Abstract
Background
There is mounting evidence for an age-dependent accumulation of somatic mutations as a result of the inherent imperfection of DNA replication and repair. A possible age-related decline in genome maintenance systems may exacerbate this age-related loss of genome integrity. A review of the current methods of mutation detection is timely in view of the lack of insight as to the magnitude of somatic mutation accumulation, the types of mutations that accumulate, and their functional consequences.
Objective
In this paper we review the current methods for measuring genome instability in organisms during aging or in relation to life span.
Methods
The review is based on established and novel concepts from the existing literature, with some examples from our own laboratory.
Results
Studies using cytogenetic assays and endogenous or transgenic mutation reporter assays provide strong evidence for age-related increases of different types of mutations in animals and humans during aging. This increase in DNA mutations is tissue-specific and also differs between species.
Conclusion
Today, our knowledge of somatic mutation profiles in aging is mainly derived from cytogenetics and the use of endogenous and transgenic mutation reporter assays. The emergence of new approaches, most notably massively parallel sequencing, will give us deeper insight into the nature of spontaneous genome instability and its possible causal relationship to aging and age-related disease.
Key Words: Somatic DNA mutation, LacZ mutation reporter assay, Genome rearrangements, Chromosomal aberrations, Aneuploidy, Next-generation sequencing
Introduction
Aging can be defined as the gradual decline of physical and mental fitness, ultimately resulting in disease and death. Instability of the genome in somatic cells has long been implicated as a causal factor in the cellular death and degeneration that underlies aging. Genome instability is driven by the incessant bombardment of genomic DNA by chemical, physical or biological insults, resulting in tens of thousands of chemical lesions per cell per day. DNA damage can result from endogenous processes, such as hydrolysis, oxidation and alkylation, or exposure to radiation or environmental mutagens [1].
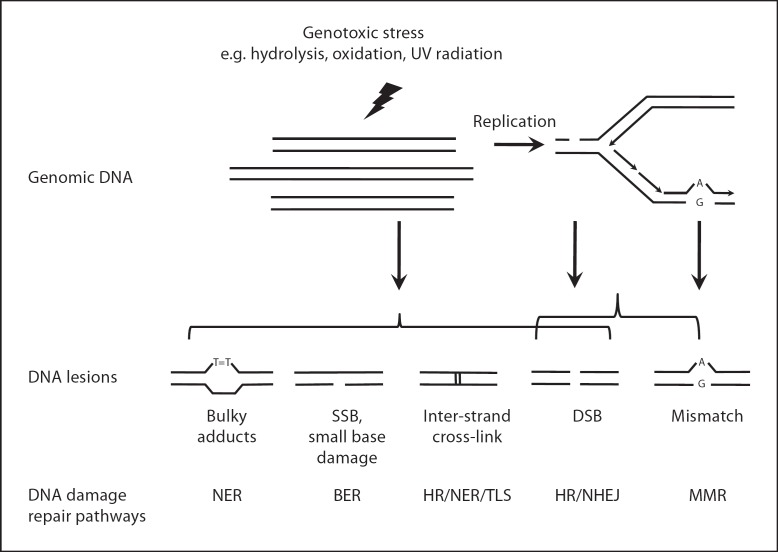
Despite the high spontaneous DNA damage rate, the conversion of DNA lesions into DNA mutations, i.e. irreversible, heritable changes in DNA sequence information, occurs at a very low frequency due to a sophisticated network of genome maintenance systems. Most DNA lesions are therefore rapidly corrected (fig. 1) [1]. For example, bulky lesions that cause DNA helix distortions are removed efficiently by nucleotide excision repair (NER). Base excision repair (BER), responsible for the repair of small base damages and single-strand breaks, is critically important for the organism as underscored by the embryonic lethality of inactivating one of its core enzymes. Mismatch repair pathways are active during replication, when they can correct nucleotide mis-incorporations and insertion/deletion loops using an excision mechanism capable of distinguishing the newly synthesized strand from the parental strand. Double-strand breaks (DSBs) are repaired by two major mechanisms, homologous recombination (HR) and non-homologous end-joining (NHEJ). HR is highly accurate because it is mainly active during the S/G2 phase when it can use the sister chromatid as template. Unlike the high fidelity HR repair, NHEJ, mainly active during G1 phase, re-anneals two broken ends in the absence of a homologous template. This can result in small deletions or (in case of multiple DSBs) chromosomal translocations. In non-dividing somatic cells, NHEJ appears to be the major pathway for DSB repair. DNA intra- or inter-strand cross-links are very toxic forms of DNA damage, which can be removed by HR-based pathways in synchrony with other pathways, such as NER as well as translesion synthesis [2].
Fig. 1.
