Abstract
The main objective of the present study was isolation, purification, and characterization of actinomycetes from soil samples, having antimicrobial activity against 12 selected pathogenic strains. Soils samples were taken from different niche habitats of Sheopur district, Madhya Pradesh, India. These samples were serially diluted and plated on actinomycete isolation agar media. Potential colonies were screened, purified, and stored in glycerol stock. Isolates were morphologically and biochemically characterized. These isolates were subjected to extraction for production of the antibacterial compound. Antibacterial activity and Minimum Inhibitory Concentration (MIC) of the purified extract of isolates were evaluated. Totally 31 actinomycete isolates were tested for antagonistic activity against 12 pathogenic microorganisms. Isolates AS14, AS27, and AS28 were highly active, while AS1 showed less activity against the pathogenic microorganisms. Isolate AS7 exhibited the highest antagonistic activity against Bacillus cereus (24 mm) and AS16 showed the highest activity against Enterococcus faecalis (21 mm). MIC was also determined for actinomycete isolates against all the tested microorganisms. MIC of actinomycete isolates was found to be 2.5 mg/ml against Shigella dysenteriae, Vancomycin-resistant enterococci, and Klebsiella pneumoniae, and was 1.25 mg/ml for Staphylococcus saprophyticus, Streptococcus pyogenes, Staphylococcus epidermidis, Methicillin-resistant Staphylococcus, Bacillus cereus, Staphylococcus xylosus, Methicillin-resistant Staphylococcus aureus, Enterococcus faecalis, and Staphylococcus aureus. All actinomycetes isolates showed antibacterial activity against S. aureus, while they showed less activity against S. dysenteriae. These isolates had antibacterial activity and could be used in the development of new antibiotics for pharmaceutical or agricultural purposes.
Keywords: Agar well diffusion method, antimicrobial activity, minimum inhibitory concentration, pathogenic microorganisms, Sheopur
INTRODUCTION
Soil microorganisms provide an excellent resource for the isolation and identification of therapeutically important products. Among them, actinomycetales are an important group.[1] The order actinomycetales is composed of approximately 80 genera, nearly all from terrestrial soils, where they live primarily as saprophytes, water and colonizing plants showing marked chemical and morphological diversity, but from a distinct evolutionary line.[2]
Actinomycetes are gram-positive bacteria with high guanine+cytosine content of over 55%[3] in their DNA, which have been recognized as sources of several secondary metabolites, antibiotics, and bioactive compounds that affect microbial growth.[4] Actinomycetes have filamentous nature, branching pattern, and conidia formation, which are similar to those of fungi. For this reason, they are also known as ray fungi.[5] Actinomycetes produce branching mycelium which may be of two types, viz., substrate mycelium and aerial mycelium. Streptomyces are the dominant of all actinomycetes.[6]
A large number of actinomycetes have been isolated and screened from soil in the past several decades, accounting for 70%-80% of relevant secondary metabolites available commercially.[7] Actinomycetes are potential source of many bioactive compounds,[8,9,10,11] which have diverse clinical effects and important applications in human medicine.[12] It has been estimated that approximately one-third of the thousands of naturally occurring antibiotics have been obtained from actinomycetes.[13]
The resistance problem demands to discover new antibacterial agents effective against resistant pathogenic bacteria and fungi. So, we need to screen more and more actinomycetes from different habitats for antimicrobial activity in the hope of getting some new actinomycetes strains that produce antibiotics, which have not been discovered yet and are active against drug-resistant pathogens.
