in ex vivo RCC cells. (a,b): Left Panel, Steady state mRNA of HIF-2alpha was examined in two independent RCC tumor tissues or normal adjacent control tissues. Right Panel, Analysis of HIF-2 alpha mRNA in polysomal fractions as described in Fig. 1D in RCC tumors and normal adjacent controls. C, Examination of eIF4E and its associated proteins using 7-methyl-GTP sepharose beads in ex vivo cultured renal tumor cells treated with buffer alone, CCI-779, or pp242.
Discussion
These studies highlight the following novel findings: 1) Rictor-associated mTORC2 complex maintain HIF-2alpha protein expression in the absence of VHL through an eIF4E-dependent mRNA translational mechanism. 2) mTORC2 complex formation/stability is redox sensitive, mediated by p22phox-based Nox oxidases. 3) p22phox-based Nox oxidases play a novel role in mediating eIF4E-dependent mRNA translation, through mTORC2. 4) Inhibition of p22phox, the pivotal Nox subunit necessary to activate both Nox1 and Nox4, inhibits HIF-2alpha-dependent gene targets in vitro and delays RCC tumor growth in vivo. These findings are outlined in Fig. 7. Here we suggest (left panel) that enhanced ROS generation in RCC cells, mediated by p22phox-based Nox oxidases, stabilize Rictor protein expression and the mTORC2 complex. We hypothesize that stabilization and activation of the mTORC2 complex enhances HIF-2alpha protein expression by maintaining eIF4E-eIF4G complex association and thus active mRNA translation. In contrast (right panel) when p22phox-based Nox oxidases are pharmacologically inhibited by DPI or p22phox is downregulated we find reduced Rictor-associated mTORC2 complexes, giving rise to enhanced eIF4E-4E-BP1 association and inhibition of mRNA translation.
Figure 7.
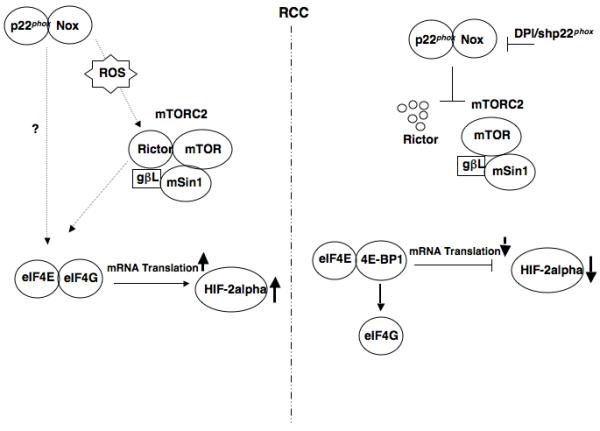
Proposed mechanism of mTORC2 regulation of HIF-2alpha protein expression through p22phox- based Nox oxidase-dependent regulation of eIF4E-dependent mRNA translation in RCC.
mTOR is found in two distinct complexes, mTORC1 and mTORC2 (13-15). Downstream targets of mTORC1 and mTORC2, including 4E-BP1 and Akt respectively, are highly phosphorylated in RCC cells suggesting both mTOR complexes are activated (4, 16, 21). The role of mTORC1 in the regulation of mRNA translation has been extensively studied. However, only recently has a role for mTORC2 in the regulation of mRNA translation been implied (20). Insulin induced eIF4E-dependent mRNA translation, determined by pull-down of eIF4E:eIF4G with 7-methyl GTP- Sepharose in treated L6 myotubes, is efficiently blocked by the mTORC1/mTORC2 inhibitor, pp242 (20). Our study supports a role for Rictor-associated mTORC2 in eIF4E-dependent mRNA translation and provides the first molecular and functional evidence of a specific endogenous target regulated in this manner, HIF-2alpha. However, the mechanisms remain under investigation. Hyperphosphorylation of 4E-BP1 by the mTORC1 complex reduces its affinity to eIF4E allowing eIF4E to direct ribsosomes to initiate mRNA translation whereas hypophosphorylation of 4E-BP1 enhances its affinity to eIF4E resulting in inhibition of mRNA translation. We do not find appreciable reduction of endogenous 4E-BP1 phosphorylation in RCC cells silenced of Rictor compared to scrambled controls (data not shown). However, silencing of Rictor enhances eIF4E association with 4E-BP1 concomitant with a shift of HIF-2alpha mRNA polysomal distribution, suggesting that inhibition of mRNA translation by Rictor-associated mTORC2 may not simply occur through mTORC1 signaling in RCC. In cells with high ERK activity, phosphorylation of 4E-BP1 may be unaffected by mTORC2 inhibition (24). RCC cells demonstrate high ERK activation (Block, unpublished observation, 25). It is likely that there remain other possible and yet unidentified mechanisms involved, which are under active investigation.