Schematic depiction of the main types of DNA damage and corresponding repair pathways. DSB = double-strand break; NER = nucleotide excision repair; BER = base excision repair; HR = homologous recombination; NHEJ = non-homologous end-joining; MMR = mismatch repair; SSB = single-strand break; TLS = translesion synthesis.
When a cell harbors such severe DNA damage that it is beyond repair, it is disposed of through apoptosis. Alternatively, DNA damage can induce cellular senescence, the irreversible cessation of mitosis. Both processes are critically dependent on p53, which is known as the guardian of the genome [3]. DNA damage may also trigger autophagy, a cellular catabolic process that maintains homeostasis [4]. It should be noted that under normal conditions cells are rarely exposed to very high doses of DNA-damaging agents, which may be the explanation why we do not age and die because we run out of cells. However, aging is associated with some atrophy [1] and it is conceivable that at older ages bursts of DNA damage, for example from free radical reactions associated with inflammation, do occur and give rise to an increasingly high rate of apoptosis or cellular senescence. While there is some evidence for increased apoptosis and cellular senescence at old age, it is doubtful that under normal conditions this would lead to a significant loss of functional cells.
Although continuous action of genome maintenance systems will keep spontaneous mutation frequency at a low level, the inevitable errors will not allow them to suppress genome instability to zero. Consequently, mutations will accumulate in the somatic genome. Age-related decline in genome maintenance may exacerbate genome instability. Several lines of evidence show that some DNA damage repair functions may become compromised and more error-prone with aging [5]. Significant decline of BER activities with aging have been reported, mainly due to the reduced activity of a variety of glycosylases and declining DNA polymerase β activity. In addition, NHEJ activities may also decline with aging. In post-mitotic somatic cells, NHEJ appears to be very important for DSB repair, since knockout of some key NHEJ genes, such as Ku70/80, causes accelerated aging [6]. Spontaneous DSBs accumulate with age in various mouse tissues [7]. Since multiple G1-phase DSBs tend to juxtapose closely [8], template-independent mis-ligation via NHEJ may promote genome rearrangements, such as chromosomal translocations [9]. Indeed, the frequency of genome rearrangements increases exponentially at old age [10]. It is also possible that cellular responses to DNA damage, such as apoptosis, are less efficient at old age. For example, when exposed to the alkylating agent methyl methanesulfonate, a remarkable decline of apoptotic response was observed in the liver of old rats in comparison with young rats [11]. This would be expected to result in an increased fraction of seriously damaged cells in tissues and organs at old age.
Thus, in youth the total repair capacity may far exceed the DNA damage rate, but this ratio is likely to decline with age making it more likely that DNA damage persist and accumulate with age. However, possibly more seriously, while DNA damage can be repaired, the errors that occasionally occur due to mistakes by the various repair pathways lead to mutations that are irreversible. Once the original template is lost the cell has no way of reverting back to the original situation. The accumulation of mutations can adversely affect cell function and lead to disease. We know that DNA mutations are a cause of cancer, a major age-related disease. However, it is not known if cellular mutation loads in aged tissues ever become high enough to cause cellular dysfunction other than those that cause cancer by clonal outgrowth. Hence, it is critically important to accurately determine DNA mutation frequency and spectrum in tissues during aging.
Because mutations are relatively rare events, with an estimated frequency as low as 10−8 to 10−10 per base [12], their detection and characterization in animals or humans is not easy. In this mini-review, we will compare some common methods used in mutation analysis, along with a brief summary of some important findings. The space limitation disallows us to comprehensively review all mutation analysis methods. Rather, a few representative methods are selected and discussed (table 1). Here we will only focus on mutations of nuclear DNA. Mutations in mitochondrial DNA and epimutations are beyond the scope of this mini-review. We will begin with the type of changes that can be detected almost since Theodor Boveri [13] proposed, in 1902, that cancers start out as a single cell in which chromosomal integrity is scrambled: chromosomal aneuploidy.
Table 1.
Comparison of various mutation analysis methods
| Method | Sample type | Reporter name | Reporter feature | Selective bias | Mutation detection | Mutation spectrum | Mutation scope |
|---|---|---|---|---|---|---|---|
| Endogenous MRA | proliferating cells | HPRT | X-linked, single allelic gene | yes | metabolic screen | PM, GR | single locus |
| proliferating cells | APRT | autosomal artificial single allele | yes | metabolic screen | PM, GR, LOH | single locus | |
| Transgenic MRA | proliferating cells post-mitotic cells | LacZ/LacI | exogenous genes integrated into host chromosomes | neutral | metabolic screen | PM, GR | single locus |
| PLAP assay | proliferating cells post-mitotic cells | G11 tract | exogenous genes integrated into host chromosomes | N/A | IHC | PM | MNR gene(s) |
| RMC | proliferating cells post-mitotic cells | TaqI site | endogenous TaqI sites | neutral | RE resistance | PM | genome wide |
| PUN assay | proliferating cells in embryos | pun locus | endogenous tandem duplication | N/A | eye-spot assay fur-spot assay | DNA deletion | single locus |
APRT = Adenine phosphoribosyltransferase; G11 = 11 continual G:C pairs; GR = genome rearrangement; IHC = immunohistochemistry; LOH = loss of heterozygosity; MNR = mononucleotide repeat; MRA = mutation reporter assay; PM = point mutation; RE = restriction enzyme; pun = pink-eyed unstable locus; RMC = random mutation capture.