MATERIALS AND METHODS
Soil Sampling and Pretreatment
Soil samples were collected from different niche habitats of Sheopur district (geographical coordinates: latitude 25.67°N and longitude 76.7°E), Madhya Pradesh, India. Soils were collected from different locations of Sheopur, such as field, near plant surface, mining area, Kaketto dam, Kamojiya poultry farm, and well. Samples were collected by inserting a sterilized polyvinyl corer into the sediments. The corer was sterilized with alcohol before sampling at each location. Each collection was made from 6-12 inches depth of the surface of ground. These samples were placed in sterile poly bags, sealed tightly, and transported immediately to the laboratory. These soil samples were air-dried for 3-4 h at 45°C, crushed, and sieved prior to use for isolation purpose.[14]
Isolation of Pure Culture of Actinomycetes
Thirty-one actinomycete strains were isolated as pure culture by using standard microbiological method.[15] One gram of dried soil was suspended in 99 ml sterile water and serially diluted in sterile water up to 10−7. An aliquot of 0.1 ml of each dilution was taken and spread evenly over the surface of actinomycete isolation agar (AIA) medium[16] supplemented with cyclohexamide (50 μg/ml) and nystatin (50 μg/ml).[17] Plates were incubated at 30°C for 7 days. After 7 days incubation, whitish pin-point colonies, characteristic of actinomycetes, with a clear zone of inhibition around them were observed. The whitish pinpoint colonies with inhibitory or clear zone of inhibition were selected and purified. The purified actinomycetes isolates were preserved in International Streptomyces Project (ISP) 1 (tryptone-yeast extract broth) at 4°C and 25% v/v glycerol stocks stored at 40°C for long time preservation.[18]
Characterization of Isolates
All the isolates were morphologically and biochemically characterized.
Gram Staining
Smear was prepared by spreading the broth culture on a glass slide followed by heat drying. The smear was covered with crystal violet for 30-60 s and washed off with water. The smear was covered with Gram's iodine for 30-60 s, decolorized with alcohol, and washed with water. Finally the smear was stained with safranin counter stain for 2 min. After washing and drying, the slides were viewed at 100× under phase-contrast microscope.[19]
Morphological Characterization
Actinomycete isolates were inoculated on seven different ISP media (ISP1-ISP7) and incubated for 5 days at 30°C. The colonies were observed under a high-power magnifying lens and colony morphology was noted with respect to color, aerial and substrate mycelium, branching, and the nature of colony.[19]
Biochemical Characterization
After preliminary studies, the isolates which were found to be positive were selected for biochemical studies. Biochemical tests generally used are gelatin hydrolysis, starch hydrolysis, urea hydrolysis, acid production from different sugars, hydrogen sulfide production test, motility test, triple sugar iron (TSI) agar test, citrate utilization test, indole test, methyl red test, Voges-Proskauer test, and catalase test, oxidase test.[20]
Test Organism
Test organisms were obtained from IMTECH, Chandigarh. The selected human pathogenic microorganisms used in antimicrobial study were Shigella dysenteriae ATCC 9754, Staphylococcus saprophyticus MTCC 6155, Streptococcus pyogenes MTCC 655, Staphylococcus epidermidis ATCC 12228, Methicillin-resistant Staphylococcus aureus ATCC 700789, Bacillus subtilis MTCC 299, Bacillus cereus MTCC 430, Staphylococcus xylosus MTCC 6149, Vancomycin-resistant enterococci VRE 912, Enterococcus faecalis ATCC 29122, Klebsiella pneumoniae MTCC 2405, and Staphylococcus aureus ATCC 25923.
Screening of Actinomycetes
The screening method of isolates consists of two steps: primary screening and secondary screening. In primary screening, the antibacterial activity of pure isolates was determined by cross-streak method[21] on Mueller Hinton Agar (MHA). Secondary screening of isolate was done by agar well diffusion method with crude extract of ethyl acetate after secondary metabolite extraction.
Extraction of Bioactive Compound
Production of bioactive compound was done by submerged fermentation.[22] Actinomycetes isolates were taken in 50 ml of ISP1 broth in a 250-ml-capacity conical flask under sterile conditions and incubated at 30°C for 7 days at 150 rpm rotation.[23] After fermentation, the medium was centrifuged at 10,000 rpm for 10 min to remove cells and debris and harvested for fermented broth. Resultant fermented broths were added to equal volume of ethyl acetate. Then the samples were shaken vigorously in a rotatory shaker. The solvent phase was collected and evaporated in a desiccator. The completely dried residues were re-dissolved in dimethyl sulfoxide (DMSO) and lyophilized, to be used for further studies.[24,25]
Determination of the Antibacterial Activity
Antibacterial activity of partially purified extracellular crude extracts; was determined by agar well diffusion method. Cell Concentration of all test microorganisms were adjusted at 0.5 McFarland turbidity standards and inoculated on MHA plates by using sterilized cotton swabs. Wells were bored by sterilized 1000 μl micro tip, and 100 μl of each crude extract was poured into wells. Plates were incubated at 37°C for 24 h.[26]
Determination of Minimum Inhibition Concentration
MIC of active ethyl acetate crude extract of isolates against test microorganisms was determined by serial dilution method. MIC was determined by enzyme-linked immunosorbent assay (ELISA) microtiter plate. The crude extract was twofold serially diluted from 1.25 mg/ml to 10 mg/ml for determining the MIC of active isolates.
