Abstract
Background
The mechanical properties of cellular microenvironments play important roles in regulating cellular functions. Studies of the molecular response of endothelial cells to alterations in substrate stiffness could shed new light on the development of cardiovascular disease. Quantitative real-time PCR is a current technique that is widely used in gene expression assessment, and its accuracy is highly dependent upon the selection of appropriate reference genes for gene expression normalization. This study aimed to evaluate and identify optimal reference genes for use in studies of the response of endothelial cells to alterations in substrate stiffness.
Methodology/Principal Findings
Four algorithms, GeNormPLUS, NormFinder, BestKeeper, and the Comparative ΔCt method, were employed to evaluate the expression of nine candidate genes. We observed that the stability of potential reference genes varied significantly in human umbilical vein endothelial cells on substrates with different stiffness. B2M, HPRT-1, and YWHAZ are suitable for normalization in this experimental setting. Meanwhile, we normalized the expression of YAP and CTGF using various reference genes and demonstrated that the relative quantification varied according to the reference genes.
Conclusion/Significance:
Consequently, our data show for the first time that B2M, HPRT-1, and YWHAZ are a set of stably expressed reference genes for accurate gene expression normalization in studies exploring the effect of subendothelial matrix stiffening on endothelial cell function. We furthermore caution against the use of GAPDH and ACTB for gene expression normalization in this experimental setting because of the low expression stability in this study.
Introduction
Change in the mechanical property of cells and their microenvironments and the relevance to biological functions have recently attracted increased attention. For example, mechanical microenvironments may regulate cellular functions relevant to development, homeostasis, and disease [1]–[3]. Many mainly pathological conditions, including aortic stiffness and liver fibrosis, result in significant mechanical changes at the whole organ, regional, and cellular levels [3]–[5]. Therefore, we need to determine the genetic and molecular basis of the mechanical changes and then identify these biomolecules and their signaling pathways for the development of future drug treatments.
Among the widely used methods, real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a rapid and sensitive method for measuring gene expression. Despite being a very powerful technique to accurately quantify gene expression, several determinants, such as input sample, RNA extraction, efficiency of reverse transcription from RNA to complementary DNA and PCR efficiency, should be taken into account. Therefore, normalization of qRT-PCR data with suitable internal reference genes (RGs) is required [6]. The ideal RG should be expressed at a constant level between samples or under different conditions. Nevertheless, there is now increasing evidence suggesting that the expression of RGs often varies significantly under different experimental conditions. Thus, identification of reliable RGs is a prerequisite for qRT-PCR experiments.
In our present study, we investigate reliable reference genes for real-time PCR in human umbilical vein endothelial cells (HUVECs) on substrates with different stiffness. It is well established that the subendothelial layers of blood vessels become stiff in cardiovascular diseases [2], [7]. The mechanical changes, therefore, may lead to dysregulate the endothelial layer and influence disease states [8], [9]. Evidence suggests that proper vessel compliance is critical for endothelial response to hemodynamic forces. Peng et al. showed that when ECs are plated on stiff-walled tubes and subjected to physiological pressures, endothelial NO synthase expression decreased in comparison to production by cells on distensible tubes [10]. Consequently, a better understanding of the response of endothelial cells to alterations in substrate stiffness could shed new light on the development of cardiovascular disease.
Previous studies demonstrated the importance of substrate stiffness on the gene expression of various cell types [11]–[18], such as Yes-associated protein (YAP) and connective tissue growth factor (CTGF) (Table 1). Endothelial cells responding to subendothelial matrix stiffening is a process of cells undergoing numerous morphological changes [19], [20], which are accompanied by substantial changes in biochemical and metabolic processes and alteration in structural proteins [10], [21]. Significant modifications in gene expression, including the widely used RGs, contribute to these cellular changes. To date, few studies dealing with how cellular gene expression responds to alterations in substrate stiffness reported on the stability of RGs prior to use in the studies. Validation of stably expressed RGs for endothelial cells on a variety of substrate stiffness is especially important.
Table 1. Some examples of genes that exhibit different expression levels as a function of substrate stiffness.
| Gene name | Cell lines | Ref |
| cyclin A, p27, and Rb | Human umbilical vein endothelial cells | [11] |
| IL-2 | Primary peripheral blood lymphocytes | [12] |
| TNF-α, IL-1β, and IL-6 | Murine bone marrow-derived primary macrophages | [13] |
| α-SMA, ET-1, and IL-1β | Colonic human myofibroblast cells | [14] |
| YAP, TAZ, TGM2, and sFRP-1, | Primary human trabecular meshwork cells | [15] |
| α-SMA and CTGF, | Primary valvular interstitial cells | [16] |
| COX-2, PGE2, MMP10, and MMP3 | Lung fibroblasts | [17] |
| Actin, Tubulin, and PFKP-1 | A549 | [18] |
The aim of this study was to identify and evaluate the expression stability of nine candidate RGs in a widely used in vitro model of the HUVECs in order to explore the effect of subendothelial matrix stiffening on endothelial cell function. Polyacrylamide gels were prepared to obtain three different stiffness values that mimic pathophysiological states (table 2), based on the currently published range of vascular basement membrane compliances of 2.5–8 kPa [22]–[24].
