Abstract
Background
Cantu syndrome is a rare condition which is characterized clinically by hypertrichosis, cardiomegaly and bone abnormalities. Inherited hypertrichoses are very rare human disorders whose incidence has been estimated as low as 1 in 1 billion. The genetic basis of hypertrichosis is largely unknown, and currently no single gene has been directly implicated in its pathogenesis, although position effects have been reported.
Methods
We analyzed the DNA of a patient with Cantu syndrome on the Affymetrix Cytogenetics Whole-Genome 2.7M array for copy number variations (CNVs). We then performed genomic copy number quantification using qPCR, and finally we performed gene expression analysis in the hair follicle for the genes lying within and around the region of the duplication.
Results
We identified a 375 kb duplication on chromosome 4q26–27. The duplication region encompassed three genes, which included MYOZ2, USP53 and FABP2. MYOZ2 and USP53 are known to be highly expressed in the cardiac muscle, and we found that USP53 is expressed in the hair follicle.
Conclusion
We propose that CNVs involving chromosome 4q26–27 may be associated with Cantu syndrome. CNVs spanning several genes may help define the molecular basis of syndromes which have unrelated clinical features.
Key Words: Cantu syndrome, Copy number variations, Hypertrichosis, Position effects
Introduction
Inherited hypertrichoses are very rare human disorders whose incidence as a group has been estimated to be as low as 1 in 1 billion [1]. Very few human syndromes have hypertrichosis as one of the main features. These syndromes include hypertrichosis universalis congenita (MIM145700), Ambras syndrome (MIM145701), gingival fibromatosis with hypertrichosis (MIM135400), Barber-Say syndrome (MIM209885), gingival fibromatosis with hypertrichosis and mental retardation (MIM605400), X-linked hypertrichosis (MIM307150), acromegaly with hypertrichosis and Cantu syndrome (MIM239850) [2]. The genetic causes of congenital hypertrichosis are largely unknown, and to date no single gene has been associated with or mutated in hypertrichosis. Therefore, several alternate genetic mechanisms, including epigenetic dysregulation, miRNA dysregulation, chromosomal translocations with position effects and copy number variations (CNVs) may play a role in the pathogenesis of hypertrichosis. Over the past two decades, there have been several reports of hypertrichosis in association with chromosomal rearrangements [3,4,5,6,7,8]. Moreover, we have previously described several patients with Ambras syndrome, secondary to position effects on TRPS1 gene [7] and recently, it has been reported that CNVs on chromosome 17 are associated with isolated hypertrichosis and hypertrichosis with gingival fibromatoses [8].
Cantu syndrome is a very rare genetic disorder of unknown etiology and is characterized clinically by congenital hypertrichosis, cardiomegaly and bone abnormalities [9]. The inheritance pattern is unclear, although autosomal dominant, autosomal recessive and sporadic occurrence have been reported [9,10,11,12]. Here, we present evidence for a spontaneous case of congenital hypertrichosis, cardiomegaly and coarse facies suggestive of Cantu syndrome resulting from a novel CNV on chromosome 4.
Methods
After obtaining informed consent, we collected peripheral blood samples from the affected individual. We performed a karyotype analysis to check for any chromosomal aberration. The chromosomes were prepared from a 72 h phytohemagglutinin-stimulated peripheral blood culture. Standard procedures for cultures, harvests, and slide preparation were modified and performed in our laboratory as previously described [13]. The chromosomes were analyzed with GTG banding, and the karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN 2009) [14].