RESULTS AND DISCUSSION
Actinomycetes have been intensively studied in several underexplored environments, niche, and extreme habitats in various parts of the world (including India) in the last few years. Yet, there is no report regarding isolation of actinomycetes from Sheopur region, MP (India). Therefore, an attempt has been made to isolate the actinomycetes from this unexplored region in order to find novel species. Totally 31 actinomycetes strains were isolated from soil samples of different areas based on the gram staining and colony morphology. All the isolates were found to be positive in gram staining and had different morphological structures. The biochemical properties of actinomycetes isolates were recorded [Table 1]. The isolates were screened for their inhibitory activity against the human pathogenic bacteria. Both the primary and secondary screening methods were used to screen the actinomycetes for antibacterial activity. The first screening was used to select the antibacterial isolates and determine the range of microorganisms that were sensitive to the antibiotics. The secondary screening method was crucial to select the isolates for further studies. The screening may be qualitative or quantitative in its approach. The qualitative approach is used to determine the range of the microorganisms that are sensitive to a potential antibiotic. The quantitative approach provides the information about the yield of antibiotic that can be expected when the organism is grown in ISP1 broth [Table 2]. The antibacterial activity of secondary metabolite extract of some isolates confirmed through secondary screening [Table 3] that were able to inhibit the extracellular growth of filaments in the test organism. All the experimental measurements were carried out in triplicates and were expressed as an average of three analyses. Isolates AS14, AS27, and AS28 were highly active [Figures 1-3], while AS1 showed less activity against pathogenic bacterial strains. Isolate AS7 exhibited the highest activity against B. cereus (zone of inhibition 24 mm) and AS16 against E. faecalis (zone of inhibition 21 mm).
Table 1.
Biochemical characterization of actinomycetes isolates
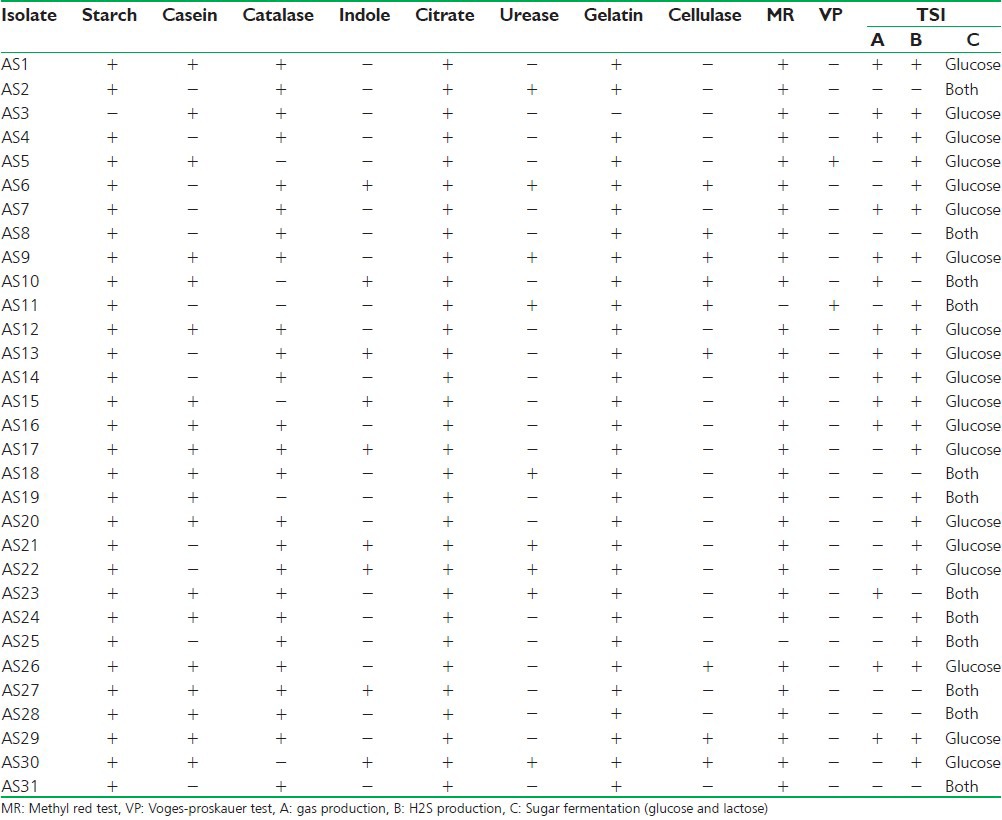
Table 2.
