1.1 ABSTRACT
A major concern for all copy number variation (CNV) detection algorithms is their reliability and repeatability. However, it is difficult to evaluate the reliability of CNV calling strategies due to the lack of gold standard data that would tell us which CNVs are real. We propose that if CNVs are called in duplicate samples, or inherited from parent to child, then these can be considered validated CNVs. We used two large family-based Genome-Wide Association Study (GWAS) datasets from the GENEVA consortium to look at concordance rates of CNV calls between duplicate samples, parent-child pairs, and unrelated pairs. Our goal was to make recommendations for ways to filter and use CNV calls in GWAS datasets that do not include family data. We used PennCNV as our primary CNV-calling algorithm, and tested CNV calls using different datasets and marker sets, and with various filters on CNVs and samples. Using the Illumina core HumanHap550 SNP (single nucleotide polymorphism) set, we saw duplicate concordance rates of approximately 55% and parent-child transmission rates of approximately 28% in our datasets. GC model adjustment and sample quality filtering had little effect on these reliability measures. Stratification on CNV size and DNA sample type did have some effect. Overall, our results show that it is probably not possible to find a CNV calling strategy (including filtering and algorithm) that will give us a set of “reliable” CNV calls using current chip technologies. But if we understand the error process, we can still use CNV calls appropriately in genetic association studies.
Keywords: evaluation, CNV calling strategies, family-based GWAS
1.2 INTRODUCTION
Most investigators performing genome-wide association studies (GWAS) would like to include association tests for CNVs, but low reliability of CNV calls has been a roadblock [Carter, 2007; Lai et al., 2005; Peiffer et al., 2006; Wineinger et al., 2008]. If a sample is genotyped twice, relatively different lists of CNVs can result, and this difference may be exacerbated if two different CNV-finding algorithms are used. Various factors are known to affect the reliability, most notably DNA quality and differences among CNV calling algorithms.
The holy grail of CNV calling for genetic association studies is a procedure that will produce “reliable” CNV calls - at least high specificity if not high sensitivity. (Note that this is somewhat different from the goals of CNV calling for clinical purposes, in which the relative value of sensitivity and specificity might be different). Such a procedure might in theory be achieved by a combination of algorithm choice, data pre-processing, sample filtering, marker sets, and CNV filtering. Typical applications currently in the literature filter by using only samples that have high quality by some metric and only CNVs of a certain length. But it has been very difficult to compare and validate such procedures because of the lack of gold-standard datasets in which CNVs have been molecularly validated. In the absence of datasets with known “right answers,” the performance of a calling algorithm on real data cannot be quantified. While simulated data can be useful for this type of investigation, particularly in the early stages of algorithm development, we believe that there is no substitute for assessing performance on fully-complex real data, which is the goal of this study.
The premise of our study is that we can use family data as a substitute for molecular validation. If a CNV is called repeatedly in duplicate samples, or transmitted from parent to child, then it can be considered validated. This type of validation is not 100% accurate and will not be appropriate for clinical use, but it is sufficient to allow us to estimate error rates and use those estimated error rates to compare CNV calling strategies. We use two large family-based GWAS datasets and compute CNV concordance rates for duplicate samples, parent-child pairs, and unrelated pairs. We then use the concordance rates to evaluate a variety of CNV calling and filtering strategies. Because previous studies have focused on comparing different software packages [Dellinger et al., 2010; Pinto et al., 2011], we focus instead on the role of filtering in CNV calling - which markers, samples, and CNV calls should be used. We primarily report results for the PennCNV package [Wang et al., 2007], which is generally acknowledged to be one of the best for the Illumina (San Diego, California) platform, although we also report some results for genoCN [Sun et al., 2009]. Our goal is to make recommendations for how CNV calls can be created and filtered for use in genetic association studies. Since these studies generally involve unrelated individuals, we do not focus on optimizing calls within families but rather we use our families to understand what the best filtering procedures are for individuals. A secondary goal is to contribute to the literature describing features and distributions of rare CNVs in the human genome.