We then analyzed the genomic DNA on the Affymetrix Cytogenetics Whole-Genome 2.7M array, which provides whole genome coverage with a high density of 2.7 million markers. Markers on this array are placed on average every 1 kb. The array includes 2.4 million non-polymorphic markers that detect CNVs. This array also includes 400,000 SNPs which enable the detection of loss of heterozygosity, uniparental disomy and regions of identity-by-descent. Coverage includes 99.1% of RefSeq, genes, 100% of cancer genes, 98.1% of X-linked genes and 99.2% of OMIM genes. Analysis of the data was then performed on the Affymetrix® Chromosome Analysis Suite. We performed genomic copy number quantification using qPCR in the duplicated region. Subsequently, we performed gene expression assays using semi-quantitative RT-PCR in the hair follicle for the genes lying within the region of the duplication and immunofluorescence. Frozen skin sections (8 mm thick) from a control were fixed in 4% paraformaldehyde for 15 min and incubated in blocking buffer (5% goat serum) for 1 h. Immunohistochemistry was performed in these sections using an anti-USP53 polyclonal antibody (Sigma-Aldrich) (1:100 dilution) raised in rabbit, followed by an Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Invitrogen) (1:800 dilution).
Results
The patient was an 18-year-old female born with generalized hypertrichosis more pronounced on the face and back associated with coarse facial features. Cardiac examination revealed a cardiac murmur. An echocardiogram was performed and revealed a patent ductus arteriosus with cardiomegaly. The patient was also found to have several bone abnormalities including bifid 5th rib, synostosis of the 6th and 7th ribs and scoliosis. She also had mild neurodevelopmental delay and severe obesity (BMI >40). The karyotype of the patient was normal 46XX. Using the Affymetrix Cytogenetics Whole-Genome 2.7M array, we detected a novel 375 kb duplication on chromosome 4q26–27. The region of duplication encompassed the three genes MYOZ2, USP53 and FABP2 (fig. 1). The duplication occurred in a region not reported to harbor normal polymorphic CNVs.
Fig. 1.
Analysis of the patient's DNA on the Affymetrix Cytogenetics Whole-Genome 2.7M array revealed a 375 kb duplication (indicated by the large blue rectangle) on chromosome 4q26–27 overlying three genes completely, these genes being MYOZ2, USP53 and FABP2. PDE5A, a phosphodiesterase enzyme involved in cardiovascular system functioning, lies in the vicinity of the duplication.
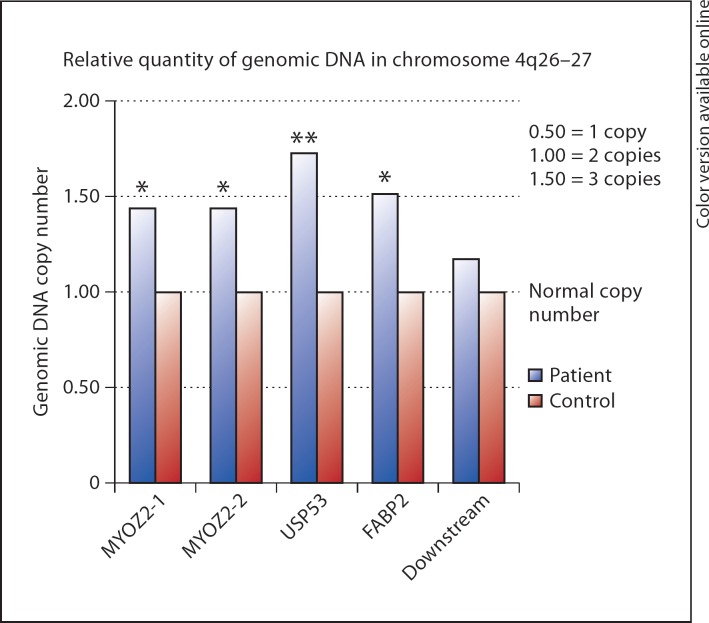
We next performed genomic PCR copy number for quantification, which confirmed that the duplication was present (fig. 2). We then performed gene expression studies for the three genes in the hair follicle and found that USP53 was also expressed in the hair follicle (fig. 3).
Fig. 2.
Semi-quantitative PCR performed for the genes within the region of duplication found on the Affymetrix Cytogenetics Whole-Genome 2.7M array confirmed that the duplication was real (MYOZ1 and MYOZ2 are within the MYOZ gene, USP53 is within the USP53 gene, FABP2 is within the FABP2 gene, and downstream lies within the region downstream of the duplication).