Table 2. Characterization of the polyacrylamide gels.
| Acrylamide % | Bisacrylamide% | Measuring elastic modulus (kPa) | |
| Soft | 3 | 0.1 | 0.91±0.73 |
| Medium | 5 | 0.15 | 2.08±1.32 # |
| Stiff | 8 | 0.48 | 12.39±7.85 &* |
Data are mean±SD from three independent experiments. Means were compared by ANOVA followed by Student-Newman-Keuls multiple range text. #P<0.05, Medium vs. Soft. *P<0.05, Stiff vs. Soft. &P<0.05, Stiff vs. Medium.
Results
Selection of Candidate RGs and Amplification Specificity
Nine candidate RGs were selected for this study. These candidates are widely used and recognized RGs, which have been described in the literature and represent several functional classes to minimize the possibility of co-regulation (Table 3).
Table 3. Symbols, names, accession numbers and functions of the candidate RGs evaluated.
| Gene symbol | Gene name | Accession number | Function |
| ACTB | Beta -actin | NM_001101 | Cytoskeletal structural protein |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046 | Carbohydrate metabolism |
| HPRT1 | Hypoxanthine phosphoribosyl-transferase 1 | NM_000194 | Purine synthesis through the purine salvage pathway |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5–monooxygenase activation protein, zeta polypeptide | NM_003406 | Protein domain in specific binding |
| TBP | TATA box binding protein | NM_003194 | Transcription initiation from RNA polymerase II promotor |
| G6PD | Glucose-6-phosphate dehydrogenase | NM_000402 | Involved in the normal processing of carbohydrates |
| RPL13A | Ribosomal protein large L13a | NM_012423 | Structural constituent of ribosome |
| 18S | RNA,18S ribosomal 1 | NR_003286 | Cytosolic small ribosomal subunit, translation |
| B2M | Beta-2-microglobulin | NM_004048 | Beta–chain of MHC class I molecules |
Their respective PCR amplification efficiencies were calculated as the first step. Table 4 lists the amplification efficiency for each of the candidate RGs that ranged from 94% to 111%. The performance of each amplification primer set was tested by qRT-PCR. Melting curve analysis confirmed the presence of a single PCR product from all samples with no primer-dimers. The amplifications were also confirmed by the presence of a single band of the expected size for each primer pair in 2% agarose gel electrophoresis (Figure S1).
Table 4. Primer Information for the selected candidate reference genes.
| Gene symbol | Forward primer | Reverse primer | Amplicon size (bp) | PCR efficiency(%) |
| ACTB | CATCGAGCACGGCATCGTCA | TAGCACAGCCTGGATAGCAAC | 211 | 94.2 |
| GAPDH | GTCAGCCGCATCTTCTTTTG | GCGCCCAATACGACCAAATC | 100 | 108.3 |
| HPRT1 | GACCAGTCAACAGGGGACAT | AACACTTCGTGGGGTCCTTTTC | 195 | 103.7 |
| YWHAZ | ACTTTTGGTACATTGTGGCTTCAA | CCGCCAGGACAAACCAGTAT | 94 | 103.8 |
| TBP | GAGCTGTGATGTGAAGTTTCC | TCTGGGTTTGATCATTCTGTAG | 118 | 107.8 |
| G6PD | CCGTCACCAAGAACATTCACG | GGACAGCCGGTCAGAGCTCT | 107 | 103.1 |
| RPL13A | CCTGGAGGAGAAGAGGAAAGAGA | TTGAGGACCTCTGTGTATTTGTCAA | 126 | 99.3 |
| 18S | CAGCCACCCGAGATTGAGCA | TAGTAGCGACGGGCGGTGTG | 252 | 99.7 |
| B2M | CACCCCCACTGAAAAAGATGAG | CCTCCATGATGCTGCTTACATG | 106 | 110.9 |
Expression Levels of Candidate RGs
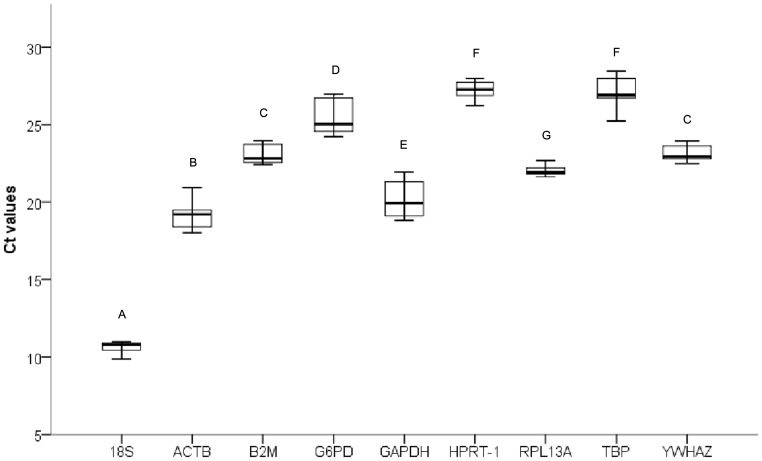
The expression levels of all nine candidate RGs were evaluated as threshold cycle (Ct) values with three biological and three technical replicates. The box plot of the Ct values of all candidate RGs show the differences in transcript levels between RGs (Figure 1). The expression levels of these RGs varied widely, with Ct values ranging from 9.87 to 28.44 cycles. 18S was the most abundantly transcribed gene, with a mean Ct value of 10.62 cycles, whereas HPRT-1 showed the lowest level of expression in all samples, with a mean Ct value of 27.21 cycles. The individual reference genes had different expression ranges across samples. Among the nine candidate RGs in this study, ACTB and GAPDH had large expression variations in their transcript levels (2.92 and 3.11 cycles, respectively), while RPL13A, 18S, and YWHAZ had much lower expression variation (1.05, 1.1, and 1.27 cycles, respectively). The wide expression range of the candidate RGs indicated the importance of accurately calculating the RGs levels with the widely used statistical algorithms described below.