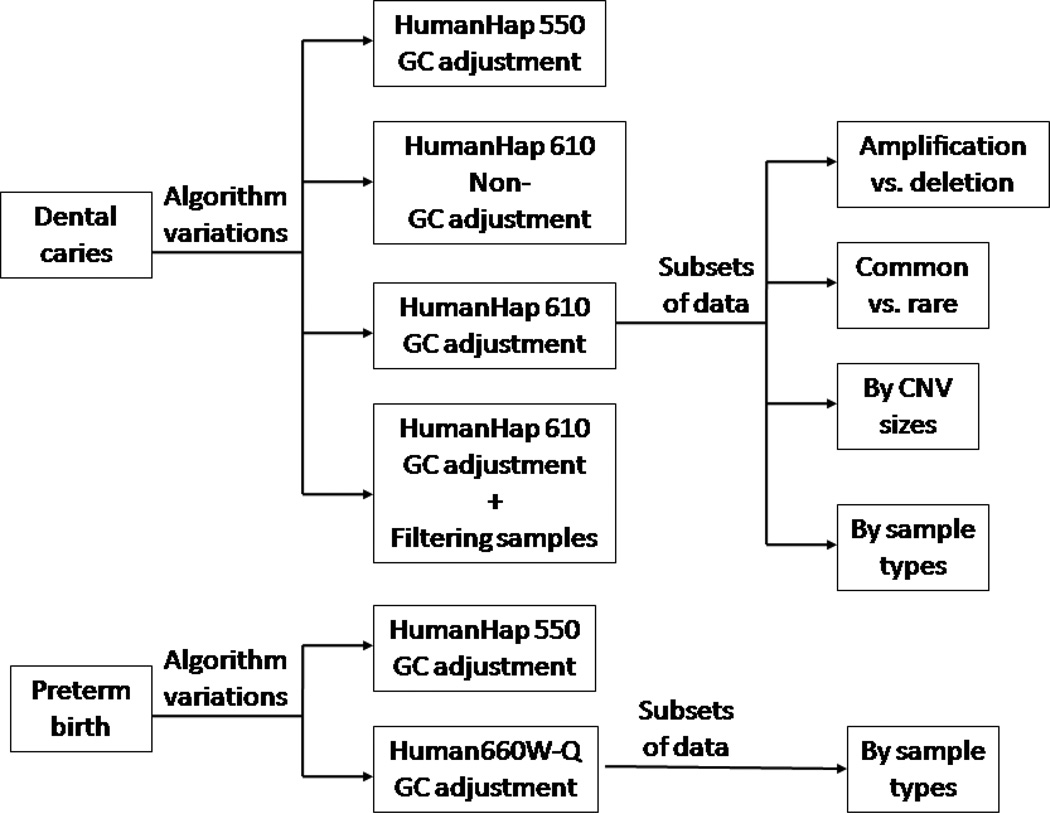
Our study design is sketched graphically in Figure 1. From the GENEVA dental caries study (http://www.ncbi.nlm.nih.gov/gap?term=geneva), which is a large community-based study of oral health genotyped on the Illumina HumanHap610 chip, we selected 91 duplicate pairs and 752 father-mother-child trios. From the GENEVA preterm delivery study (http://www.ncbi.nlm.nih.gov/gap?term=geneva), we used almost all samples −1782 mother-child pairs genotyped on Illumina Human660W-Quad chip. Of these, 943 pairs were cases of pre-term delivery, 779 pairs were controls and the remaining 60 pairs were neither cases nor controls. All subjects in the preterm delivery study were from the Danish National Birth Cohort. Since the chips used in these two studies share a core set of 550K SNPs, we started by calculating and comparing the CNV concordance rates in the two datasets using that shared SNP set. We then looked at the concordance rates for the full sets of SNP and CNV markers on each chip. We also looked at the effects of using PennCNV’s GC adjustment and filtering out high-variability samples in the dental caries dataset only. Finally, we looked at subsets of data such as amplifications vs. deletions, common vs. rare CNVs, different CNV sizes, and different DNA sample types.
Figure 2-1.
Summary of study design.
1.3 MATERIALS AND METHODS
1.3.1 Study Populations
Both the dental caries and preterm delivery datasets are part of the GENEVA consortium. In both datasets, GWAS data was used to verify all parent-child relationships. Detailed information on both studies is available from study documents in dbGAP (http://www.ncbi.nlm.nih.gov/gap) [Mailman et al., 2007]. The full dental caries study included four different community-based samples from Western Pennsylvania, West Virginia, and Iowa. Individuals were selected without regard to phenotype, and then were extensively phenotyped for oral health and related traits. We used a subset of the full study: 91 pairs of duplicate samples and 752 complete trio-family samples from two of the four recruitment sites. The pre-term delivery study is a case-control study within a cohort of approximately 1000 mother-child case pairs (cases were defined as infants<37 weeks of gestation), and 1000 mother-child controls pairs (controls were defined as infants=40 weeks of gestation) from the Danish National Birth Cohort study [Olsen et al., 2001]. 1782 mother/child pairs with complete genotype information were used in this study.