Importantly, upstream of Rictor-mTORC2, we find a novel role for p22phox-based Nox oxidases in these processes. Our study provides the first evidence that p22phox-based Nox oxidases play a role in eIF4E-dependent mRNA translation, at least in part through Rictor-associated mTORC2 activity, and suggests this is the mechanism by which Nox oxidases maintain HIF-2alpha in the absence of VHL. Other putative roles for and targets of p22phox-based Nox oxidases in the regulation of mRNA translation, independent of mTORC2, are under investigation. mTORC2 is a heterocomplex consisting of mTOR, mLST8, Rictor and mSin1 (14). The activity and function of mTORC2 depends on the presence of each of these components in the complex (22-23). Therefore, alteration of the expression of subunits associated with mTORC2 will directly affect the activity of the system (22-23). Our data suggest that the regulation of mTORC2 complex formation/stability is redox sensitive, as knockdown of p22phox inhibits NADPH-dependent superoxide generation and reduces Rictor-associated mTORC2 complex without affecting Raptor-associated mTORC1 complex. The mechanism is likely mediated through regulation of Rictor expression. The role of the p22phox-dependent oxidase is confirmed by the fact that overexpression of p22phox leads to enhanced Rictor expression in HK2 cells. We are currently investigating the mechanisms of p22phox-based Nox oxidase regulation of Rictor expression. Enhanced Rictor expression has been reported in ovarian and hepatocellular carcinomas (26, 27), it remains to be determined if p22phox-based Nox oxidases play a role in Rictor expression in these tumors or if Rictor is regulated by multiple mechanisms. We do not observe obvious increases of Rictor protein expression in human RCC tumors (data not shown); however, mTORC2 activity is upregulated in RCC tumors as examined by enhanced phosphorylation of Akt 473. Importantly, enhanced phosphorylation of Akt 473 is correlated with increased p22phox expression (16).
The biological downstream consequence of p22phox in HIF-2alpha gene targeting as well as in tumor growth has not been previously examined. p22phox is the pivotal subunit that enhances Nox1- and Nox4-derived ROS, both of which have been implicated in proliferation and angiogenesis of various cancers. In this study, we show that downregulation of p22phox, in vitro, blocks transcriptional activity of a general HIF-dependent responsive element (HRE) in addition to HIF-alpha dependent targets, TGF-alpha and VEGF. Stable inhibition of p22phox in VHL-deficient cells results in a significant reduction, ~70-80%, of tumor growth when injected subcutaneously to nude mice compared to vector controls. We hypothesize that this was a delayed response as no tumors were observed in the shp22phox mice until ~6 weeks at which point vector controls show marked tumor growth. It remains unknown if p22phox-based Nox oxidases are signaling exclusively through HIF-2alpha to delay tumor growth, via downregulation of HIF-2alpha target genes, or if other pathways may be involved. These questions remain under investigation.
The Von Hippel-Lindau tumor suppressor protein is inactivated in a large number of human clear cell renal tumors. In the absence of VHL, HIF-2alpha expression is stabilized through pathways that require ongoing mRNA translation (4, 16). HIF-2alpha has been defined as the critical downstream target for renal tumorigenesis (1). Clinically, Rapamycin analogues (CCI-779) have been utilized for the treatment for advanced RCC. However, objective response rates to these drugs remain low (28). Several studies, including this study, show that rapamycin is not an effective mTORC2 inhibitor, does not demonstrate the ability alone to switch eIF4E association to 4E-BP1, and has no effect on reducing HIF-2alpha expression (here and 17 REF). In support of this and clinically relevant is our finding that in human RCC tumors the distribution of HIF-2alpha mRNA is found in all polysomal fractions suggesting HIF-2alpha is actively translated in RCC tumors whereas this is not the case in normal adjacent control tissue. Moreover, isolation of ex vivo cells from an RCC tumor demonstrate that pharmacological inhibition of mTORC2/mTORC1, but not mTORC1 alone, can only shift eIF4E complex assembly to 4E-BP1. Together, our findings may provide an explanation to the disappointing outcome of clinical trials using Rapamycin in RCC.
Collectively, our data couple upstream p22phox-based Nox oxidase activation to downstream mTORC2 signaling and demonstrate for the first time their roles in initiation processes of cap-dependent mRNA translation and RCC tumorigenesis.