Figure 1. Distribution of qRT-PCR quantification cycle values for the candidate RGs.
Box plot graphs of Ct values for each RG tested in all samples. Vertical lines indicate range of values, and the median Ct value is represented as black horizontal line within the box plot. The box indicates the 25th and 75th percentiles. Letters indicate a significant difference in average Ct value. Average Ct values that have the same letter are not significantly different (P>0.05).
Analysis of Candidate RGs Stability
Four distinct algorithms, GeNormPLUS, Normfinder, Bestkeeper and the Comparative ΔCt method, were employed to further evaluate the expression stability of the candidate RGs individually.
GeNormPLUS Analysis
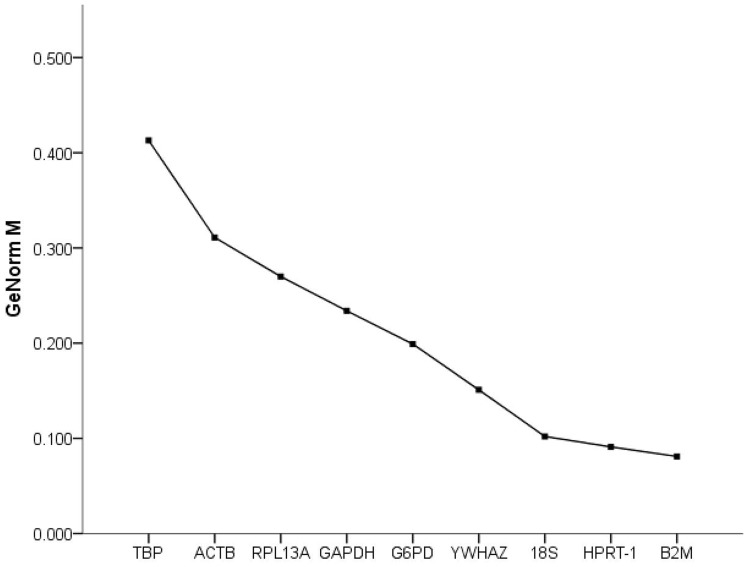
The GeNorm algorithm applies a statistical algorithm to calculate the average stability measure (M). A lower value of average expression stability M indicated more stable expression [6]. The Pairwise variation (V) parameter was calculated to determine the optimal number of RGs required for normalizing the expression of genes of interest. Generally, 0.15 was used as a cutoff value to determine the optimal number of RGs [6]. Below this value, the addition of extra RGs does not improve the expression stability of the RG set and is therefore not recommended.
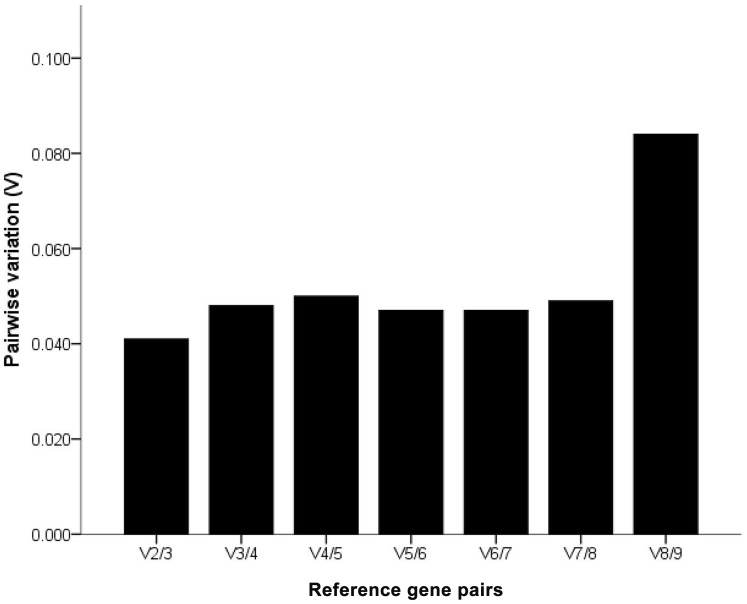
All candidates were ranked based on M values (Figure 2). The nine selected candidate genes all reached the high expression stability criterion, with M<0.42, which is well below the default limit of 1.5 suggested by GeNormPLUS. Of the candidates, the B2M gene had the lowest M value, followed by HPRT-1. Interestingly, ACTB, although frequently used for gene expression, had the lowest expression stability in this study. Moreover, the V parameter calculated by GeNormPLUS recommended the use of two RGs for reliable normalization in this experimental setting (Figure 3), and the addition of a third gene is optional.
Figure 2. Expression stability values (M) of candidate RGs calculated by GeNormPLUS.
M values of the remaining candidate citrus RGs during stepwise exclusion of the least stable citrus RG in the different subsets. A lower average M value indicates more stable expression.
Figure 3. Determination of the optimal number of RGs for qRT-PCR normalization by GeNormPLUS.
The pairwise variation (V) calculated using GeNormPLUS to determine the optimal number of RGs for accurate qRT-PCR normalization in different experimental settings. V2/3 exhibited the value below the cut-off value of 0.15, indicating that use of 2 RGs for normalization is necessary, whereas addition of a third RG is optional.
