Abstract
The past decade has seen fundamental advances in our understanding of the ageing process and raised optimism that interventions to slow ageing may be on the horizon. Studies of budding yeast have made immense contributions to this progress. Yeast longevity factors have now been shown to modulate ageing in invertebrate and mammalian models, and studies of yeast have resulted in some of the best candidates for anti-ageing drugs currently in development. The first interventions to slow human ageing may spring from the humble yeast.
The budding yeast Saccharomyces cerevisiae is one of the most important model organisms used in ageing-related research. In comparison with other systems, the relative ease and rapidity with which longevity can be quantified in yeast has allowed rapid progress is defining the molecular mechanisms of ageing in this organism and the identification of dozens of factors that modify its longevity. How much of what we learn about ageing in yeast is relevant to people has become an important question. We do not yet know the answer, but the evidence so far suggests that although some aspects of ageing in yeast are specific to this organism, many of the most important features have been evolutionarily conserved in invertebrate species and rodents.
Two ageing models have been established in yeast: replicative ageing and chronological ageing. Replicative ageing is a model of the ageing of mitotically active cells in which the lifespan of a mother cell is defined by the number of daughter cells produced before senescence. Replicative lifespan (RLS) is measured by physical removal of daughter cells, which are easily distinguished from mother cells under an optical microscope1. Chronological ageing is a model of the ageing of post-mitotic cells in which lifespan is defined by how long a yeast cell can survive in a non-dividing, quiescence-like state2. Chronological lifespan (CLS) has been most commonly measured by culturing cells in liquid media, where they enter a non-dividing state once the carbon source has been exhausted. Viability over time, measured as the ability to resume mitotic growth in fresh medium, is then monitored.
A central aspect of ageing-related research is determining the nature of the damage that contributes to ageing and age-associated disease. In the replicative model, damage must be asymmetrically inherited by the mother cell to result in ageing (Fig. 1). Daughter cells produced by aged mother cells retain full replicative capacity until near the end of the mother cell's RLS, demonstrating that a healthy mother cell is generally able to retain most of the age-associated damage3,4. In the chronological model, by contrast, damage accumulates over time within a non-dividing cell until a threshold is reached beyond which the cell is no longer able to re-enter the cell cycle (Fig. 1). Despite these differences, there are some interesting parallels between the two types of yeast ageing. For example, both ageing models are strongly influenced by nutrient availability, and it has been shown that the chronological age of a yeast cell has a negative impact on RLS, even for cells that have not yet produced a single daughter5.
Figure 1. Damage accumulation in yeast ageing.
In chronologically ageing yeast, damage accumulates in non-dividing cells. In the external medium, ethanol initially accumulates and is converted to acetic acid, which induces an apoptosis-like response and cell death. Inside the chronologically ageing cell, damaged mitochondria and oxidized proteins also accumulate and probably contribute to chronological senescence. In replicatively ageing yeast cells, damage is asymmetrically inherited by the mother cell and removed from the daughter cell. Nuclear extrachromosomal ribosomal DNA circles, cytoplasmic oxidized proteins and damaged mitochondria contribute to replicative senescence. In very old mother cells, asymmetry breaks down and the daughter cell can inherit sufficient damage to become prematurely aged.
In this Review, I focus on recent advances in understanding both replicative and chronological ageing in yeast. Potential molecular causes of yeast ageing are discussed, including both yeast-specific mechanisms and those that seem to be conserved in multicellular organisms. Particular focus is placed on key longevity factors shared between yeast and mammals, including dietary restriction, the target of rapamycin (TOR) nutrient response pathway, and sirtuins.
Chronological ageing and acid toxicity
The standard CLS assay used in the majority of studies in this area consists of growing cells in synthetically defined liquid medium with 2% glucose as the carbon source2. Under these conditions, cells initially ferment the glucose to ethanol, which accumulates in the extracellular environment (Fig. 2). After glucose depletion, cells begin to metabolize ethanol by mitochondrial respiration6. Several studies indicate that damage arising from mitochondrial production of reactive oxygen species contributes to the chronological ageing process7. Consistent with this hypothesis, oxidative damage to proteins and mitochondria accumulates with chronological age, and many interventions that increase CLS also promote resistance to oxidative stress2,8.
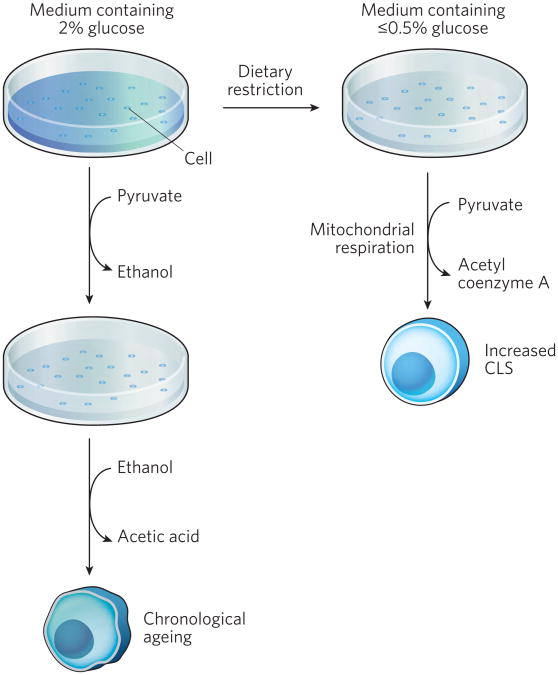
Figure 2. Carbon metabolism and yeast ageing.