Measuring Mutations at the Chromosomal Level: Aneuploidy
In the early 1960s, age-dependent aneuploidy, i.e. the loss or gain of whole chromosomes resulting in an abnormal numerical karyotype, was first described for human lymphocytes [14]. However, at the time only few cell types were amenable to analysis and methodological limitations precluded a systematic, quantitative analysis. Originally, aneuploidy could only be detected by cytogenetic banding analysis of stained metaphase chromosomes with a variety of dyes such as Giemsa or quinacrine. Unfortunately, conventional cytogenetic banding analysis is very labor-intensive and initial sample sizes were small, precluding more detailed analysis. Hence, while a high frequency of aneuploidy in tumor cells was rapidly accepted [15], the lack of large-scale data on a wide variety of normal cell types in aging precluded any conclusions on the situation in normal tissues at young or old age. In the 1980s, the invention of FISH (fluorescent in situ hybridization) analysis, which conjugates chromosome-specific DNA probes with fluorochromes, ushered in the era of molecular cytogenetics and remarkably improved the efficiency of cytogenetic analysis. Modification of FISH, such as computer-assisted spectral karyotyping, also known as chromosome painting or multiplex FISH (M-FISH), now makes it possible to examine the entire karyotype of a human or mouse genome simultaneously [16]. Furthermore, interphase FISH was introduced to examine the karyotype of not only dividing cells but also non-dividing cells, which for the first time allowed the study of gross chromosomal changes in post-mitotic tissues.
Using the more advanced FISH-based methods, a dramatically more severe picture of aneuploidy levels was obtained. For example, up to 15–20% of aged human oocytes have chromosomal abnormalities, mainly aneuploidy [17]. In comparison, paternal age only causes a modest increase in the frequency of sex chromosomal aneuploidy in sperm cells [18]. Interestingly, this is the other way around for small DNA mutations, such as basepair substitutions. Virtually all genetic diseases based on point mutations are inherited from the father, most likely because such small mutations can arise through replication errors and sperm cells undergo many more rounds of replication than oocytes [19]. Indeed, the so-called ‘paternal age effect’, as observed first by Weinberg in achondroplasia, indicates that the high incidence of sporadic genetic diseases found among the youngest children in a family may reflect accelerating mutagenesis in sperms as men age [19].
High levels of aneuploidy were also observed in normal tissues, such as liver and brain [20], and in buccal mucosa cells aneuploidy frequency was found to be increased in aged people and individuals with Alzheimer's disease [21]. Of note, recent evidence obtained with yeast indicates that aneuploidy in that organisms leads to an increased frequency of point mutations, mitotic recombination and aneuploidy itself [22].
Age-related polyploidization for certain tissues, such as the liver in humans and rodents, has been well documented [23]. The phenomenon has been interpreted as the creation of redundant genetic information to make the cell more resistant to mutation. By contrast, aneuploidy is deemed to cause gene dosage imbalances and reduced fitness. However, the high prevalence of aneuploidy in somatic tissues, such as brain, raises the question of whether aneuploidy has any selective advantage. In liver cells, the intermixed euploid, polyploid and aneuploid hepatocytes could provide a pool of functional diversity, best fit for adapting to severe environmental stress, such as the pressure of liver repopulation after hepatectomy [23]. In this regard, it is analogous to the versatility of a pool of aneuploid tumor cells to survive and thrive under various conditions. Using the host of novel cytogenetic methods of today, we may gain deeper insight into the nature of somatic aneuploidy and its relevance for aging in the near future.
Measuring Small Mutations
Mutational events at sub-chromosomal levels are no longer directly detectable at the microscopic level. The lack of methods to directly assess mutation loads in a genome-wide manner has thus far prevented us from obtaining more detailed insight into the landscape of somatic mutations in aging tissues. Direct assays for detecting DNA sequence variation in DNA from tissues are usually not capable of detecting unknown mutations present in only a small fraction of cells. Bielas and Loeb [12] developed an assay capable of selectively enriching a DNA sample for fragments containing a mutated restriction site. This method, termed random mutation capture, is capable of detecting mutations at a frequency of 1 in 108 basepairs [12]. However, a major disadvantage is its very small target size, i.e. the 4 basepairs of the TaqI restriction site (TCGA) distributed over the whole genome. This comprises only about 0.4% of the whole genome.