NormFinder Analysis
NormFinder is another algorithm used to determine the optimal RGs for qRT-PCR normalization. This algorithm accounts for intra- and intergroup variation in the normalization factor calculations and calculates a stability value (S) for the RGs. The lowest S value indicates the most stable RG expression.
According to NormFinder analysis, the best ranked RG was B2M, followed by HPRT-1, YWHAZ, and RPL13A (Table 5). TBP and GAPDH were the least stable RGs in the present study.
Table 5. Gene stability (S) values calculated by Normfinder.
| Rank | Gene symbol | Stability Value |
| 1 | B2M | 0.122 |
| 2 | HPRT-1 | 0.180 |
| 3 | YWHAZ | 0.226 |
| 4 | RPL13A | 0.468 |
| 5 | ACTB | 0.504 |
| 6 | G6PD | 0.532 |
| 7 | 18S | 0.592 |
| 8 | TBP | 0.621 |
| 9 | GAPDH | 0.656 |
BestKeeper Analysis
BestKeeper calculates the gene expression variation for candidate genes based on standard deviation (SD), coefficient of correlation (r) and percentage covariance (CV). The lowest SD value indicates the most stable RG expression.
As shown in Table 6, the BestKeeper analysis highlighted RPL13A and 18S as the most stable genes with the lowest SD (0.22 and 0.29, respectively), followed by YWHAZ, HPRT-1, B2M, ACTB, TBP, G6PD and GAPDH. The differences between the GeNormPLUS and BestKeeper results were expected because their statistical algorithms were distinct.
Table 6. Expression stability evaluated by BestKeeper.
| Candidate Genes | ACTB | GAPDH | HPRT-1 | YWHAZ | TBP | G6PD | RPL13A | 18S | B2M |
| n | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 |
| geo Mean [Ct] | 19.04 | 20.01 | 27.21 | 23.30 | 27.08 | 25.42 | 22.01 | 10.62 | 23.07 |
| ar Mean [Ct] | 19.06 | 20.03 | 27.21 | 23.30 | 27.09 | 25.44 | 22.01 | 10.63 | 23.08 |
| Min [Ct] | 18.01 | 18.83 | 26.23 | 22.67 | 25.25 | 24.23 | 21.63 | 9.87 | 22.40 |
| Max [Ct] | 20.93 | 21.94 | 27.98 | 23.94 | 28.44 | 26.97 | 22.68 | 10.97 | 23.97 |
| std dev [+/− Ct] | 0.60 | 0.93 | 0.44 | 0.41 | 0.75 | 0.91 | 0.22 | 0.29 | 0.52 |
| CV [% Ct] | 3.17 | 4.64 | 1.62 | 1.76 | 2.76 | 3.58 | 0.98 | 2.74 | 2.27 |
Comparative ΔCt Method Analysis
The Comparative ΔCt method assesses the most stable RGs by comparing the relative expression of “pairs of genes” within each tissue sample or each treatment [25]. This algorithm highlighted B2M and HPRT-1 as the most stable genes, followed by YWHAZ and RPL13A (Table 7); these findings are highly consistent with the GeNormPLUS and Normfinder results.
Table 7. Ranking of RGs stability.
| Rank | GeNorm | NormFinder | BestKeeper | ΔCt method | Final ranking |
| 1 | B2M | B2M | RPL13A | B2M | B2M |
| 2 | HPRT-1 | HPRT-1 | 18S | HPRT-1 | HPRT-1 |
| 3 | 18S | YWHAZ | YWHAZ | YWHAZ | YWHAZ |
| 4 | YWHAZ | RPL13A | HPRT-1 | RPL13A | RPL13A |
| 5 | G6PD | ACTB | B2M | G6PD | 18S |
| 6 | GAPDH | G6PD | ACTB | ACTB | G6PD |
| 7 | RPL13A | 18S | TBP | 18S | ACTB |
| 8 | ACTB | TBP | G6PD | GAPDH | GAPDH |
| 9 | TBP | GAPDH | GAPDH | TBP | TBP |
Final Ranking of Candidate Reference Genes
RG rankings obtained with all four algorithms (GeNormPLUS, Normfinder, BestKeeper, and the Comparative ΔCt method) were compared (Table 7). While RG rankings vary slightly by algorithm, a method previously described [26] was used to give an overall ranking of the best candidate RGs. The geometric means of the four ranking numbers were calculated, and the gene with a smaller geometric mean is the most stable RG. The recommended comprehensive rankings were also given in Table 7. Using the results from all four algorithms, an overall ranking of candidate RGs was obtained. B2M, HPRT-1, and YWHAZ represent the most reliable RGs in this experimental setting. The conventional RGs, GAPDH and ACTB were found to be less reliable and are not the good choices for RGs in this experimental setting.