Dry weight of antibiotic as actinomycete isolates
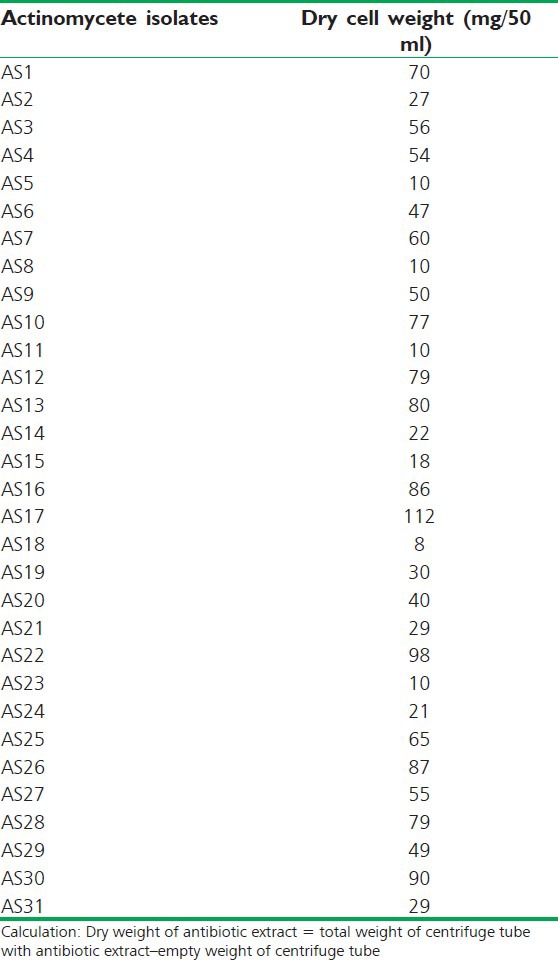
Table 3.
Results of antibacterial activity of actinomycetes isolates, which was checked on mueller hinton agar media by agar well diffusion method [zone of inhibition recorded in millimeter]
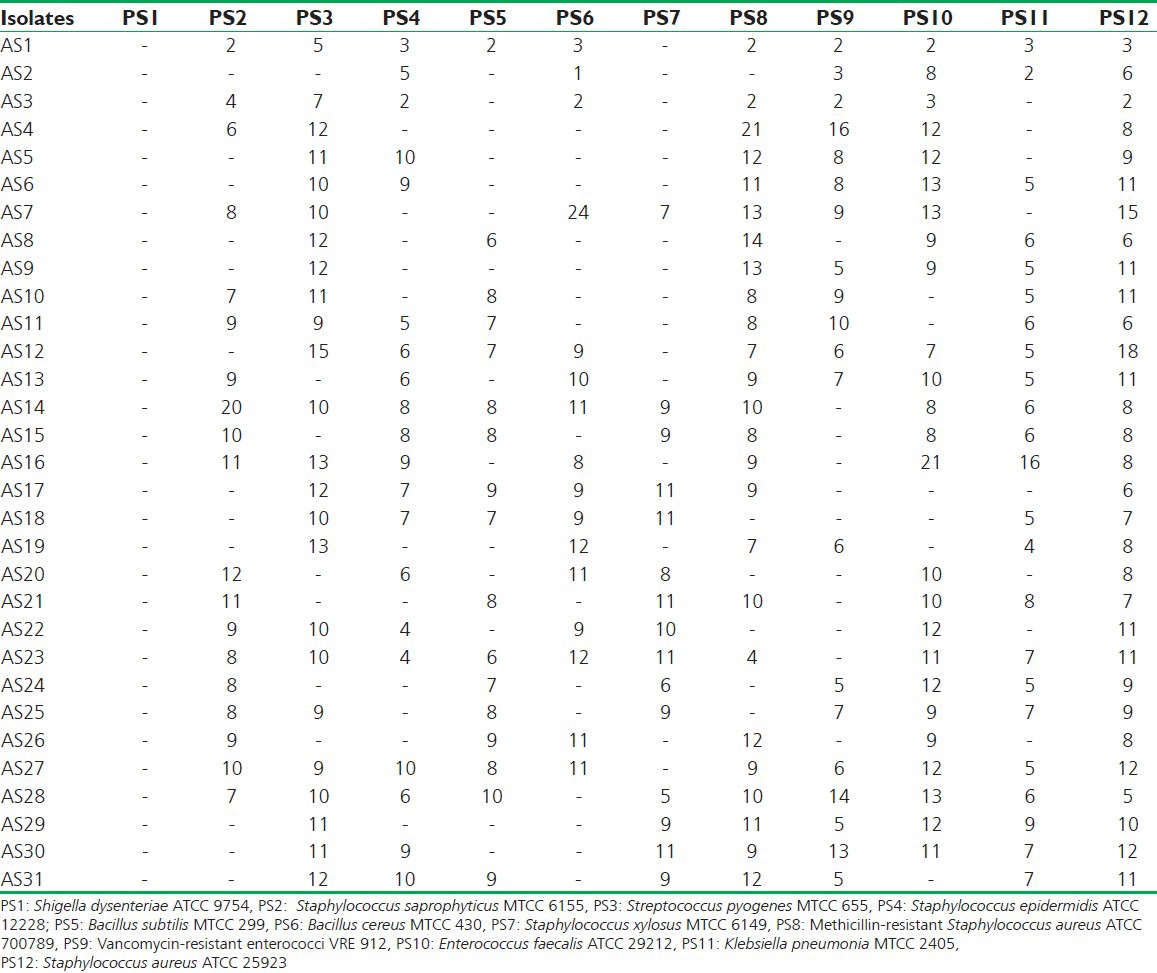