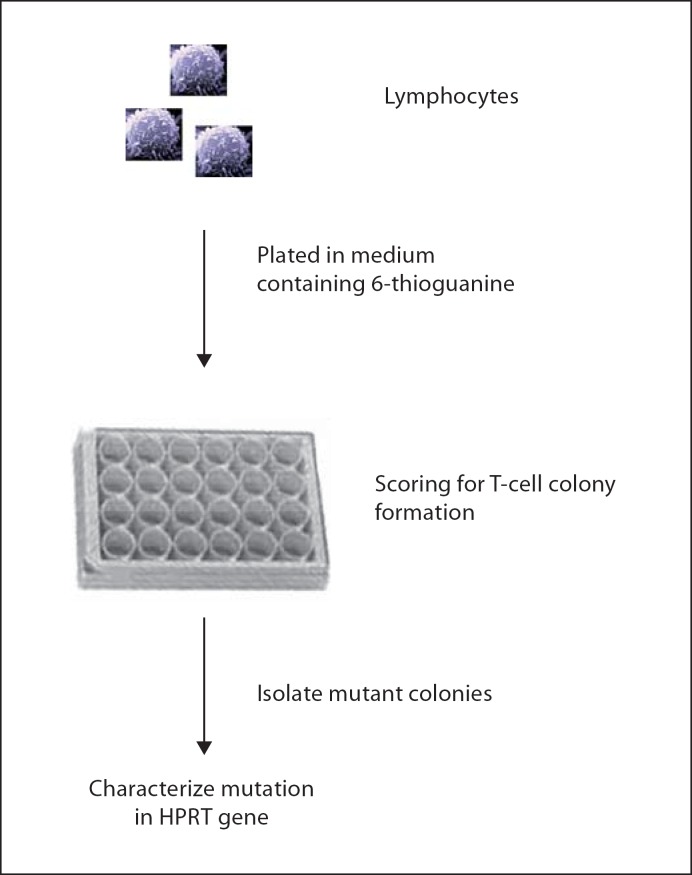
To detect all inactivating mutations in an entire gene, a selection-based system is the most straightforward solution, and many such assays have been designed, including those using a number of endogenous selectable genes. Successful mutation analyses were first achieved in some proliferating human cells cultured in vitro. One classic selectable marker is the hypoxanthine phosphoribosyltransferase (HPRT) gene, identified as the culprit in the X-linked Lesch-Nyhan syndrome [24]. In both human and mouse, the functional HPRT gene is a single-copy gene located on the active X chromosome. This is essential for its use as a mutational reporter. Indeed, inactivation of the only copy of a functional HPRT gene in a cell is sufficient to be detected by a metabolic screen. HPRT catalyzes purine biosynthesis from hypoxanthine and guanine, but also accounts for the metabolic activation of pro-toxins, such as the purine analogue 6-thioguanine. Based on these biochemical properties, a metabolic screening method was devised to positively select HPRT-deficient mutants based on their resistance to 6-thioguanine, which is lethal to the HPRT-proficient wild-type cells. The mutation frequency is the ratio of the number of clones in the presence of 6-thioguanine versus the total number of cells plated, normalized over the plating efficiency. Furthermore, different types of mutations, such as point mutations and genome rearrangements, can be characterized by the mutation spectral analysis based on DNA sequencing, Southern blotting or PCR [24] (fig. 2).
Fig. 2.
HPRT assay. Mitotically active cells (fibroblasts, mononuclear cells from blood or spleen) are counted and plated in 6-thioguanine-containing medium. After a period of incubation the wells are inspected for growing colonies. Visible colonies are considered to be mutants. The mutant frequency is the ratio between the cloning efficiency in selective medium and in non-selective medium. HPRT-deficient cells are resistant to 6-thioguanine toxicity and positively selected. Each colony can be analyzed for the spectrum of mutations [for details, see 24].
Using the HPRT assay on T lymphocytes revealed an age-dependent increase in mutation frequency from 0.3 × 10−5 to 2.5 × 10−5[25]. In cultured human kidney tubular epithelial cells, mutation frequency was found to increase with aging from 0.5 × 10−4 to 2.5 × 10−4[26]. It is very interesting to notice that the mutation frequency measured from solid tissue cells, such as kidney cells [26], is one order of magnitude higher than in blood lymphocytes. This tissue-specific difference may reflect the selective elimination of HPRT mutant cells from blood. Indeed, while obviously not lethal, inactivation of HPRT may lead to a selective disadvantage of the cell. This is the reason why mutation reporter genes should preferably be neutral.