Validation of Reference Genes
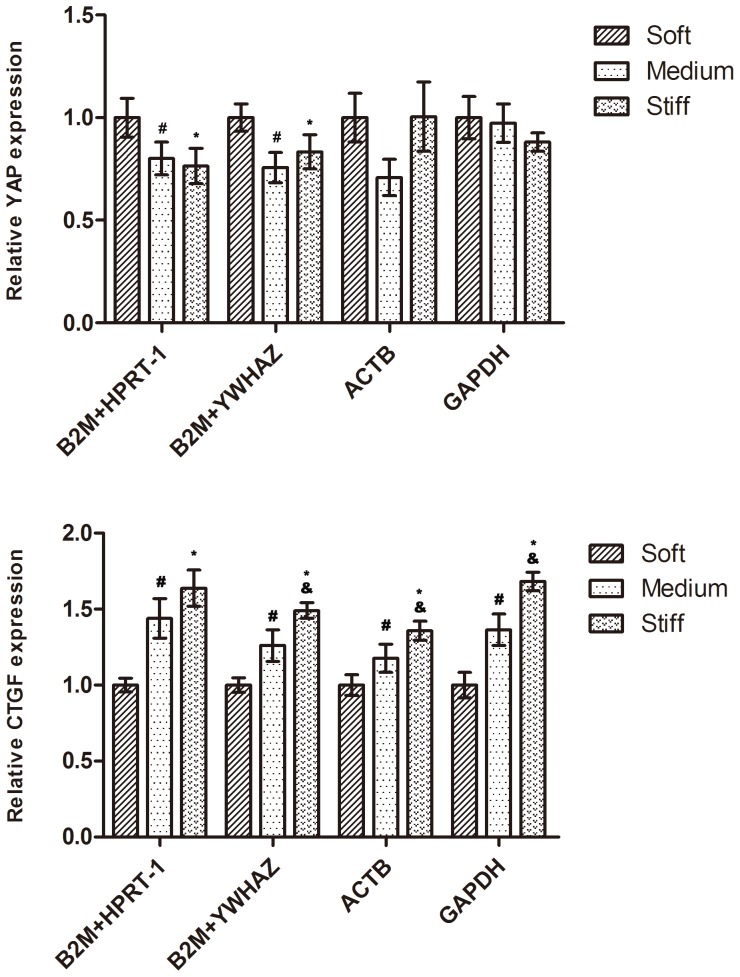
To show the effect of a reference gene on the outcome of a practical experiment, we evaluated the expression patterns of two genes, YAP and CTGF, using different normalization strategies. In previous studies, the transcription level of YAP was downregulated on stiffer substrates (18S as the RG) [15], and CTGF was significantly upregulated (18S as the RG) [16]. The 2 representative least stable RGs (ACTB and GAPDH) and a combination of 2 of the 3 most stable RGs (B2M, HPRT-1, and YWHAZ) were used as RGs for expression normalization (Figure 4). When B2M and HPRT-1 or YWHAZ were used as RGs, the expression of YAP decreased with substrate stiffness. Whereas when the least stable reference genes, ACTB and GAPDH, were used for normalization, YAP exhibited a similar expression level between different substrate stiffness. The CTGF exhibited a similar expression pattern between different normalization strategies. In addtion, we also evaluated the expression patterns of two functional genes, PAI-1 and tPA, using different normalization strategies (Figure S2). Both of these genes are known to play a crucial role in EC-mediated fibrinolytic activity. When B2M and YWHAZ or HPRT-1 were used as RGs, the expression of PAI-1 decreased with substrate stiffness. Whereas when the least stable reference genes, ACTB and GAPDH, were used for normalization, PAI-1 showed a different result when compared to the stable reference genes for normalization. Thus, these results reinforce the importance of validating reference genes prior to experimental applications.
Figure 4. Expression levels of YAP and CTGF in endothelial cells on a variety of substrate stiffness.
Genes were normalized to individual and/or combined RGs. Results are mean±SD, n = 3; Means were compared by ANOVA followed by Student-Newman-Keuls text. #P<0.05, Medium vs. Soft. *P<0.05, Stiff vs. Soft. &P<0.05, Stiff vs. Medium.
Discussion
Analysis of gene expression using qRT-PCR, which is a powerful method that combines high specificity and sensitivity, is a commonly used technology for gene expression analysis in response to different environmental conditions [27]. However, numerous studies have demonstrated that the performance of this technique is strongly dependent on a normalization strategy through the selection of appropriate RGs [28]–[30]. Thus, it is advisable to validate the expression stability of candidate RGs under specific experimental settings prior to use in qRT-PCR normalization. In this study, we sought to identify the appropriate RGs for normalizing qRT-PCR gene expression data in HUVECs responding to subendothelial matrix stiffening in vitro. Of nine candidate RGs, we identified B2M, HPRT-1, and YWHAZ as the most suitable RGs, using four widely recognized algorithms (GeNormPLUS, Normfinder, BestKeeper, and the Comparative ΔCt method). We further found that both ACTB and GAPDH, which are frequently used for gene expression normalization in many experiments, were not suitable normalization controls in this experimental setting. The results from GeNormPLUS, NormFinder, and the Comparative ΔCt method assessment were more consistent with each other than with the BestKeeper method.
The three most stably expressed genes in our experimental setting, B2M, HPRT-1, and YWHAZ, encode proteins with independent physiological functions. The protein encoded by B2M is a component of the major histocompatibility complex (MHC) class I [31]. HPRT-1 encodes an enzyme that plays a central role in the generation of purine nucleotides through the purine salvage pathway [32]. YWHAZ encodes the protein that mediates signal transduction by binding to phosphoserine-containing proteins [33]. Thus, these genes represent several distinct functional classes so as to minimize the possibility of co-regulation when combining these genes as RGs for qRT-PCR normalization in future studies.