Materials and methods
Chemicals and reagents
Diphenyleneoidonium (DPI) was purchased from Sigma and used at a final of 5 μM. CCI779 and pp242 (pp242, a kind gift from Dr. Shokat, UCSF) and both were purchased from ChemieTek (Indianapolis, IN) and used at a final of 20 nM and 2.5 μM respectively. 7-Methyl-GTP Sepharose™ 4B beads were purchased from GE Health Care Life Sciences.
Cell culture and pharmacological treatments
All cell lines were maintained as outlined (4, 16). Experiments were carried out in complete media.
Cellular transfections, for transient assays and stable selection
For transient transfection assays or generation of stable clones, HK2, 786-O, or A498 cells were transfected by nucleofection using Amaxa biosystems. 2.0×106 cells were pulsed with 5 μg of DNA or 3 μM siRNA smart pool. 36-72 hours post-transfection, cells were harvested using the indicated buffers (detailed below) for the specific assay to be tested. In some experiments, transient transfection was carried out using lipofectamine 2000 (Invitrogen) (4). shp22phox plasmid was generated as outlined in (Maranchie) and used for transient transfection of A498 cells using lipofectamine. Generation of shp22phox stable single cell knockdown clones was achieved using MISSION Lentiviral Transduction Particles (Sigma) for shp22phox or vector control (Sigma). 786-O cells were infected with Lentiviral shp22phox or shVector control as per manufacturers instructions. 24 hr post-transfection, cells were passed into 150mm plates and single cells were selected and passed into 96 well-plates in Puromycin (750ng/mL) antibiotic for selection and maintained in Puromycin (350ng/mL). Small interference RNAs (siRNAs) were carried out with SMARTpool siRNAs (Dharmacon) transfected at 100 nM as described above. Scrambled non-targeting RNA was used as a control.
Antibodies
Anti-HIF-2α and GAPDH (Novus Biologicals), anti-Actin (Sigma). p22phox (Santa Cruz), pAkt, Akt p4E-BP1, 4E-BP1 pAkt S473, peIF4E, eIF4G, Rictor, and Raptor (Cell Signaling), mSin1 (Bethyl) and mTOR (Abcam). Immunoblotting was performed as described (4,16).
Preparation of cell lysates, Immunoprecipitation-Western (IP-Western), 7-Methyl pulldown
For Western blot analysis, cells were lysed in a modified RIPA buffer (4,16). For mTORC immunoprecipitations, cellular lysates were prepared in mTOR lysis buffer as described (18). Cell lysates (~200μg) for 7-Methyl pull down assays were prepared in buffer A (10 mM KH3PO4, 1 mM EDTA, 10 mM MgCl2, 50 mM Glycerol 2-phosphate, 5 mM EGTA, 0.5% NP40, 1 mM Orthovanadate, 0.1% sodium deoxycholate). eIF4E and associated proteins were immunoprecipitated overnight at 4°C using 50uL of 7-methyl beads (GE Healthcare UK Limited), washed twice in buffer A and once in ST buffer (50 mM Tris.Cl, 5 mM Tris base, 150 mM NaCl). Bound proteins were eluted by boiling in 1x Laemmli sample buffer and analyzed by SDS-PAGE.
NADPH oxidase assay
NADPH-dependent superoxide anion generation in cell homogenates was measured by lucigenin enhanced chemiluminencence as previously described (4, 16).
Polysome isolation and fractionation and Quantitative Real time PCR
The polysome assay was performed as described (19). Briefly, indicated RCC 786-O, A498 cells, RCC tumors or normal adjacent tissue was lysed in polysome resuspension buffer containing and centrifuged for 10 min at 14,000 rpm. The cytosolic supernatants ( 500 μl) were over laid on top of a 15-40% sucrose gradient and centrifuged for 90 min at 200,000 g. The gradients were separated into 10 fractions of 200 μl. RNA was extracted from each fraction using TRIzol (Invitrogen). Real-time (RT-PCR) amplification was performed in polyribosomal fractions using the Superscript One-Step RT-PCR kit from Invitrogen. Real Time-PCR Sybr Green was performed on 50 ng of RNA using the SuperArray kit (SuperArray Bioscience Corporation) and the Eppendorf realplex machine. Primers for HIF-2alpha, VEGF, TGF-α and GAPDH were purchased from SuperArray. Reactions were done in triplicate and normalized to GAPDH. Sedimentation analysis was examined in parallel of RCC 786-O cells by measuring optical density at 254 nm.
Xenograft animal model
Stable shp22phox or vector control RCC 786-O cells were mixed with matrigel and injected into nude mice as described in (4).