Figure 1.
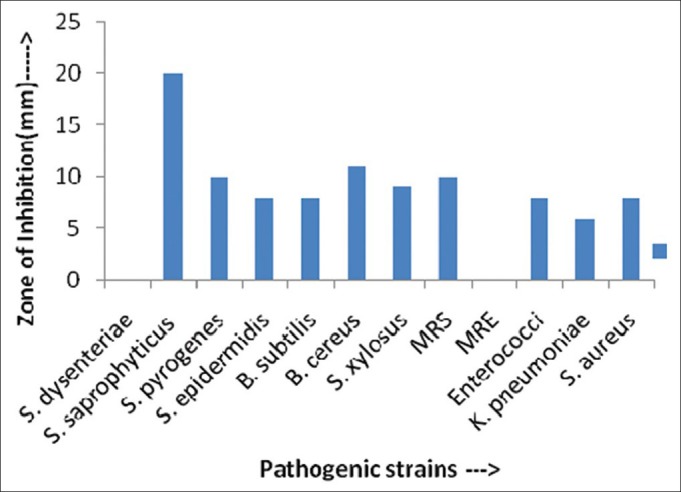
Antimicrobial activity (in ZOI) against pathogenic bacterial strains shown by AS14 actinomycetes isolates
Figure 3.
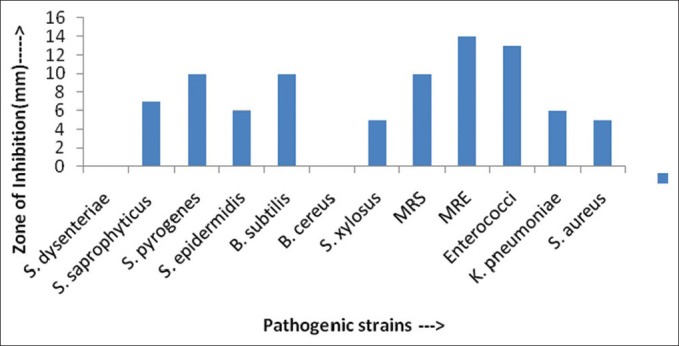
Antimicrobial activity (in ZOI) against pathogenic bacterial strains shown by AS28 actinomycetes isolates
Figure 2.