Numerous morphological changes, which are accompanied by substantial changes in biochemical processes and metabolism and alteration in structural proteins, occur when cells adapt to substrates with different stiffness. Substrate stiffness regulates actin organization, cellular metabolism and protein synthesis in cells [21], [34], [35]. Byfield et al. showed that endothelial cells in stiffer gels exhibited more pronounced stress fibers and 1.5-fold greater actin staining [21]. Tilghman et al. demonstrated several of the proteins involved in the metabolic state are sensitive to changes in stiffness in cancer cells by measuring the rates of protein synthesis [36]. Thus, it is not that surprising to find that both ACTB and GAPDH are not suitable qRT-PCR normalization controls for these experiments. GAPDH and G6PD encode the key enzymes in the glycolytic pathway [37], [38], and ACTB encodes β-actin, which is involved in cell motility, structure and integrity.
Due to the original intention of our research, it is impossible in this study to research all human cell lines, and thus, these conclusions should be corroborated prior to application in other human cell lines. Furthermore, optimization of reference genes for real-time PCR associated with mechanical environments allows us to conduct more standardized biomechanopharmacology studies [39] in various patho/physiological stations.
In conclusion, we validated a stably expressed RG set for use in endothelial cells on a variety of substrate stiffness. B2M, HPRT-1, and YWHAZ were identified as the most stable RGs. Indeed, a combination of 2 genes out of these 3 genes is sufficient to provide accurate qRT-PCR normalization. Our results also demonstrated that special attention must be given to the choice of suitable RGs during the studies of the cell behaviors responding to the mechanical changes of their microenvironments.
Materials and Methods
Polyacrylamide Gel Preparation and Characterization
Polyacrylamide gels were prepared as described previously [40], [41]. Briefly, glass coverslips were treated with 3-aminopropyltrimethoxysilane and 0.5% glutaraldehyde after plasma cleaning (Harrick Plasma, Ithaca, NY). The PA gel premix solutions of acrylamide and bis-acrylamide (Bio-Rad, Hercules, CA) were mixed with tetramethylethylenediamene and ammonium persulfate, and the polymerizing solution was immediately added onto chloro-silanated glass slide after placing the glass coverslips on a drop of solution. The gels were allowed to polymerize at room temperature to a thickness of approximately 500 µm. Gel stiffness was manipulated by varying the acrylamide and bis-acrylamide ratio. Next, the gels were functionalized by UV irradiation of Sulfo-SANPAH in 50 mM HEPES buffer (pH 8.5) and then coated with 0.1 mg/ml type I collagen overnight at 4°C.
Force curves were acquired with an atomic force microscope (AFM, Agilent Technologies, CA). Silicon nitride cantilever (spring constant, 0.031 N/m; NT-MDT) with a spherical tip, of which the diameter was 16.46 µm, was used for measurements of Soft and Medium PA gels. For Stiff PA gels measurement, the silicon nitride cantilever (spring constant, 0.292 N/m; NT-MDT) with a borosilicate sphere of 11.9 µm in diameter was used. Force curves were obtained on at least seven different locations of each sample and twenty force curves were collected at each sample. Each force curve was taken at a rate of 1 Hz.
The force curves were analyzed using the Hertz model for a sphere in contact with a flat surface. In this model, the Young’s modulus E are calculated according to the following equation [42]:
.Where F is the loading force,  is Poisson’s ratio (assumed to be 0.3),
is Poisson’s ratio (assumed to be 0.3),  is the indentation depth (50 nm), E is the Young’s modulus, R is the radius of the silica sphere. Loading force was calculated by using Hook’s law,
is the indentation depth (50 nm), E is the Young’s modulus, R is the radius of the silica sphere. Loading force was calculated by using Hook’s law, 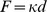 , where
, where  is the spring constant of the cantilever and d is the cantilever deflection. The characterization of the substrates is shown in table 2.
is the spring constant of the cantilever and d is the cantilever deflection. The characterization of the substrates is shown in table 2.
Cell Culture and Treatments
Primary HUVECs were obtained from ScienCell Research Laboratories (ScienCell, Carlsbad, CA) and cultured on plastic flasks coated with 1% gelatin in endothelial cell growth medium-2 (EGM-2) supplemented with 2% FCS and growth factors (Lonza, Walkersville, MD) at 37°C under 5% CO2. Cells between passages 2 and 6 were used for all experiments. The cells were plated onto the previously prepared PA gels and grown for 48 hours. The medium was replaced once after 24 h.
RNA Isolation and cDNA Synthesis
Total RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. The quantity and quality of RNA samples were determined using a Helios beta spectrometer (Thermo Scientific, Milford, MA). RNA samples with 260/280 ratio from 1.9 to 2.1 were used for further analysis.
First strand cDNA was synthesized by reverse transcribing 500 ng of total RNA with the RevertAid™ first strand cDNA synthesis kit (Fermentas life sciences, Vilnius, Lithuania) in a 20 µl reaction using random primers according to manufacturer’s instructions. The RT reaction sequence consisted of incubation at 25°C for 5 minutes, followed by 60 minutes at 42°C. The reaction was terminated by heating to 70°C for 5 minutes. The cDNA was stored at −20°C until the PCR experiments.
Quantitative Real-time PCR Procedure
The qRT-PCRs were performed in 96-well plates with an ABI StepOne Plus Sequence Detection System (Applied Biosystems, Foster City, CA) using the SYBR® Green Realtime PCR Master Mix (Toyobo, Osaka, Japan). Thermocycling was performed using the following conditions: 94°C for 1 minutes, followed by 40 cycles of 94°C for 20 s, 59°C for 20 s, and 72°C for 25 s. The Ct values were automatically calculated using commercial software (StepOne Software V2.1, Applied Biosystems). The details of primer sequences are given in Table 4 and Table S1.