Yeast cells are typically aged in synthetic medium containing 2% glucose. Under these conditions, cells ferment pyruvate to ethanol. After glucose depletion, ethanol is metabolized leading to the production of acetic acid, which is toxic to yeast cells and induces an apoptosis-like response that limits CLS. Under conditions of dietary restriction, the glucose concentration of the growth medium is reduced to 0.5% or lower, resulting in direct use of pyruvate by mitochondrial respiration, decreased acetic acid production and increased CLS.
A new perspective on the conjecture that oxidative stress limits CLS has been offered by the finding that acetic acid is a primary molecular factor limiting the lifespan of yeast cells under these standard conditions6. As cells proceed to use ethanol as a secondary carbon source, acetic acid and other organic acids are secreted into the extracellular milieu, leading to acidification of the growth medium (Fig. 2). Buffering the ageing culture to a higher pH or removing acetic acid from the expired medium is sufficient to extend CLS6. Transferring cells to water, rather than allowing them to age in expired medium, has also been shown to increase CLS, demonstrating that extracellular factors limit CLS9. Transferring post-mitotic yeast to water containing physiologically relevant concentrations of acetic acid, but not other acids, suppresses this lifespan extension, indicating that acetic acid is both necessary and sufficient to cause chronological ageing6. These findings are consistent with evidence that ageing yeast cells undergo an apoptosis-like process induced by acetic acid8,10,11 and with the observation that addition of ethanol to the medium can shorten CLS12.
The identification of acetic acid as a limiting factor for chronological survival under standard conditions has led to questions regarding the validity of this system as a model for human ageing6,7. Although it seems unlikely that acetic-acid-induced apoptosis has an important role in human ageing, the chronological ageing model may still provide a reasonable description of some aspects of ageing in people. For example, there is evidence that acetic acid increases the production of reactive oxygen species and causes mitochondrial dysfunction in yeast, suggesting that although the factor inducing chronological senescence (acetic acid) may be specific to yeast, the resulting damage and cellular responses may be shared7. In addition, some interventions that increase CLS also increase RLS, as well as increasing lifespan in invertebrates and even mammals. For example, inhibition of the TOR kinase complex 1 (TOR; in yeast both Tor1 and Tor2 can act in this complex) with rapamycin was first shown to increase yeast CLS13 and has recently been found to increase lifespan in mice14. Likewise, deletion of the gene encoding the ribosomal S6 kinase homologue in yeast, SCH9, was first shown to increase CLS15 and has since been shown to increase RLS similarly, and to increase longevity in nematodes, flies and mice16. Furthermore, both TOR and Sch9 act in a pathway downstream of nutrient availability to modulate CLS, and similar results hold in other systems.
In future work, it will be important for researchers to explore alternative culture conditions in the chronological ageing assay to identify additional molecular factors that modulate survival of non-dividing yeast cells. One simple approach would be to buffer the culture medium so that acetic acid toxicity no longer limits chronological survival. Yeast cells cultured in buffered media still lose viability over time, and hence age6. It will be instructive to learn which interventions that increase CLS under standard conditions also increase CLS when the medium is buffered, and whether oxidative stress and apoptosis-like cell death still contribute to senescence under these conditions. It may be that the yeast chronological ageing model will be an even more informative model for ageing in multicellular eukaryotes when accumulation of acetic acid no longer limits cellular survival.
Asymmetrical damage inheritance during replicative ageing
The best-characterized type of molecular damage associated with replicative ageing is the mother-cell-specific accumulation of extra-chromosomal ribosomal DNA circles1 (ERCs). ERCs are formed by homologous recombination within the rDNA, resulting in the formation of a self-replicating circular DNA molecule that is asymmetrically retained in the mother-cell nucleus during cell division. Proteins that modulate rDNA recombination and ERC formation also influence RLS. For example, the histone deacetylase Sir2 (discussed in detail below) represses ERC formation by inhibiting rDNA recombination, and the replication fork block protein Fob1 promotes rDNA recombination and ERC formation. Thus, deletion of FOB1 or overexpression of SIR2 increases RLS, whereas deletion of SIR2 decreases RLS1.