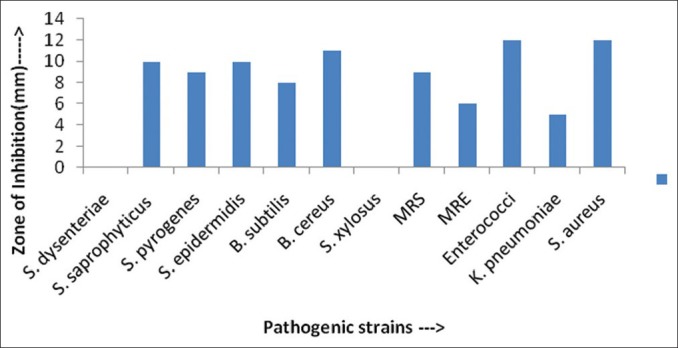
Antimicrobial activity (in ZOI) against pathogenic bacterial strains shown by AS27 actinomycetes isolates
The MIC evaluation of the active actinomycetes isolates was performed against all the tested microorganisms by the broth dilution method. MIC 2.5 mg/ml was exhibited by isolates AS23, AS33, and AS34 against E. coli, Vancomycin-resistant enterococci, K. pneumoniae, respectively. While MIC 1.25 mg/ml was shown by isolates AS25 and AS32 against S. saprophyticus; by isolates AS33 and AS36 against S. pyogenes; by isolates AS35 and AS36 against S. epidermidis; by isolates AS29, AS31, and AS36 against Methicillin-resistant Staphylococcus (MRS906); by isolate AS21 against B. cereus; by isolates AS22, AS26, and AS35 against S. xylosus; by AS31and AS36 against Methicillin-resistant S. aureus; by isolates AS21 and AS33 against E. faecalis; and by isolates AS35 and AS36 against S. aureus. The similar antimicrobial activity had been also reported against selected multidrug resistant bacteria.[27]
CONCLUSION
This study shows that the test actinomycetes isolates have the potential to act as sources of new antibacterial compounds against pathogenic microorganisms to humans. Here, we found that Sheopur is a good region of biodiversity and has been adequately acceptable due to its vast floral diversity and also microbial diversity. The results showed that all the isolates were able to inhibit the extracellular growth of filaments in the test organism. The isolates were not able to inhibit intracellular growth of mycelium. The reason might be the non-reaching of the isolate extract to intracellular cell of the test organism and non-denaturation of the bacterial cell wall by the isolate extract. Although the isolate AS7, AS16 exhibited the highest activity against B. cereus and E. faecalis respectively.
Mycelium color and diffusible pigment of actinomycetes isolates were determined on the basis of morphological characterization, and most of the actinomycetes isolates showed positive results for catalase, starch utilization, and casein utilization and others also, while the isolates showed negative results for indole and Voges–Proskauer tests. Our studies will establish the rich actinomycetes diversity of the region. So, further intensive studies are required on the actino-bacterial diversity of exclusive biotopes in Sheopur, which could form an important input into pharmaceutical industries.
ACKNOWLEDGMENT
We wish to deeply thank Director, MITS, Gwalior, for providing the necessary facilities and support for this study.
Footnotes
Source of Support: In house facility of Madhav Institute of Technology and Science, Gwalior.,
Conflict of Interest: Nil.
REFERENCES
- 1.Bérdy J. Bioactive microbial metabolites: A personal view. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Goodfellow M, O’Donnell AG. Search and discovery of industrially significant Actinomycetes. In: Baumberg S, Hunter IS, Rhodes PM, editors. Microbial Products: New Approaches, Society for General Microbiology Symposium No. 44. Cambridge: Cambridge University Press; 1989. pp. 343–83. [Google Scholar]
- 3.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida N, Miyazaki K, Kumagai K, Rikimaru M. Neocarzinostatin, an antitumor antibiotic of high molecular weight, isolation, physiochemical properties and biological activities. J Antibiot (Tokyo) 1965;18:68–76. [PubMed] [Google Scholar]
- 5.Wang Y, Zhang ZS, Ruar TS, Wang YM, Ali SM. Investigation of Actinomycetes diversity in the tropical rainforests of Singapore. J Ind Microbiol Biotechnol. 1999;23:178–87. [Google Scholar]
- 6.Okami Y, Okazaki T. Studies on marine microorganisms isolation from the sea. J Antibiot. 1972;25:456–60. doi: 10.7164/antibiotics.25.456. [DOI] [PubMed] [Google Scholar]