After completion of standard qRT-PCR, melting curve analysis demonstrated a single PCR amplicon for each reaction. The PCR efficiency was evaluated by the dilution series method using a mix of sample cDNAs as the template. A standard curve was generated using linear regression based on the threshold cycle (Ct) values for all dilution points in a series. The correlation coefficients (R2) and slope values were obtained from the standard curve, and the corresponding PCR amplification efficiencies (E) were calculated using the slope of the calibration curve according to the following equation: 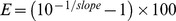 .
.
Determination of RG Expression Stability and Minimum Number of RGs Required
To assess the stability of candidate RGs, four widely recognized RG normalization algorithms were used, GeNormPLUS [6], NormFinder [43], BestKeeper [44], and the Comparative ΔCt method [25]. Four different algorithms were applied to data on three different stiffness and the same RGs were detected in all three different conditions.
Statistical Analysis
Data are expressed as mean±standard deviation (SD) and analyzed using the Statistical Analysis System (SAS) software. Means of different groups were compared using one way analysis of variance followed by Student-Newman-Keuls text. P values <0.05 were considered significant.
Supporting Information
Specificity of qRT-PCR amplification. (a) Amplified fragments were separated by 2% agarose gel. BM2000 represented DNA size marker. The fifth marker band corresponds to 250bp, and the last marker band corresponds to 100bp. (b) Dissociation curves of the nine amplicons showing single peaks.
(TIF)
Expression levels of PAI-1 and tPA in endothelial cells on a variety of substrate stiffness. Genes were normalized to individual and/or combined RGs. Results are mean±SD, n = 3; Means were compared by ANOVA followed by Student-Newman-Keuls text. #P<0.05, Medium vs. Soft. *P<0.05, Stiff vs. Soft. &P<0.05, Stiff vs. Medium.
(TIF)
Primer Information for YAP, CTGF, tPA, and PAI-1.
(DOC)
Acknowledgments
We thank Dr Dou Guifang and Gan Hui, Laboratory of pharmacology, Institute of Transfusion Medicine, Academy of Military Medical Sciences, for allowing us to use their facilities and providing technical support to this work.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (No. 31271001) and the National High Technology Research and Development Program of China (No. 2012AA021902). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kolahi KS, Donjacour A, Liu X, Lin W, Simbulan RK, et al. (2012) Effect of Substrate Stiffness on Early Mouse Embryo Development. PLoS ONE 7: e41717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duprez DA (2010) Arterial Stiffness and Endothelial Function. Hypertension 55: 612–613. [DOI] [PubMed] [Google Scholar]
- 3. Wells RG (2008) The role of matrix stiffness in regulating cell behavior. Hepatology 47: 1394–1400. [DOI] [PubMed] [Google Scholar]
- 4. Cohn JN (2006) Arterial Stiffness, Vascular Disease, and Risk of Cardiovascular Events. Circulation 113: 601–603. [DOI] [PubMed] [Google Scholar]
- 5. Schram MT, Henry RMA, van Dijk RAJM, Kostense PJ, Dekker JM, et al. (2004) Increased Central Artery Stiffness in Impaired Glucose Metabolism and Type 2 Diabetes. Hypertension 43: 176–181. [DOI] [PubMed] [Google Scholar]
- 6.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al.. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.0031 - research0034.0011. [DOI] [PMC free article] [PubMed]
- 7. Wood JA, Shah NM, McKee CT, Hughbanks ML, Liliensiek SJ, et al. (2011) The role of substratum compliance of hydrogels on vascular endothelial cell behavior. Biomaterials 32: 5056–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wood JA, Liliensiek SJ, Russell P, Nealey PF, Murphy CJ (2010) Biophysical Cueing and Vascular Endothelial Cell Behavior. Materials 3: 1620–1639. [Google Scholar]
- 9. Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, et al. (2004) Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol 166: 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng XQ, Haldar S, Deshpande S, Irani K, Kass DA (2003) Wall stiffness suppresses Akt/eNOS and cytoprotection in pulse-perfused endothelium. Hypertension 41: 378–381. [DOI] [PubMed] [Google Scholar]
- 11. Yeh YT, Hur SS, Chang J, Wang KC, Chiu JJ, et al. (2012) Matrix Stiffness Regulates Endothelial Cell Proliferation through Septin 9. PLoS ONE 7: e46889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Connor RS, Hao X, Shen K, Bashour K, Akimova T, et al. (2012) Substrate rigidity regulates human T cell activation and proliferation. J Immunol 189: 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blakney AK, Swartzlander MD, Bryant SJ (2012) The effects of substrate stiffness on the in vitro activation of macrophages and in vivo host response to poly(ethylene glycol)-based hydrogels. J Biomed Mater Res A 100A: 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson LA, Sauder KL, Rodansky ES, Simpson RU, Higgins PDR (2012) CARD-024, a vitamin D analog, attenuates the pro-fibrotic response to substrate stiffness in colonic myofibroblasts. Exp Mol Pathol 93: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raghunathan VK, Morgan JT, Dreier B, Reilly CM, Thomasy SM, et al. (2013) Role of substratum stiffness in modulating genes associated with extracellular matrix and mechanotransducers YAP and TAZ. Invest Ophthalmol Vis Sci 54: 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H, Haeger SM, Kloxin AM, Leinwand LA, Anseth KS (2012) Redirecting