The mechanism by which ERCs cause senescence in yeast remains unknown. One idea is that essential replication or transcription factors that act on the rDNA interact physically with the accumulating ERCs in ageing cells and are no longer available to carry out their normal cellular functions. Another possibility is that the increasing abundance of DNA coding for ribosomal RNA with age leads to an imbalance between rRNA and ribosomal proteins and impairs ribosome biogenesis and function. Also, a new report has suggested that ERCs may limit RLS by inducing rDNA instability during ageing, and argues that this is the primary cause of senescence17. This idea is based on the observation that a strain engineered to have a reduced ability to replicate the rDNA also has increased rDNA instability and reduced concentrations of ERCs, but is shorter lived than the wild-type strain17. Although this is a reasonable hypothesis, further examination is required because the mutation introduced to reduce rDNA replication activity may limit RLS through a mechanism that is not relevant to ageing of wild-type cells.
Recent efforts have provided insight into how ERCs, and other circular DNA molecules, are asymmetrically inherited by the mother cell. Shcheprova et al.18 have described a septin-dependent nuclear diffusion barrier that inhibits the passage of nuclear pores from the mother cell to the daughter cell. Circular DNA molecules lacking a centromeric sequence, such as ERCs, seem to be physically associated with nuclear pores such that they are also retained in the mother cell by the diffusion barrier. Loss of the septin protein Bud6 leads to symmetrical segregation of nuclear pores and ERCs18. Interestingly, bud6Δ mother cells also have an increased lifespan, presumably due to dilution of ERCs, and perhaps other forms of age-associated damage, through partitioning to the daughter cells18.
Abundant evidence points to ERCs' being important contributors to replicative ageing of yeast cells; however, several additional types of damage are also likely to have a role. For example, oxidatively damaged proteins and dysfunctional mitochondria have both been shown to be asymmetrically segregated to the mother cell and to contribute to replicative ageing. Segregation of damaged cytoplasmic proteins is mediated by the actin cytoskeleton and requires the histone deacetylase Sir2 (ref. 19), resulting in an enhanced capacity to respond to oxidative stress in the daughter cell20. Atp2, a component of the mitochondrial ATPase, is required for proper segregation of active mitochondria to daughter cells21, and mitochondrial dysfunction leads to activation of the retrograde response, which has also been shown to modulate RLS22.
Recently, an interesting link has been suggested between asymmetrical segregation of damaged mitochondria in the mother cell and age-associated genomic instability23. In comparison with young cells, old mother cells show a 100-fold higher frequency of loss of heterozygosity, which is a measure of genomic instability24. Veatch et al.23 proposed that this genomic instability results from a defect in iron–sulphur-cluster biogenesis arising from loss of mitochondrial DNA in ageing cells. How a defect in iron–sulphur-cluster biogenesis leads to genomic instability remains unknown, but it may result from a decrease in function of one or more cluster-binding proteins involved in promoting genomic stability23. It also remains to be determined whether age-associated loss of mitochondrial DNA drives mitochondrial dysfunction or vice versa, as well as the degree to which this contributes to replicative senescence.
Dietary restriction and conserved longevity pathways
Many notable advances in ageing-related research have come from studying interventions that slow ageing across evolutionarily divergent species. Dietary restriction is the most-studied such intervention, and has been shown to increase longevity in organisms ranging from yeast to mammals25. The most common dietary restriction protocol in yeast involves reducing the concentration of glucose in the growth medium from 2% to 0.5% or lower, which has been shown to increase both RLS and CLS26–29. An alternative dietary restriction protocol, in which amino-acid availability is reduced, has also been found to increase RLS30 but has not been studied in detail. Several genetic models of dietary restriction have also been described in which mutations in glucose-sensing pathways lead to reduced activity of nutrient responsive kinases, including protein kinase A, TOR and Sch9 (ref. 1).
Decreased glucose availability leads to several physiological changes that are likely to be relevant for ageing, including a metabolic shift from fermentation to respiration, increased autophagy, decreased messenger RNA translation and enhanced stress resistance. With respect to chronological ageing, the shift away from fermentative metabolism seems to be of primary importance, as this prevents the accumulation of acetic acid and acidification of the culture medium6. In addition, improved clearance of damaged macromolecules by means of autophagy has also been shown to have a role in CLS extension by dietary restriction31. The mechanism of RLS extension by dietary restriction remains an area of ongoing debate and has been proposed to involve activation of Sir2 as well as inhibition of TOR.
Sirtuins
Sirtuins are a family of NAD+-dependent protein deacetylases named for their founding member, the yeast histone deacetylase Sir2 (ref. 32). The importance of sirtuins in ageing was established in studies showing that deletion of Sir2 decreases RLS, whereas overexpression of Sir2 increases RLS33,34. With the subsequent discoveries that activation of Sir2 orthologues can increase lifespan in both nematodes and flies, the idea that sirtuins may mediate lifespan extension in many different organisms has received much attention, and sirtuin activators are being developed and studied for beneficial effects in a variety of mammalian disease models32.