The HPRT assay has served as a prototype for the development of other new methods of mutation analysis based on endogenous reporter genes. One example is the APRT (adenine phosphoribosyltransferase) gene, which like HPRT plays a role in purine biosynthesis from adenine. Like the HPRT assay, APRT mutations can also be detected by metabolic screening. However, unlike HPRT, APRT is located on chromosome 16 in humans and on chromosome 8 in the mouse. Since the autosomal gene does not have the convenience of a single-copy gene as X-linked HPRT, one copy of the APRT gene needed to be knocked out in the mouse [27]. The heterozygotes, containing one disabled and one wild-type Aprt allele in each cell, allow the detection of mutations at this locus in a similar way as in the HPRT assay. However, while the second Aprt copy is dysfunctional, it is still physically present. Therefore, in contrast to HPRT, complete loss of the aprt function can now also result from mitotic recombination allowing the detection of loss of heterozygosity (LOH) events, a major type of spontaneous mutation [27].
In B6D2F1 mice, heterozygous for Aprt, an age-dependent increase of the mutation frequency was observed in kidney epithelial cells and ear mesenchymal cells [28]. Using the APRT assay, the spontaneous LOH frequency was also found higher in solid tissues than in blood cells [27], reminiscent of the difference in mutation frequency between blood cells and solid tissue cells as detected by the HPRT assay.
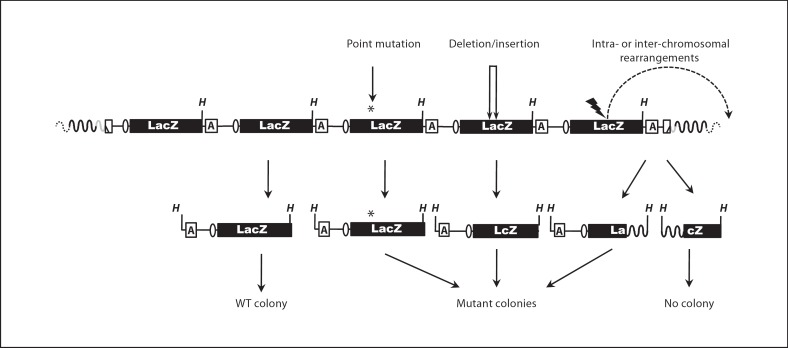
Although the HPRT approach paved the way for quantitative mutation frequency measurement and mutant characterization, a major limitation of this assay is its dependence on cells that can still proliferate in culture. This changed with the emergence of transgenic mice harboring bacterial reporter genes, such as LacZ or LacI, as part of a bacteriophage or plasmid vector that can be excised from chromosomal DNA and subsequently screened for mutations in a bacterial host [29,30,31] (fig. 3). More recently, this was extended to Drosophila melanogaster[32]. All these selectable markers are integrated into the host chromosomes where they remain unexpressed and therefore neutral and not subject to selection pressure.
Fig. 3.
Example of a transgenic mutation reporter assay. Multiple lacZ-containing plasmids (only five are shown) are integrated in a head-to-tail orientation at one or more chromosomal locations of C57BL/6 mice. The LacZ-plasmids can be retrieved from host genomic DNA (black waved lines) by HindIII (H) digestion and cloned in Escherichia coli. A variety of inactivating mutations, included point mutations (*), small deletions or insertions (doublearrowed lines) and intra- and inter-chromosomal rearrangements (dashed curve), can be detected by positive selection for inactivation of the lacZ gene [for details, see 30, 32]. Ellipse: origin of replication. A = Ampicillin resistance gene. Gray-colored curve: the 5′ end integration site of the LacZ reporter.
Like the HPRT assay, both mutation frequency and the types of mutations can be analyzed in a transgenic mutation reporter assay. Because there is no need for cell culture, this type of analysis is much easier than performing a HPRT assay. More importantly, post-mitotic cells, tissues and organs, which cannot be analyzed through the HPRT assay, are readily examinable by the transgenic mutation reporter assay. However, their disadvantage is that they cannot be used to study mutagenesis in humans.
Like the HPRT and APRT assays, the transgenic mutation reporter assays also revealed an age-dependent accumulation of somatic mutation loads. Using the lacZ-plasmid model, results from our laboratory indicated an age-related increase in mutation frequency in a variety of organs and tissues, including liver, brain, heart, small intestine, and spleen [10,33,34,35]. The rate of this increase, however, differed greatly from organ to organ. While in small intestine mutations accumulated rapidly to a very high level at old age [35], brain [10] and testes [36] proved almost resistant to mutation accumulation. Interestingly, in brain point mutations were found to accumulate rapidly in hypothalamus and hippocampus [34]. Since there is persistent neurogenesis in the hippocampus in adulthood [37], the observed accumulation of point mutations may be caused by replication errors.
Also the mutation spectra were very different from tissue to tissue. While most mutations found to accumulate in small intestine were point mutations [34,35], probably also a consequence of replication errors in this highly proliferative organ, mutations in heart [35] and liver [10] included genome rearrangements. Such rearrangements could involve large deletions, inversions and chromosomal translocations [38]. The spectra of point mutations, including various types of basepair substitutions and 1-bp deletions, were also very different from organ to organ [33] and appear to reflect the cell type or organ-specific function and proliferative history.