Valvular Myofibroblasts into Dormant Fibroblasts through Light-mediated Reduction in Substrate Modulus. PLoS ONE 7: e39969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, et al. (2010) Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol 190: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tilghman RW, Blais EM, Cowan CR, Sherman NE, Grigera PR, et al. (2012) Matrix Rigidity Regulates Cancer Cell Growth by Modulating Cellular Metabolism and Protein Synthesis. PLoS ONE 7: e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saunders R, Hammer D (2010) Assembly of Human Umbilical Vein Endothelial Cells on Compliant Hydrogels. Cell Mol Bioeng 3: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sieminski AL, Hebbel RP, Gooch KJ (2004) The relative magnitudes of endothelial force generation and matrix stiffness modulate capillary morphogenesis in vitro . Exp Cell Res 297: 574–584. [DOI] [PubMed] [Google Scholar]
- 21. Byfield FJ, Reen RK, Shentu TP, Levitan I, Gooch KJ (2009) Endothelial actin and cell stiffness is modulated by substrate stiffness in 2D and 3D. J Biomech 42: 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stroka KM, Aranda-Espinoza H (2011) Endothelial cell substrate stiffness influences neutrophil transmigration via myosin light chain kinase-dependent cell contraction. Blood 118: 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stroka KM, Levitan I, Aranda-Espinoza H (2012) OxLDL and substrate stiffness promote neutrophil transmigration by enhanced endothelial cell contractility and ICAM-1. J Biomech 45: 1828–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peloquin J, Huynh J, Williams RM, Reinhart-King CA (2011) Indentation measurements of the subendothelial matrix in bovine carotid arteries. J Biomech 44: 815–821. [DOI] [PubMed] [Google Scholar]
- 25. Silver N, Best S, Jiang J, Thein S (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen D, Pan X, Xiao P, Farwell MA, Zhang B (2011) Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. J Cell Physiol 226: 2469–2477. [DOI] [PubMed] [Google Scholar]
- 27. Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50: 227–230. [DOI] [PubMed] [Google Scholar]
- 28. Ferguson BS, Nam H, Hopkins RG, Morrison RF (2010) Impact of Reference Gene Selection for Target Gene Normalization on Experimental Outcome Using Real-Time qRT-PCR in Adipocytes. PLoS ONE 5: e15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MaeSZ M, Sendelbach S, Lorkowski S (2010) Selection of reliable reference genes during THP-1 monocyte differentiation into macrophages. BMC Mol Biol 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR – a perspective. J Mol Endocrinol 34: 597–601. [DOI] [PubMed] [Google Scholar]
- 31. Bouvier M (2003) Accessory proteins and the assembly of human class I MHC molecules: a molecular and structural perspective. Mol Immunol 39: 697–706. [DOI] [PubMed] [Google Scholar]
- 32. Stout JT, Caskey CT (1985) HPRT: Gene Structure, Expression, and Mutation. Annu Rev Genet 19: 127–148. [DOI] [PubMed] [Google Scholar]
- 33. Aitken A (2006) 14–3-3 proteins: A historic overview. Semin Cancer Biol 16: 162–172. [DOI] [PubMed] [Google Scholar]
- 34. Schlunck G, Han H, Wecker T, Kampik D, Meyer-ter-Vehn T, et al. (2008) Substrate Rigidity Modulates Cell–Matrix Interactions and Protein Expression in Human Trabecular Meshwork Cells. Invest Ophth Vis Sci 49: 262–269. [DOI] [PubMed] [Google Scholar]
- 35. Rao SS, Bentil S, DeJesus J, Larison J, Hissong A, et al. (2012) Inherent Interfacial Mechanical Gradients in 3D Hydrogels Influence Tumor Cell Behaviors. PLoS ONE 7: e35852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tilghman RW, Blais EM, Cowan CR, Sherman NE, Grigera PR, et al. (2012) Matrix Rigidity Regulates Cancer Cell Growth by Modulating Cellular Metabolism and Protein Synthesis. PLoS ONE 7: e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, et al. (1998) Importance of Glucose-6-phosphate Dehydrogenase Activity for Cell Growth. J Biol Chem 273: 10609–10617. [DOI] [PubMed] [Google Scholar]
- 38. Sirover MA (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. BBA - Protein Struct Mol Enzym 1432: 159–184. [DOI] [PubMed] [Google Scholar]
- 39. Liao F, Li M, Han D, Cao J, Chen K (2006) Biomechanopharmacology: a new borderline discipline. Trends Pharmacol Sci 27: 287–289. [DOI] [PubMed] [Google Scholar]
- 40.Tse JR, Engler AJ (2010) Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol Chapter 10: Unit 10.16. [DOI] [PubMed]
- 41. Pelham RJ Jr, Wang Y (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A 94: 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Radmacher M, Fritz M, Hansma PK (1995) Imaging soft samples with the atomic force microscope: gelatin in water and propanol. Biophysical journal 69: 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 44. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specificity of qRT-PCR amplification. (a) Amplified fragments were separated by 2% agarose gel. BM2000 represented DNA size marker. The fifth marker band corresponds to 250bp, and the last marker band corresponds to 100bp. (b) Dissociation curves of the nine amplicons showing single peaks.
(TIF)
Expression levels of PAI-1 and tPA in endothelial cells on a variety of substrate stiffness. Genes were normalized to individual and/or combined RGs. Results are mean±SD, n = 3; Means were compared by ANOVA followed by Student-Newman-Keuls text. #P<0.05, Medium vs. Soft. *P<0.05, Stiff vs. Soft. &P<0.05, Stiff vs. Medium.
(TIF)
Primer Information for YAP, CTGF, tPA, and PAI-1.
(DOC)