A perplexing aspect of the function of Sir2 in promoting RLS in yeast is that the mechanism by which Sir2 is thought to act (inhibition of rDNA recombination and ERC formation) does not seem to be relevant to ageing in multicellular eukaryotes, despite the fact that Sir2 orthologues promote longevity in other species. This suggests either that sirtuins have evolved to modulate longevity by different downstream mechanisms in different organisms, or that Sir2 acts by additional non-rDNA mechanisms to promote RLS in yeast. In support of the second possibility, sir2Δ fob1Δ mother cells have a lifespan shorter than fob1Δ single-mutant cells, despite the fact that both strains have comparably low concentrations of ERCs33. One non-rDNA function of Sir2 that may contribute to ageing is its requirement for asymmetrical retention of damaged cytoplasmic proteins in the mother cell19,20. This hypothesis is attractive, because oxidatively damaged proteins are known to accumulate with age in invertebrates and mammals, and failure to maintain protein homeostasis is likely to have a contributing role in multiple age-associated diseases35.
A second non-rDNA function of Sir2 that may contribute to its role in promoting RLS is maintenance of telomeric chromatin. Dang et al.36 observed that Sir2 abundance declines in replicatively ageing yeast and that this loss of Sir2 is accompanied by a decrease in histone acetylation and histone abundance near telomeres. The idea that stabilization of telomeres could have a role in RLS extension by overexpression of Sir2 is appealing, because telomeric dysfunction has been suggested as a potential contributing factor in human ageing37,38. Unlike in human cells, however, telomere length does not generally decrease during replicative ageing of yeast cells, and mutations that shorten telomere length have been found to increase RLS39. Thus, if telomeric Sir2 is important in promoting RLS, it is probably by a mechanism distinct from regulation of telomere length. Such a mechanism could involve maintaining the structural integrity of telomeres during ageing or promoting transcriptional silencing of one or more subtelomeric genes involved in the ageing process.
Sirtuins have been proposed to mediate the longevity and health benefits of dietary restriction in yeast, worms, flies and mice40. This hypothesis has proven controversial (Box 1) (see page 480), and several groups have reported Sir2-independent RLS extension by dietary restriction in both yeast and Caenorhabditis elegans, indicating that additional dietary-restriction-responsive longevity factors must exist in these organisms41. In mice, SIRT1 seems to be required for lifespan extension by dietary restriction, and there is accumulating evidence that it has an important role in modulating mitochondrial function in response to reduced nutrient availability32. Thus, although sirtuins are clearly important conserved longevity factors, whether they are directly involved in the lifespan extension that results from dietary restriction remains unclear.
Box 1. Points of contention about sirtuins.
The hypothesis that Sir2 activation accounts for RLS extension from dietary restriction has been controversial. Standard yeast medium contains 2% glucose, and the two most common methods of dietary restriction are to reduce glucose from 2% to either 0.5% (moderate dietary restriction) or 0.05% (extreme dietary restriction). On the basis of the NAD+-dependent nature of the Sir2 deacetylation reaction, Lin et al.27 proposed that dietary restriction alters the NAD+/NADH ratio in a Sir2-activating manner and that this is how dietary restriction increases RLS. In support of this hypothesis, Lin et al.27 reported that Sir2 activity is increased by moderate dietary restriction, as measured by silencing of a MET15 (also known as MET17) marker integrated into the rDNA array, and that lifespan extension by moderate dietary restriction is blocked by deletion of SIR2.
A complicating factor in the study by Lin et al.27 is that the strain background used does not show increased RLS in response to overexpression of Sir2 (ref. 26). In a strain background that responds to both dietary restriction and overexpression of Sir2, combining these two interventions results in an additive increase in RLS26. This led to the hypothesis that Sir2 and dietary restriction might act in parallel pathways to modulate RLS. To test this possibility directly, Kaeberlein et al.26 examined whether dietary restriction could increase RLS in a strain lacking both Sir2 and Fob1. Deletion of FOB1 suppresses the short lifespan and rDNA recombination defect of the sir2Δ mutant33, preventing death caused by abnormally high concentrations of ERCs from masking the longevity effects of dietary restriction26. In this context, both extreme and moderate dietary restriction increased RLS, demonstrating that dietary restriction does not always require Sir2. Lamming et al.69 proposed that the sirtuin Hst2 can compensate for deletion of SIR2 by relocalizing to the rDNA, and that activation of Hst2 accounts for RLS extension by dietary restriction in sir2Δ fob1Δ cells. By contrast, Kaeberlein et al.70 and Tsuchiya et al.71 reported that all of the sirtuins, including Hst2, are dispensable for RLS extension by moderate or extreme dietary restriction. This issue has yet to be resolved.
The hypothesis that Sir2 mediates RLS extension by dietary restriction is based on the assumption that Sir2 activity is increased by dietary restriction27. Despite recent studies reporting changes in NAD+ metabolism predicted to activate Sir2 (ref. 72), the direct evidence for activation of Sir2 has remained unconvincing. The assay initially used by Lin et al.27 to measure Sir2 activity in dietary-restricted cells requires detection of a colour change caused by MET15 expression when cells are grown on media containing Pb2+. Two recent reports using either the same assay as Lin et al.27 or a more sensitive, survival-based assay for silencing of URA3 integrated into the rDNA52,56 indicate that moderate dietary restriction does not alter rDNA silencing. Smith et al.56 reported that extreme dietary restriction does cause a colour change using the MET15 assay, but this is Sir2 independent and is probably an artefact of the MET15 marker. Other studies have also reported that dietary restriction does not increase Sir2 activity, on the basis of silencing of ADE2 and URA3 marker genes integrated near the telomeres45,73. Taken together, these studies suggest that Sir2 activity is unlikely to be significantly increased by either moderate or extreme dietary restriction.