- 7.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–63. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Vining LC. Secondary metabolism, inventive evolution and biochemical diversity: A review. Gene. 1992;115:135–40. doi: 10.1016/0378-1119(92)90551-y. [DOI] [PubMed] [Google Scholar]
- 9.Edwards C. Isolation, properties and potential applications of thermophilic actinomycetes. Appl Biochem Biotechnol. 1992;42:161–79. [Google Scholar]
- 10.Demain A. Cambridge: Cambridge University Press; 1995. Why do microorganisms produce antimicrobial? Proceeding of symposium on society of General Microbiology; pp. 205–28. [Google Scholar]
- 11.Xu LH, Jiang Y, Li WJ, Wen ML, Li MG, Jiang CL. Streptomyces roseoalbus sp. nov., an Actinomycete isolated from soil in Yunnan, China. Antonie Van Leeuwenhoek. 2005;87:189–94. doi: 10.1007/s10482-004-3720-y. [DOI] [PubMed] [Google Scholar]
- 12.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by genus Streptomyces? Arch Microbiol. 2001;176:386–90. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 13.Takizawa M, Colwell RR, Hill RT. Isolation and diversity of actinomycetes in the chasapeake bay. Appl Environ Microbiol. 1993;59:997–1002. doi: 10.1128/aem.59.4.997-1002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saadoun I, Hameed KM, Moussauui A. Characterization and analysis of antibiotic activity of some aquatic actinomycetes. Microbios. 1999;99:173–9. [PubMed] [Google Scholar]
- 15.Rahman MA, Islam MZ, Islam MA. Antibacterial activities of Actinomycete isolates collected from soils of Rajshahi, Bangladesh. Biotechnol Res Int. 2011;2011:857925. doi: 10.4061/2011/857925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pine L, Watson SJ. Evaluation of an isolation and maintenance medium for Actinomyces species and related organisms. J Lab Clin Med. 1959;54:107–14. [PubMed] [Google Scholar]
- 17.Waksman SA. Vol. 2. Baltimore: The Williams and Wilkins Company; 1961. The Actinomycetes, classification, identification and description of genera and species; pp. 61–292. [Google Scholar]
- 18.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, et al. Norwich, UK: The John lnnes Foundation; 1985. Genetic Manipulation of Streptomyces. A Laboratory Manual. [Google Scholar]
- 19.Pepper IL, Garba CP. Vol. 2. San Diego, CA: Elsevier Acadamic Press; 2004. Environmental Microbiology; pp. 37–49. [Google Scholar]
- 20.Nonomura H. Key for classification and identification of 458 species of the Streptomycetes included in ISP. J Ferment Technol (formerly known) 1974;52:78–92. [Google Scholar]
- 21.Holt JG. UK: Cambridge University Press; 1989. p. 4. [Google Scholar]
- 22.Egorov NS. Antibiotics a Scientific Approach. Moscow: Mir Publishers; 1985. Antibiotic properties of microorganism cultivated in the Laboratory; pp. 170–7. [Google Scholar]
- 23.Romankova AG, Zurabova ER, Fursenko MV, Sukharevich VI, Pronina MI. Selection of strains of some antibiotic producing Actinomycetes during repeated passages in submerged cultures. Antibiotiki. 1971;16:579–83. [PubMed] [Google Scholar]
- 24.Lakshmipatipathy D, Krishnan K. Antibacterial and antifungal activity of Streptomyces sp. VITDDK3 isolated from ennore coast, Tamil Nadu, India. J Pharm Res Health Care. 2010;2:186–96. (New name of journal is asian journal of pharmaceutical research and health care) [Google Scholar]
- 25.Raja A, Prabakaran P. Preliminary screening of antimycobacterial effect of psychrophillic actinomycetes isolated from Manali ice point Himachal Predesh. J Microbiol Antimicrob. 2011;3:41–6. [Google Scholar]
- 26.Selvameenal L, Radhakrishnan M, Balagurunathan R. Antibiotic pigment from desert soil Actinomycetes; biological activity, purification and chemical screening. Indian J Pharm Sci. 2009;71:499–504. doi: 10.4103/0250-474X.58174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S, Kumar P, Gopalan N, Shrivastava B, Kuhad RC, Chaudhary HS. Isolation and partial characterization of actinomycetes with antimicrobial activity against multidrug resistant bacteria. Asian Pac J Trop Biomed. 2012:S1147–50. [Google Scholar]