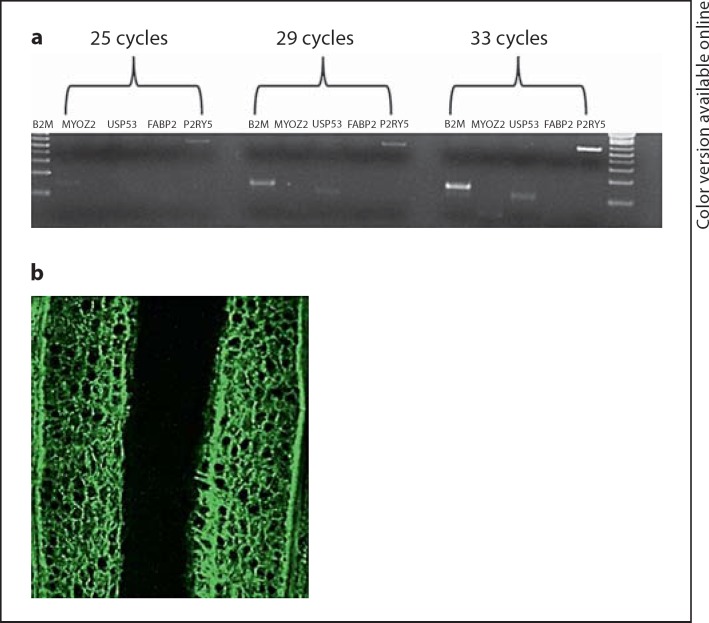
Fig. 3.
a Semi-quantitative RT-PCR done on hair follicles from normal individuals revealed that USP53 is expressed in the hair (B2M is a housekeeping gene and P2RY5 is a positive control). bUSP53 is mainly expressed in the outer root sheath of the hair follicle.
Discussion
Hypertrichosis is rarely reported as an isolated finding and is usually part of a more complex syndrome. Craniofacial malformations and cardiac defects [15,16,17] are frequently associated with congenital hypertrichosis. Thus, genes implicated in hypertrichosis may be clustered close to those involved in craniofacial and cardiac development, or alternatively, genes which are involved in cardiac and craniofacial formation might also be involved in hypertrichosis. The observation that many syndromes related to hypertrichosis with cardiac or craniofacial anomalies are associated with chromosomal aberrations and no definite gene mutations may suggest that mutations in these genes are lethal early in embryonic life, whereas chromosomal alterations affecting gene dosage or expression and CNVs may result in less severe phenotypes [7,15,16].
Contiguous gene syndromes are characterized by unrelated clinical features due to the deletion of a series of nearby genes. CNVs lead to similar syndromes due to differential expression of several genes lying in the vicinity of the variation. Here, we identified a 375 kb duplication which completely contained three genes. None of these genes have been previously reported to be associated with any human disease. Of note, MYOZ2 is expressed in cardiac muscle. Myozenins may serve as intracellular binding proteins involved in linking Z-disk proteins such as alpha-actinin [18]. MYOZ2–/– mice lead to the activation of a hypertrophic gene involved in cardiomegaly [19]. USP53 has also previously been shown to be expressed in the human cardiac muscle and most likely functions as a ubiquitin-specific protease, though its function is yet unclear. Interestingly, we found that USP53 is also expressed in the hair follicle. These data might suggest the presence of a contiguous gene syndrome in our case, where the duplication overlying several genes could have resulted in differential expression of these genes, resulting in cardiac abnormality and hypertrichosis.
Recently, CNVs were shown to be associated with hypertrichosis and gingival fibromatoses on chromosome 17q24.2–24.3 [8]. Moreover, a role of CNVs has been shown in the pathogenesis of several diseases, including psoriasis, inflammatory bowel disease, chronic obstructive pulmonary disease, Charcot Marie Tooth disease and schizophrenia, to name a few [20,21]. Generally, CNVs of >100 kb are considered pathogenic. CNVs can result in disease through effecting gene dosage within the CNV or through differential gene expression as a result of a position effect.