TOR signalling and mRNA translation
In recent years, increasing attention has been paid to TOR signalling as a conserved longevity pathway and a possible component in the response to dietary restriction. TOR kinases are highly conserved in organisms ranging from yeast to humans and function to control growth, metabolism and stress resistance in response to nutrient and growth factor cues42. Decreased TOR activity has been found to increase lifespan in both models of yeast ageing, nematodes, flies and mice, and, consistent with its having a role in dietary restriction, TOR signalling is reduced by dietary restriction in each of these organ-isms14,43. Genetic studies have placed TOR in the same pathway as dietary restriction with respect to yeast RLS and CLS, and a similar relationship has been reported in both nematodes and flies43.
The importance of mRNA translation downstream of TOR signalling as a conserved longevity pathway was demonstrated in a comparative analysis of ageing in yeast and nematodes by Smith et al.44. They used a bioinformatic approach to identify yeast homologues of previously identified nematode ageing genes and measured the RLS of 264 single-gene deletion strains corresponding to these homologues. On the basis of a previous study of 564 randomly selected deletion strains45, Smith et al.44 expected to identify about six long-lived mutants. Instead, 25 homologue pairs that modulate longevity in both S. cerevisiae and C. elegans were found, providing the first quantitative evidence that genetic control of ageing has been conserved between divergent eukaryotic species. Interestingly, eight of these 25 homologue pairs could be definitively assigned as functioning in the TOR signalling pathway to control mRNA translation: the TOR kinase Tor1 (yeast)/LET-363 (nematode), the ribosomal S6 kinase Sch9 (yeast)/RSKS-1 (nematode), three translation initiation factors and three ribosomal proteins44. Several additional studies have reinforced the idea that regulation of mRNA translation is a key component of TOR signalling involved in modulating longevity in invertebrates35, and this has recently been extended to mammals with the finding that knockout of the gene coding for the Sch9 homologue RPS6KB1 also increases lifespan in mice46.
The study of ribosomal protein mutants in yeast has proven informative regarding potential mechanisms of longevity control by TOR and mRNA translation machinery. The yeast ribosome consists of 78 ribosomal proteins, encoded by 137 ribosomal protein genes, that function in the small (40S) or large (60S) ribosomal subunits. RNA interference knockdown of both 40S-subunit and 60S-subunit ribosomal proteins has been found to increase nematode lifespan35. By contrast, yeast RLS seems to be influenced much more robustly by proteins of the 60S sub-unit than by proteins of the 40S subunit. Twenty ribosomal protein gene deletions are known to increase RLS, of which 18 encode 60S-subunit proteins45,47–49. The greater importance of 60S function in RLS determination can be explained in part by differential expression of the Gcn4 stress-responsive and nutrient-responsive transcription factor49. Gcn4 abundance is regulated translationally by the presence of inhibitory open reading frames in the 5′ untranslated region of the GCN4 mRNA. GCN4 is not translated efficiently under conditions favouring growth; however, specific depletion of the 60S subunit leads to increased translation of GCN4, despite a global decrease in protein synthesis. This increase in Gcn4 abundance is required for full lifespan extension by deletion of 60S-subunit ribosomal protein genes. Deletion of GCN4 also partly blocks lifespan extension by dietary restriction, deletion of TOR1 or deletion of SCH9, consistent with a model placing ribosomal proteins downstream of dietary restriction, TOR and Sch9 in a conserved longevity pathway49.
The fact that loss of Gcn4 does not completely block RLS extension indicates that additional longevity modifiers must exist downstream of dietary restriction and TOR. One possibility is that other longevity-modulating mRNAs are also differentially translated, as has been suggested for components of the electron transport chain in flies subjected to dietary restriction50. RLS-extending mutations in ribosomal proteins also suppress mitochondrial degeneration51, although whether this is related to altered translation of mitochondrial proteins remains unknown. Deletion of TOR1 or SCH9 also results in decreased rDNA recombination by a Sir2-independent mechanism52, suggesting that ERC formation may also be dependent on TOR activity. Finally, because reduced TOR activity results in increased autophagic degradation, it is reasonable to speculate that accumulation of damaged proteins and mitochondria during replicative ageing could also be alleviated (Fig. 3).
Figure 3. Modulation of yeast ageing by TOR and Sir2.
Decreased TOR activity increases both RLS and CLS by several possible mechanisms. Reduced rDNA recombination and reduced mRNA translation probably contribute to increased RLS, whereas decreased production of acetic acid increases CLS. Resistance to oxidative stress, improved mitochondrial function and enhanced clearance of damaged proteins probably contributes to both RLS and CLS. Increased Sir2 activity extends RLS but not CLS. Sir2 represses rDNA recombination, promotes asymmetrical segregation of oxidized proteins to the mother cell and maintains chromatin structure near telomeres and possibly other loci.