A lacZ-plasmid reporter model, similar to the mouse model, was constructed in D. melanogaster[32]. Interestingly, the results obtained when comparing the two models indicate a threefold higher spontaneous mutation frequency in flies than in mice [39]. Also the fraction of genome rearrangements in flies was found to be much larger than in mice [39]. In these assays the mutation frequency is determined per locus, as the number of mutated transgenes versus the total number of transgenes recovered from a DNA sample. Flies have a much smaller genome, which is also much more compact than that of mammals, with only 10% comprised of repeat elements as compared to 50% for mammals. Therefore, the question comes up how Drosophila cells can function with such a high mutation load and an increased likelihood for a mutation to affect a gene. The explanation may be attributed to the lack of long-distance regulatory interactions between genes in fly genome [39].
In agreement with the data obtained from transgenic mice, significantly higher somatic mutation loads were also found in old flies (4 weeks) in comparison with young flies (5 days) [39]. Tissue-specific mutation profiles were also observed, with the highest mutation frequency in the thorax, as compared with the head and abdomen. Since the thorax is mainly composed of flight muscles, metabolic DNA damage inflicted by free radicals is hypothesized as a putative cause [39].
Recently, some mutation reporter assays of novel designs have been reported. A human placental alkaline phosphatase (PLAP) gene was introduced into the mouse genome. In this transgenic PLAP mouse, the PLAP allele possesses a tract of 11 G:C basepairs (G11) which cause a frame-shifting inhibition of PLAP translation. The deletion or insertion of one or more basepair(s) can disrupt the G:C track, attenuating the inhibition and consequently restoring PLAP expression [40]. The advantage of the PLAP assay is that the mutant cells can be directly detected in situ using histochemical examination. Age-dependent accumulation of somatic mutations was also observed in PLAP mice from 1 week to 24 months old in heart, kidney and brain [40]. An elevated mutation rate was observed in proliferating cells such as kidney epithelial cells, as well as in post-mitotic cells such as cardiomyocytes and neurons [40]. A somewhat similar mouse model, also capable of visualizing mutations directly in tissue, is the so-called PUN assay [41]. This assay measures HR between two 70-kb tandem repeats at the pink-eyed unstable locus, resulting in a deletion of one of the repeats. The deletion is visualized as a black-pigmented cell or a clone thereof on the unpigmented retinal pigment epithelium of the eye.
Conclusions and Future Perspectives
While we have learned a lot since Boveri, the current rich palette of assays to detect and characterize somatic mutations in organs, tissues and cells of animals during aging is still insufficient to draw major conclusions about a possible causal role of genome instability in aging and disease, apart from cancer.
As we have seen, the great improvements made in cytogenetic analysis have now led to the realization that large-scale chromosomal aberration, most notably aneuploidy, are much more common (with a cumulative frequency at 10−3−10−1 aneuploidy per cell) in normal somatic cells than we previously expected. Furthermore, since the resolution of these assays is very low, these studies may show us only the very tip of what could be an iceberg. Indeed, the development of reporter-based assays, including HPRT and APRT, but also the transgenic mice and flies with their neutral reporters that can be recovered from any organ or tissue, suggest the picture of a genome undergoing frequent changes during aging of a tissue. However, in spite of a wealth of data obtained with these methods, definite conclusions of a causal role of somatic mutations in aging cannot be drawn yet.
We are confronted with at least three questions that need to be addressed in the near future. First, we need to expand the assay systems discussed above to the epigenome, i.e. the structural information that controls gene expression but does not reside in the primary DNA sequence. The epigenome includes DNA methylation and histone protein modifications. During DNA replication or repair the layers of epigenomic information are removed and must afterwards be re-installed. Like copying DNA sequence information, also the restoration of epigenomic information is imperfect. Hence, ‘epimutations’ occur and most likely accumulate with age. In the absence of assays that can detect low-abundance epigenomic alterations, we are ignorant about the potential functional impact of such changes.
Second, we need to move away from reporter assays and develop tools that allow us to directly detect mutations everywhere in the genome. The mutation reporter assays are single-locus assays, which may not be representative for the genome overall. Therefore, precautions must be taken when calculating the genome-wide mutation rate by extrapolation. The advent of massively parallel sequencing (MPS) or next-generation sequencing may provide a solution to these seemingly insurmountable disadvantages of conventional mutation assay methods.
Thus far, the high cost of sequencing essentially constrained widespread application in mutation analysis. However, with the emergence of MPS whole genomes can now be sequenced at a fraction of the costs associated with the original Sanger sequencing technology that gave us the first complete draft of the human genome sequence. Soon, the costs of sequencing a whole mammalian genome will be less than USD 1,000.