We have previously found that Ambras syndrome is associated with a position effect on the TRPS1 gene on chromosome 8 [7]. A position effect is defined as an alteration in gene expression caused by a change in the position of a gene relative to its native chromosomal surroundings [22]. Gene expression can be affected by a variety of mechanisms, for example disruption of transcriptional regulation in cis and/or modification of the surrounding chromatin structure. The resulting phenotype may be attributed to separation of the gene from a nearby tissue- or temporal-specific modifier of gene expression, such as an enhancer or repressor element (Korean Ambras) [22,23].
Cantu syndrome is a condition which is defined according to phenotypic features since the molecular basis is not yet defined. The diagnosis of Cantu syndrome is made clinically, whenever patients present with abnormalities affecting the heart, the hair distribution pattern and the bone. Here, we presented a patient with hypertrichosis, cardiomegaly and coarse facies, in whom we found a duplication on chromosome 4q26–27. We suggest that abnormalities involving chromosome 4q26–27 and affecting several genes in the vicinity may be associated with additional cases of Cantu syndrome. Other regions in the genome might also be associated with Cantu syndrome, since additional regions may contain nearby genes which are implicated in hair morphogenesis and cardiac development. CNVs may be associated with several of the syndromes whose pathogenesis is yet unknown. Searching for CNVs may be useful in the setting of syndromes that contain several unrelated clinical features, especially in de novo cases and in the setting of unclear Mendelian inheritance patterns.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgements
We are grateful to the family members for their participation in this study. Supported in part by USPHS/NIH grant RO1AR44924 from NIAMS (to A.M.C.) and NIH Institutional Research Training Grant T32AR007605 (P.I. David Bickers, Postdoctoral Fellow, Department of Dermatology, Columbia University). The work was performed at Columbia University.
References
- 1.Macias-Flores MA, Garcia-Cruz D, Rivera H, Escobar-Luján M, Melendrez-Vega A, Rivas-Campos D, Rodríguez-Collazo F, Moreno-Arellano I, Cantú JM. A new form of hypertrichosis inherited as an X-linked dominant trait. Hum Genet. 1984;66:66–70. doi: 10.1007/BF00275189. [DOI] [PubMed] [Google Scholar]
- 2.Irvine AD, Dolan OM, Hadden DR, Stewart FJ, Bingham EA, Nevin NC. An autosomal dominant syndrome of acromegaloid facial appearance and generalised hypertrichosis terminalis. J Med Genet. 1996;33:972–974. doi: 10.1136/jmg.33.11.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balducci R, Toscano V, Tedeschi B, Mangiantini A, Toscano R, Galasso C, Cianfarani S, Boscherini B. A new case of Ambras syndrome associated with a paracentric inversion (8) (q12; q22) Clin Genet. 1998;53:466–468. doi: 10.1111/j.1399-0004.1998.tb02596.x. [DOI] [PubMed] [Google Scholar]
- 4.Baumeister FA, Egger J, Schildhauer MT, Stengel-Rutkowski S. Ambras syndrome: delineation of a unique hypertrichosis universalis congenita and association with a balanced pericentric inversion (8) (p11.2; q22) Clin Genet. 1993;44:121–128. doi: 10.1111/j.1399-0004.1993.tb03862.x. [DOI] [PubMed] [Google Scholar]
- 5.Tadin-Strapps M, Warburton D, Baumeister FA, Fischer SG, Yonan J, Gilliam TC, Christiano AM. Cloning of the breakpoints of a de novo inversion of chromosome 8, inv (8)(p11.2q23.1) in a patient with Ambras syndrome. Cytogenet Genome Res. 2004;107:68–76. doi: 10.1159/000079573. [DOI] [PubMed] [Google Scholar]