Another hypothesis that has been proposed is that Sir2 functions downstream of TOR and is activated in response to reduced TOR sig-naling53. This model is based on the observation that TOR inhibition leads to activation of two stress-responsive transcription factors, Msn2 and Msn4 (ref. 54) These in turn promote expression of the nicotinamidase Pnc1, which could lead to activation of Sir2 by reducing the cellular abundance of nicotinamide55. Although this is an interesting possibility, there is little direct evidence that inhibition of TOR activates Sir2. In fact, deletion of TOR1 has bee n reported to have no effect on Sir2 activity in vivo45. Also arguing against this model is evidence that dietary restriction does not activate Sir2 in vivo and that lifespan extension by either dietary restriction or deletion of TOR1 does not require Sir2 (refs 45, 52, 56) (Box 1). It may be that TOR signalling can modulate Sir2 activity under certain conditions, but the bulk of the current data support the idea that TOR and Sir2 act in distinct genetic pathways to modulate RLS.
Like RLS, reduced TOR signalling also acts downstream of dietary restriction to increase CLS13; however, control of mRNA translation does not seem to have a significant role in this response. Instead, a shift in metabolism from fermentation to respiration in response to reduced TOR activity has been proposed to mediate increased CLS57. Such a shift could mimic the effect of dietary restriction by reducing the amount of acetic acid produced from metabolism of ethanol6. Induction of autophagy in response to TOR inhibition has also been shown to be important for CLS58, and a recent report implicates upregulation of glycerol biosynthesis in response to deletion of TOR1 or SCH9 as a contributing factor for CLS extension59.
Emerging yeast models
Although S. cerevisiae is the most widely used yeast ageing model, additional yeast species are also providing valuable insights. The best-studied alternative yeast ageing model pertains to the fission yeast Schizosaccharomyces pombe. CLS is measured in S. pombe identically to that in S. cerevisiae, and many of the same factors have been found to modulate CLS in this organism. These include CLS extension by dietary restriction in the form of decreased glucose availability and by mutations that reduce signalling through glucose-sensing pathways involving the S. pombe homologues of protein kinase A and Sch9 (refs 60, 61). Accumulation of oxidatively damaged proteins with chronological age has also been observed in S. pombe, suggesting that similar molecular processes occur during ageing in both fission and budding yeasts. An important question for future studies will be to determine whether acetic acid toxicity is a primary cause of chronological senescence in S. pombe. If this is not the case, then S. pombe could prove to be a particularly informative model for characterizing additional processes leading to oxidative damage and mitochondrial dysfunction during ageing.
Unlike S. cerevisiae, cell division in young S. pombe is morphologically symmetrical, resulting in the formation of two apparent sister cells rather than easily identifiable mother and daughter cells. By carefully following individual cells for multiple generations, however, aged mother cells can be morphologically differentiated from daughter cells, allowing measurement of RLS62. Similar to the case in S. cerevisiae, oxidatively damaged proteins seem to be asymmetrically segregated to the mother cell in a Sir2-dependent manner63. This conservation suggests that the asymmetrical retention of damage has been highly conserved between these two divergent yeast species.
A second example of an alternative yeast model for ageing studies is Kluyveromyces lactis. Unlike S. cerevisiae, K. lactis metabolizes glucose primarily by respiration, rather than by alcoholic fermentation. Dietary restriction by glucose restriction does not extend CLS in this species64. Thus, the different evolutionary strategies taken by K. lactis, S. pombe and other yeast species are likely to provide new insights into the ageing process.
Future directions and perspectives
Studies of yeast ageing have had a significant impact on ageing-related research. Some of the most promising longevity factors and potential anti-ageing drugs (Box 2) were first identified and characterized in yeast. In two cases, those of rapamycin and resveratrol, compounds first reported to slow ageing in yeast have progressed to clinical trials for ageing-related diseases65. In the future, it will be important to continue to clarify which aspects of ageing are conserved between yeast and mammals and which are specific to yeast. Thus far, the approach of using an invertebrate species, such as C. elegans, as an intermediate model system has proven fruitful in differentiating conserved mechanisms of ageing from yeast-specific mechanisms of ageing.
Box 2. Three anti-ageing drugs from yeast.
Resveratrol is a polyphenolic compound found in red wine. It began to be studied as an anti-ageing drug after it was identified in 2003 from an in vitro screen for Sir2 activators74. Resveratrol was first reported to extend yeast RLS74 and then to extend lifespan in nematodes, flies and a short-lived fish75,76. Subsequent studies have failed to reproduce lifespan extension by resveratrol in yeast and flies, and have shown that it activates sirtuins in a substrate-specific manner77–80. Feeding mice a diet supplemented with resveratrol protects them against toxicity associated with a high-fat diet, enhances metabolic function and leads to gene expression changes similar to those caused by dietary restriction81–83. Thus far, dietary supplementation with resveratrol has not resulted in increased lifespan in mice84. Resveratrol and additional substrate-specific sirtuin activators are currently being studied for safety and efficacy against type 2 diabetes in human clinical trials.