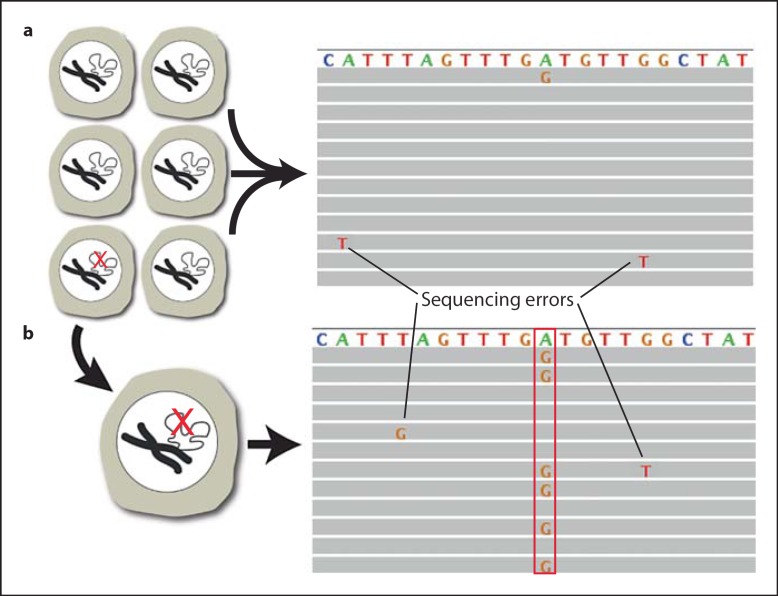
However, current MPS protocols for mutation detection are based on consensus, i.e. finding the same event in multiple, independent reads from the same section of the genome. This allows the detection of clonally amplified mutations present, for example in most or all cells of a tumor. But it will not reveal low-abundant mutations present in one or few cells in a tissue. In principle, one could sequence a particular segment of the genome tens of thousands of times to discover variants due to rare mutations. Unfortunately, the error rate of current MPS systems precludes distinguishing real mutations from sequencing errors (fig. 4a). Therefore, current MPS systems are unsuitable for the direct analysis of a normal tissue sample for randomly occurring mutations, for example during aging or after genotoxic exposure.
Fig. 4.
Single-cell mutation analysis by massively parallel sequencing. a The A→G mutation of interest (red check mark in one of the cells in the whole tissue) cannot be distinguished from the sequencing errors. b When sequencing the genome of the single cell containing the mutation (red check mark in the single cell) after whole genome amplification, the A→G mutation can now be distinguished from the sequencing errors because it shows up in approximately 50% of the reads (box), corresponding to one allele [42].
There are two ways to circumvent this problem and account for the mutational heterogeneity within normal organs and tissues during aging. First, new filtering tools can be developed to distinguish sequencing errors from genuine mutations. This will be greatly facilitated by some of the new, single-molecule sequencing systems [43] that can greatly increase accuracy and will be able to directly assess mutational diversity, provided sequencing is conducted to a very great depth. Alternatively, it is possible to sequence entire genomes of a representative number of single cells. In the single-cell approach, the rare but real somatic mutations will be consistently amplified whereas the randomly occurring sequencing errors will be filtered out (fig. 4b).
Finally, the third and to some extent the most important question is how we can test if increased random mutation loads can give rise to the functional impairment of organs and tissues as observed during aging. To address this question it will be necessary to develop new model systems that will allow us to measure phenotypic effects of random, low-abundant mutations on a cell population. Mathematical modeling should then be applied to gain information as to how random mutations can affect functional pathways and gene networks, how they interact and how they can synergistically give rise to an age-related decline of function and an increase in disease risk.
Acknowledgements
We sincerely thank Alex Maslov and Anastasia Bendebury for critically reading the text. This work was supported by the National Institute of Health (grants R01AG034421, P01AG17242, and R21ES019520). Funding for open access charge: R01AG034421.