- 6.Tadin M, Braverman E, Cianfarani S, Sobrino AJ, Levy B, Christiano AM, Warburton D. Complex cytogenetic rearrangement of chromosome 8q in a case of Ambras syndrome. Am J Med Genet. 2001;102:100–104. doi: 10.1002/1096-8628(20010722)102:1<100::aid-ajmg1396>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 7.Fantauzzo KA, Tadin-Strapps M, You Y, Mentzer SE, Baumeister FA, Cianfarani S, Van Maldergem L, Warburton D, Sundberg JP, Christiano AM. A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum Mol Genet. 2008;17:3539–3551. doi: 10.1093/hmg/ddn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun M, Li N, Dong W, Chen Z, Liu Q, Xu Y, He G, Shi Y, Li X, Hao J, Luo Y, Shang D, Lv D, Ma F, Zhang D, Hua R, Lu C, Wen Y, Cao L, Irvine AD, McLean WH, Dong Q, Wang MR, Yu J, He L, Lo WH, Zhang X. Copy-number mutations on chromosome 17q24.2–q24.3 in congenital generalized hypertrichosis terminalis with or without gingival hyperplasia. Am J Hum Genet. 2009;84:807–813. doi: 10.1016/j.ajhg.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantu JM, Garcia-Cruz D, Sanchez-Corona J, Hernandez A, Nazara Z. A distinct osteochondrodysplasia with hypertrichosis: individualization of a probably autosomal recessive entity. Hum Genet. 1982;60:36–41. doi: 10.1007/BF00281261. [DOI] [PubMed] [Google Scholar]
- 10.Lazalde B, Sanchez-Urbina R, Nano-Arana I, Bitar WE, Ramirez-Duenas ML. Autosomal dominant inheritance in Cantu syndrome (congenital hypertrichosis, osteochondrodysplasia and cardiomegaly) Am J Med Genet. 2000;94:421–427. doi: 10.1002/1096-8628(20001023)94:5<421::aid-ajmg15>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Grange DK, Lorch SM, Cole PL, Singh GK. Cantu syndrome in a woman and her two daughters: further confirmation of autosomal dominant inheritance and review of the cardiac manifestations. Am J Med Genet A. 2006;140:1673–1680. doi: 10.1002/ajmg.a.31348. [DOI] [PubMed] [Google Scholar]
- 12.Tan TY, Bankier A, Slater HR, Northrop EL, Zacharin M, Savarirayan R. A patient with monosomy 1p36, atypical features and phenotypic similarities with Cantu syndrome. Am J Med Genet A. 2005;139:216–220. doi: 10.1002/ajmg.a.31013. [DOI] [PubMed] [Google Scholar]
- 13.Park TS, Lee ST, Song J, Lee KA, Kim J, Kim SJ, Lee JH, Song S, Choi JR. A tandem triplication, trp(1)(q21q32), in a patient with follicular lymphoma: a case study and review of the literature. Cancer Genet Cytogenet. 2009;189:127–131. doi: 10.1016/j.cancergencyto.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Shaffer L, Slovak M, Campbell L. An International System for Human Cytogenetic Nomenclature. Basel: Karger; 2009. [Google Scholar]
- 15.Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HG, Higgins AW, Herrick SR, Kishikawa S, Nicholson L, Kutsche K, Ligon AH, Harris DJ, MacDonald ME, Bruns GA, Morton CC, Quade BJ, Gusella JF. Candidate loci for Zimmermann-Laband syndrome at 3p14.3. Am J Med Genet A. 2007;143:107–111. doi: 10.1002/ajmg.a.31544. [DOI] [PubMed] [Google Scholar]
- 17.Herman TE, McAlister WH. Cantu syndrome. Pediatr Radiol. 2005;35:550–551. doi: 10.1007/s00247-004-1386-2. [DOI] [PubMed] [Google Scholar]
- 18.Frey N, Olson EN. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J Biol Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 19.Frey N, Barrientos T, Shelton JM, Frank D, Rütten H, Gehring D, Kuhn C, Lutz M, Rothermel B, Bassel-Duby R, Richardson JA, Katus HA, Hill JA, Olson EN. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 20.Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffin KC, Woodcock J, Krueger GG. Genetic variations associated with psoriasis and psoriatic arthritis found by genome-wide association. Dermatol Ther. 2010;23:101–113. doi: 10.1111/j.1529-8019.2010.01303.x. [DOI] [PubMed] [Google Scholar]
- 22.Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 23.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]