1.3.2 Genotyping and Quality Control
Complete genotyping and data cleaning reports for both studies are available in dbGAP (http://www.ncbi.nlm.nih.gov/gap).The level of genotyping quality was extremely high.
1.3.3 CNV Calls by PennCNV
We generated CNV calls using the PennCNV software (2009Aug27 verion) [Wang et al., 2007]. Each sample was called individually, regardless of family relationships. PennCNV is a Hidden Markov Model (HMM) based method. It uses the log R ratio (LRR) and B allele frequency (BAF) measures computed from the signal intensity files by BeadStudio. To limit analyses to the core HumanHap550 (550K) marker sets in the HumanHap610 and Human660W-Quad chips, we used the hg18 (NCBI 36) “hh550” Population Frequency of B allele (PFB) file during CNV calling. For algorithms with GC model adjustment, we implemented the GC model wave adjustment procedure in PennCNV. For sample filtering, after GC model adjustment, we excluded samples meeting the criterion lrrsd > 0.3. All analysis was restricted to autosomes. The PennCNV trio-based CNV calling feature was not used, since we were interested in assessing quality of calls in individuals. PennCNV did not find any loss-of-heterozygosity in our samples.
1.3.4 CNV Calls by genoCN
We generated the CNV calls using the genoCN package (version genoCN 1.08) in R [Sun et al., 2009]. genoCN is also a HMM-based method using the log R ratio (LRR) and B allele frequency (BAF) measures from the signal intensity files by Beadstudio. Unlike PennCNV, which assumes that the mean value and SD of LRR and BAF for each HMM state are known, genoCN estimates HMM parameters from data. All procedures followed the user guidelines of genoCN.
1.3.5 Calculation of Overlap Quantities
Overlap quantities calculated include duplicate concordance, transmission and inheritance rates and unrelated pair concordance rates. “Overlap” of CNVs was defined as follows (both criteria must be met): a) the overlap length in base pairs is larger than 50% of the length in base pairs of the smaller size CNV, b) copy number (cn) state must be either both deletion or both amplification.
For concordance rate, in each sample pair, say sample A and sample B, we first used sample A as a “template” and counted how many CNV calls in sample A overlapped with ones in sample B. Then we used sample B as a “template” and counted how many overlapped with those called in sample A. We summed the numbers of overlapping CNVs in the two comparisons, and then divided it by sum of CNV calls in sample A and sample B. We restricted the maximum number of overlaps for each CNV in a template sample to one. For example, if a single CNV in sample A overlapped with two different CNVs in sample B, only one overlap was counted; this avoided overcounting of larger CNVs that were broken into smaller pieces by the algorithm. For the dental caries dataset, the unrelated pair concordance rate was computed from father-mother pairs. For the preterm delivery dataset, the unrelated pair concordance rate was computed separately in mothers and children. The unrelated-pair concordance rate derived from children was highly consistent with the one from mothers, so only the rate from mothers was reported. The concordance rate in table 2-12 is also calculated in the same way.
Table 1-12.
Concordance rate between PennCNV and genoCN for each person.
| Subject id | Num of CNVs/Sample |
concordance rate | |
|---|---|---|---|
| PennCNV | genoCN | ||
| 175040850 | 88 | 540 | 0.19 |
| 9942 | 23 | 616 | 0.07 |
| 175043297 | 37 | 129 | 0.29 |
| 9950 | 37 | 101 | 0.33 |
| 175192256 | 64 | 195 | 0.52 |
| 175049618 | 63 | 507 | 0.19 |
| 175133191 | 58 | 322 | 0.26 |
| 175097605 | 51 | 304 | 0.25 |
For the transmission rate, in each parent-child pair, we used CNV calls in the parent as a “template,” counted how many of them were also called in the child, and then divided by the total number of CNV calls in the parent. For the inheritance rate, we used CNV calls in the child as a “template,” counted how many CNVs in the child overlapped with those in either parent, and then divided by the total number of CNVs in the child.
All concordance and transmission rates were calculated as the average over all pairs, so each pair contributed equally to the mean overlap rate and pairs with especially high or low rates were not excessively influential. All calculation was done in R (version 2.10.1) [R Development Core Team. 2009].