References
- 1.Vijg J. Aging of the Genome. New York: Oxford University Press; 2007. [Google Scholar]
- 2.Ho TV, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 3.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 4.Vicencio JM, Galluzzi L, Tajeddine N, Ortiz C, Criollo A, Tasdemir E, Morselli E, Ben Younes A, Maiuri MC, Lavandero S, Kroemer G. Senescence, apoptosis or autophagy? When a damaged cell must decide its path – a mini-review. Gerontology. 2008;54:92–99. doi: 10.1159/000129697. [DOI] [PubMed] [Google Scholar]
- 5.Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 8.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 9.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 10.Dollé ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- 11.Suh Y, Lee KA, Kim WH, Han BG, Vijg J, Park SC. Aging alters the apoptotic response to genotoxic stress. Nat Med. 2002;8:3–4. doi: 10.1038/nm0102-3. [DOI] [PubMed] [Google Scholar]
- 12.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 13.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci. 2008;121(suppl 1):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs PA, Court Brown WM, Doll R. Distribution of human chromosome counts in relation to age. Nature. 1961;191:1178–1180. doi: 10.1038/1911178a0. [DOI] [PubMed] [Google Scholar]
- 15.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 17.Pellestor F, Anahory T, Hamamah S. Effect of maternal age on the frequency of cytogenetic abnormalities in human oocytes. Cytogenet Genome Res. 2005;111:206–212. doi: 10.1159/000086891. [DOI] [PubMed] [Google Scholar]
- 18.Sloter E, Nath J, Eskenazi B, Wyrobek AJ. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil Steril. 2004;81:925–943. doi: 10.1016/j.fertnstert.2003.07.043. [DOI] [PubMed] [Google Scholar]
- 19.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 20.Faggioli F, Vijg J, Montagna C. Chromosomal aneuploidy in the aging brain. Mech Ageing Dev. 2011;132:429–436. doi: 10.1016/j.mad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas P, Fenech M. Chromosome 17 and 21 aneuploidy in buccal cells is increased with ageing and in Alzheimer's disease. Mutagenesis. 2008;23:57–65. doi: 10.1093/mutage/gem044. [DOI] [PubMed] [Google Scholar]
- 22.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albertini RJ, Nicklas JA, O’Neill JP, Robison SH. In vivo somatic mutations in humans: measurement and analysis. Annu Rev Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 25.Cole J, Skopek TR. International Commission for Protection Against Environmental Mutagens and Carcinogens. Working paper No. 3. Somatic mutant frequency, mutation rates and mutational spectra in the human population in vivo. Mutat Res. 1994;304:33–105. doi: 10.1016/0027-5107(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 26.Martin GM, Ogburn CE, Colgin LM, Gown AM, Edland SD, Monnat RJ., Jr Somatic mutations are frequent and increase with age in human kidney epithelial cells. Hum Mol Genet. 1996;5:215–221. doi: 10.1093/hmg/5.2.215. [DOI] [PubMed] [Google Scholar]
- 27.Van Sloun PP, Wijnhoven SW, Kool HJ, Slater R, Weeda G, van Zeeland AA, Lohman PH, Vrieling H. Determination of spontaneous loss of heterozygosity mutations in Aprt heterozygous mice. Nucleic Acids Res. 1998;26:4888–4894. doi: 10.1093/nar/26.21.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turker MS, Lasarev M, Connolly L, Kasameyer E, Roessler D. Age-related accumulation of autosomal mutations in solid tissues of the mouse is gender and cell type specific. Aging Cell. 2007;6:73–86. doi: 10.1111/j.1474-9726.2006.00264.x. [DOI] [PubMed] [Google Scholar]
- 29.Gossen JA, de Leeuw WJ, Tan CH, Zwarthoff EC, Berends F, Lohman PH, Knook DL, Vijg J. Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proc Natl Acad Sci USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boerrigter ME, Dollé ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 31.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia AM, Derventzi A, Busuttil R, Calder RB, Perez E, Jr, Chadwell L, Dollé ME, Lundell M, Vijg J. A model system for analyzing somatic mutations in Drosophila melanogaster. Nat Methods. 2007;4:401–403. doi: 10.1038/NMETH1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dollé ME, Snyder WK, Dunson DB, Vijg J. Mutational fingerprints of aging. Nucleic Acids Res. 2002;30:545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busuttil RA, Garcia AM, Reddick RL, Dollé ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS One. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dollé ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc Natl Acad Sci USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin SL, Hopkins CL, Naumer A, Dollé ME, Vijg J. Mutation frequency and type during ageing in mouse seminiferous tubules. Mech Ageing Dev. 2001;122:1321–1331. doi: 10.1016/s0047-6374(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 37.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 38.Dollé ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002;12:1732–1738. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia AM, Calder RB, Dollé ME, Lundell M, Kapahi P, Vijg J. Age- and temperature-dependent somatic mutation accumulation in Drosophila melanogaster. PLoS Genet. 2010;6:e1000950. doi: 10.1371/journal.pgen.1000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer JM, Stringer JR. Mutation in aging mice occurs in diverse cell types that proliferate postmutation. Aging Cell. 2008;7:667–680. doi: 10.1111/j.1474-9726.2008.00416.x. [DOI] [PubMed] [Google Scholar]
- 41.Reliene R, Bishop AJ, Aubrecht J, Schiestl RH. In vivo DNA deletion assay to detect environmental and genetic predisposition to cancer. Methods Mol Biol. 2004;262:125–139. doi: 10.1385/1-59259-761-0:125. [DOI] [PubMed] [Google Scholar]
- 42.Gundry M, Li W, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic Acids Res 2011, DOI: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed]
- 43.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]