Rapamycin (also known as sirolimus) is a macrolide antibiotic that specifically inhibits TOR activity. The first longevity study with rapamycin was published in 2006 and showed that the drug increases yeast CLS13. Rapamycin has also been reported to increase RLS53, and genetic models of reduced TOR activity increase lifespan in yeast, nematodes and flies43. In 2009, lifespan extension by feeding mice a diet supplemented with an encapsulated form of rapamycin was reported14. Initiating rapamycin supplementation at as late as 600 days of age, which is roughly comparable to 60 years in humans, was sufficient to increase mouse lifespan14. Rapamycin is used clinically to treat certain forms of cancer, to prevent coronary restenosis and as an immunosuppressant43.
Spermidine is a naturally occurring polyamine that modulates diverse cellular processes, including DNA stability, transcription, translation and apoptosis85. In 2009, supplementation with spermidine was reported to increase yeast CLS and lifespan in nematodes and flies86. The mechanism by which spermidine promotes longevity is proposed to involve induction of autophagy86.
The field of yeast ageing is likely to continue to advance rapidly as new methods and technologies are applied. Network biology, for example, has been used to predict ‘longevity networks’ and identify new longevity genes on the basis of known interactions with ageing-related proteins48,66. New metabolomic and proteomic technologies also have great potential to enhance our understanding of ageing and are easily applied to the yeast system. High-throughput methods for quantifying lifespan are also likely to continue to improve, providing quantitative data for RLS and CLS for thousands of genetic variants and perhaps allowing small molecule screens to identify new anti-ageing drugs. Recently, a promising high-throughput method for measuring RLS based on the selective killing of daughter cells, rather than manual micromanipulation, has been described and may accelerate studies in this area significantly67.
An aspect of yeast ageing that remains relatively unstudied is the interplay between RLS and CLS. Although the reduction in RLS caused by chronological ageing of cells was described more than a decade ago5, the mechanistic details underlying the phenomenon remain a mystery. It was recently found that, in addition to shortening RLS, chronological ageing leads to elevated genomic instability and loss of asymmetry when cells resume mitotic growth68. This phenotype seems unlikely to be related to acetic acid toxicity and may represent an aspect of chronological ageing that is currently masked by acetic-acid-induced cell death. A hybrid ageing assay that combines features of both chronological and replicative ageing might be fertile ground for exploring shared aspects of cellular ageing.
A major challenge in future will be to understand how multiple types of damage in different cellular compartments cause senescence in yeast. As discussed above, at least three different types of damage are known to be asymmetrically inherited by mother cells and contribute to replicative ageing: nuclear ERCs, damaged cytoplasmic proteins and defective mitochondria. It will be important to clarify how these different types of damage interact to ultimately block cell division and whether they act in an independent, additive or synergistic manner. Understanding how these different types of age-associated damage contribute to cellular dysfunction in yeast is likely to prove informative about mammalian ageing, where, although many different types of damage have been proposed to cause ageing, there is little consensus regarding which are most relevant.
Despite the vast differences in complexity between yeast and humans, the study of ageing in yeast has provided key insights into pathways that modulate ageing in mammals. The pioneering efforts that Leonard Guarente, Michal Jazwinski and others made in establishing yeast as an accepted ageing model in the face of strong scepticism deserve recognition. Without this perseverance, the field of ageing-related research would be poorer. Undoubtedly, S. cerevisiae and its fungal brethren will continue to have a role at the forefront of efforts to understand the basic biology of ageing.