Human Tumor Specimens
Clear cell renal tumor samples and normal corresponding tissue from patients with clear cell RCC were obtained from the Department of Urology at the UTHSCSA according to an approved Institutional Review Board IRB protocol #HSC 20070777N (16).
Statistical analysis
Results are expressed as the mean±s.e.m. Statistical analyses were performed using the ANOVA (analysis of variance) followed by a Fisher’s test. A P-value of 0.05 was used to define statistical significance.
Supplementary Material
Acknowledgements
The authors acknowledge Dr. Goutam Gosh-Choudhury for the HRE responsive element, Cynthia Galindo for technical contributions and Dr. Hanna E. Abboud for helpful discussions and critical reading of the manuscript.
grant support: Supported by Veterans Administration Career Development Award and NIH R01 NCI CA131272 (KB), and NIH K08 CA138774 and Voelcker Fund Young Investigator Award (SS). Supported in part by the Cancer Center Support Grant, National Cancer Institute, 5 P30 CA054174-18 (DJP).
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003 Dec;1(3):E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr. The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007 Jan 15;13(2 Pt 2):680s–684s. doi: 10.1158/1078-0432.CCR-06-1865. [DOI] [PubMed] [Google Scholar]
- 3.Baldewijns MM, van Vlodrop IJ, Vermeulen PB, Soetekouw PM, van Engeland M, de Bruïne AP. VHL and HIF signalling in renal cell carcinogenesis. J Pathol. 2010 Jun;221(2):125–38. doi: 10.1002/path.2689. Review. [DOI] [PubMed] [Google Scholar]
- 4.Block K, Gorin Y, Hoover P, Williams P, Chelmicki T, Clark RA, et al. NAD(P)H oxidases regulate HIF-2alpha protein expression. J Biol Chem. 2007 Mar 16;282(11):8019–26. doi: 10.1074/jbc.M611569200. [DOI] [PubMed] [Google Scholar]
- 5.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990 Jun 7;345(6275):544–7. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 6.Pandolfi PP. Aberrant mRNA translation in cancer pathogenesis: an old concept revisited comes finally of age. Oncogene. 2004 Apr 19;23(18):3134–7. doi: 10.1038/sj.onc.1207618. [DOI] [PubMed] [Google Scholar]
- 7.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004 May;10(5):484–6. doi: 10.1038/nm1042. Epub 2004 Apr 18. [DOI] [PubMed] [Google Scholar]
- 8.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004 Mar 18;428(6980):332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 9.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004 Jun;5(6):553–63. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004 Apr 19;23(18):3189–99. doi: 10.1038/sj.onc.1207545. Review. [DOI] [PubMed] [Google Scholar]
- 11.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004 Dec 1;64(23):8639–42. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 12.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010 Aug 10;107(32):14134–9. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004 Aug 15;18(16):1926–45. doi: 10.1101/gad.1212704. Review. [DOI] [PubMed] [Google Scholar]
- 14.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007 Jul;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009 Oct 15;122(Pt 20):3589–94. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Block K, Gorin Y, New DD, Eid A, Chelmicki T, Reed A, et al. The NADPH oxidase subunit p22phox inhibits the function of the tumor suppressor protein tuberin. Am J Pathol. 2010 May;176(5):2447–55. doi: 10.2353/ajpath.2010.090606. Epub 2010 Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem. 2008 Dec 12;283(50):34495–9. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008 Jun;28(12):4104–15. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feliers D, Lee MJ, Ghosh-Choudhury G, Bomsztyk K, Kasinath BS. Heterogeneous nuclear ribonucleoprotein K contributes to angiotensin II stimulation of vascular endothelial growth factor mRNA translation. Am J Physiol Renal Physiol. 2007 Aug;293(2):F607–15. doi: 10.1152/ajprenal.00497.2006. [DOI] [PubMed] [Google Scholar]
- 20.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009 Feb 10;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003 May 13;13(10):797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 22.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008 Jul 23;27(14):1919–31. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009 Feb 3;15(2):148–59. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010 Jul 13;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HJ, Kim DI, Kang GH, Kwak C, Ku JH, Moon KC. Phosphorylation of ERK1/2 and prognosis of clear cell renal cell carcinoma. Urology. 2009 Feb;73(2):394–9. doi: 10.1016/j.urology.2008.08.472. [DOI] [PubMed] [Google Scholar]
- 26.Foster H, Coley HM, Goumenou A, Pados G, Harvey A, Karteris E. Differential expression of mTOR signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010 Sep;30(9):3529–34. [PubMed] [Google Scholar]
- 27.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008 Dec;135(6):1972–83. doi: 10.1053/j.gastro.2008.08.008. 1983.e1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009 Apr 21;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