Acknowledgments
Studies related to this topic in the Kaeberlein laboratory have been supported by US National Institutes of Health (NIH) grant R21AG031965, a Pilot Project grant from the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH grant P30AG013280) and a New Scholar in Aging Award from the Ellison Medical Foundation.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints. The author declares competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. In this paper, acetic acid toxicity was shown to be the primary cause of chronological senescence under standard growth conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burhans WC, Weinberger M. Acetic acid effects on aging in budding yeast: are they relevant to aging in higher eukaryotes? Cell Cycle. 2009;8:2300–2302. doi: 10.4161/cc.8.14.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rockenfeller P, Madeo F. Apoptotic death of ageing yeast. Exp Gerontol. 2008;43:876–881. doi: 10.1016/j.exger.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabrizio P, et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herker E, et al. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrizio P, Longo VD. Chronological aging-induced apoptosis in yeast. Biochim Biophys Acta. 2008;1783:1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabrizio P, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 13.Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 16.Kaeberlein M, Kapahi P. Aging is RSKy business. Science. 2009;326:55–56. doi: 10.1126/science.1181034. [DOI] [PubMed] [Google Scholar]
- 17.Ganley AR, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell. 2009;35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. This study describes the existence of a septin-dependent diffusion barrier required for asymmetrical inheritance of nuclear pores and extrachromosomal rDNA circles by the mother cell during division. [DOI] [PubMed] [Google Scholar]
- 19.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. This study showed that oxidatively damaged cytoplasmic proteins are asymmetrically segregated to the mother cell during ageing in a Sir2-dependent manner. [DOI] [PubMed] [Google Scholar]
- 20.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:e296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 28.Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. J Gerontol A. 2008;63:113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- 29.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 31.Alvers AL, et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy BK, et al. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy BK, Kaeberlein M. Hot topics in aging research: protein translation, 2009. Aging Cell. 2009;8:617–623. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. In this paper, a specific chromatin modification, H4K16 acetylation, was shown to be an important Sir2-regulated function in determining RLS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path frommaize, Tetrahymena and yeast to human cancer and aging. Nature Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 38.Shawi M, Autexier C. Telomerase, senescence and ageing. Mech Ageing Dev. 2008;129:3–10. doi: 10.1016/j.mad.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Austriaco NR, Jr, Guarente LP. Changes of telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:9768–9772. doi: 10.1073/pnas.94.18.9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guarente L. Calorie restriction and SIR2 genes — towards a mechanism. Mech Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Kaeberlein M, Powers RW., III Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith ED, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. This study demonstrated that genetic control of longevity has been conserved between yeast and nematodes and identified 25 homologue pairs that modulate longevity in both species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 46.Selman C, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian lifespan. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiocchetti A, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Managbanag JR, et al. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS ONE. 2008;3:e3802. doi: 10.1371/journal.pone.0003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steffen KK, et al. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. This study defined the function of the large ribosomal subunit as a key determinant of yeast ageing and identified the transcription factor Gcn4 as a translationally regulated longevity factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zid BM, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Zuo X, Kucejova B, Chen XJ. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nature Cell Biol. 2008;10:1090–1097. doi: 10.1038/ncb1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riesen M, Morgan A. Calorie restriction reduces rDNA recombination independently of rDNA silencing. Aging Cell. 8:624–632. doi: 10.1111/j.1474-9726.2009.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 55.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith DL, et al. Calorie restriction effects on silencing and recombination at the yeast rDNA. Aging Cell. 2009;8:633–642. doi: 10.1111/j.1474-9726.2009.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvers AL, et al. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei M, et al. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roux AE, et al. Pro-aging effects of glucose signaling through a G protein-coupled glucose receptor in fission yeast. PLoS Genet. 2009;5:e1000408. doi: 10.1371/journal.pgen.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roux AE, Quissac A, Chartrand P, Ferbeyre G, Rokeach LA. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- 62.Barker MG, Walmsley RM. Replicative ageing in the fission yeast Schizosaccharomyces pombe. Yeast. 1999;15:1511–1518. doi: 10.1002/(sici)1097-0061(199910)15:14<1511::aid-yea482>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 63.Erjavec N, Cvijovic M, Klipp E, Nystrom T. Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proc Natl Acad Sci USA. 2008;105:18764–18769. doi: 10.1073/pnas.0804550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oliveira GA, Tahara EB, Gombert AK, Barros MH, Kowaltowski AJ. Increased aerobic metabolism is essential for the beneficial effects of caloric restriction on yeast life span. J Bioenerg Biomembr. 2008;40:381–388. doi: 10.1007/s10863-008-9159-5. [DOI] [PubMed] [Google Scholar]
- 65.Kaeberlein M. Resveratrol and rapamycin: are they anti-aging drugs? BioEssays. 2010;32:96–99. doi: 10.1002/bies.200900171. [DOI] [PubMed] [Google Scholar]
- 66.Lorenz DR, Cantor CR, Collins JJ. A network biology approach to aging in yeast. Proc Natl Acad Sci USA. 2009;106:1145–1150. doi: 10.1073/pnas.0812551106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin H, Lu M, Goldfarb DS. Genomic instability is associated with natural life span variation in Saccharomyces cerevisiae. PLoS ONE. 2008;3:e2670. doi: 10.1371/journal.pone.0002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamming DW, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 70.Kaeberlein M, et al. Comment on ‘HST2 mediates SIR2-independent life-span extension by calorie restriction’. Science. 2006;312:1312. doi: 10.1126/science.1124608. [DOI] [PubMed] [Google Scholar]
- 71.Tsuchiya M, et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 72.Lu SP, Lin SJ. Regulation of yeast sirtuins by NAD+metabolism and calorie restriction. Biochim Biophys Acta. doi: 10.1016/j.bbapap.2009.09.030. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaeberlein M, et al. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 75.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 76.Valenzano DR, et al. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 77.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 78.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 79.Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev. 2007;128:546–552. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 80.Beher D, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 81.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 83.Barger JL, et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pearson KJ, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaeberlein M. Spermidine surprise for a long life. Nature Cell Biol. 2009;11:1277–1278. doi: 10.1038/ncb1109-1277. [DOI] [PubMed] [Google Scholar]
- 86.Eisenberg T, et al. Induction of autophagy by spermidine promotes longevity. Nature Cell Biol. 2009;11:1277–1278. doi: 10.1038/ncb1975. This study identified spermidine as a compound that increases lifespan in yeast, nematodes and flies. [DOI] [PubMed] [Google Scholar]