1.3.6 Stratification of CNV calls
Deletion vs. duplication CNVs were defined as CNV calls with cn <2 vs. >2 respectively. Common CNVs were defined as a frequency greater than 2%. Frequencies of CNVs were derived from unrelated individuals. Each CNV was compared with CNVs in other individuals; its frequency was defined as the overlap rate. We restricted the maximum number of overlaps from a pair of samples for each CNV to one.
1.4 RESULTS AND DISCUSSION
1.4.1 CNV Concordance Rates Using the Illumina HumanHap550 SNP Set
In an ideal dataset, duplicate concordance rates would be 100%, transmission rates would be 50%, and inheritance rates would be 100%. Several major factors, however, potentially cause datasets to deviate from this ideal. Most importantly, falsely detected CNVs will cause all of these rates to be below their ideal levels. Failure to detect CNVs (false negatives) will have a similar effect. For both false negatives and false positives, we should consider the possibility that the error is not random - that it could be repeated in duplicate samples or even in parent-child pairs because of sample or sequence similarity. A third important factor is de novo mutations in children, which will not affect duplicate concordance rates or transmission rates, but will affect inheritance rates. Finally, there is the possibility of somatic mutation with age (essentially de novo mutations in parents), which would affect apparent transmission rates but not inheritance or duplicate concordance rates. Because all of these factors are acting simultaneously, it is not possible to estimate them from this type of dataset, but some qualitative conclusions can be drawn, as discussed below, in particular if we are willing to assume that de novo mutations and somatic mutations are rare compared to CNV-calling errors.
The first column of Table 2-1 shows the results for the dental caries dataset using the common HumanHap550 SNP set. The average parent-child transmission rate is 28%, and the duplicate concordance rate is 55%. Father-child transmission rates and mother-child transmission rates are essentially identical. The fact that parent-child transmission rates are just about half of duplicate concordance rates suggests that we are probably not seeing repeated false calls in duplicates due to sample issues – repeated calls in duplicates are likely to be real. The average unrelated pair concordance rate is 5%, which is presumably primarily accounted for by common CNVs, although a small amount of concordance by chance of rare CNVs and systematic error would also be included. Under simple but very conservative assumptions (such as that almost all CNV calls are false positives) we estimate a completely random concordance rate of about 0.3%. The fact that the inheritance rate of 42% is much less than twice the transmission rate implies that de novo CNVs may account for a non-ignorable proportion of the child CNVs. We note that the average child inheritance rate (42%) in our study is lower than what was reported by K. Wang et al [2007]. They examined “the fraction of CNVs inferred in offspring but not detected in parents (CNV-NDPs)”, and found 25.2% of offspring CNVs from HumanHap550 were CNV-NDPs. This may due to differences in sample size, sample quality and sample populations. K. Wang et al. examined CNV-NDPs in the HapMap CEU + YRI offspring, which is a much smaller dataset.
Table 1-1.
Mean overlap quantities (± SEM) in the dental caries dataset.
| HumanHap550 |
HumanHap610 |
|||
|---|---|---|---|---|
| GC* | GC | non-GC | GC+filtering** | |
| Num of duplicate samples (avg. CNV/sample)*** | 182 (18.6) | 182 (79.4) | 182 (61.1) | 162 (54.1) |
| Num of non-duplicate samples(avg. CNV/sample) | 1736 (26.6) | 1736 (68.5) | 1736 (92.9) | 1512 (54.9) |
| Duplicate concordance rate | 0.55(±0.02) | 0.45(±0.02) | 0.43(±0.02) | 0.48(±0.02) |
| Unrelated pair concordance rate**** | 0.05(±0.003) | 0.13(±0.004) | 0.11(±0.004) | 0.14(±0.004) |
| Father-child transmission rate | 0.28(±0.006) | 0.28(±0.005) | 0.27(±0.005) | 0.31(±0.005) |
| Mother-child transmission rate | 0.28(±0.006) | 0.27(±0.005) | 0.26(±0.005) | 0.31(±0.005) |
| Child inheritance rate | 0.42(±0.009) | 0.40(±0.008) | 0.36(±0.008) | 0.45(±0.008) |
All mean overlap quantities were calculated as the average over pairs.
GC model adjustment procedure in PennCNV.
Samples were filtered by the criterion: LRR standard deviation (sd) > 0.3.
Number of duplicate samples (average number of CNVs per sample).
The unrelated pair concordance rate was calculated among father-mother pairs.
The first column of Table 2-2 shows the corresponding results for the preterm delivery dataset. The concordance rates are very similar to those in the dental caries dataset: duplicate concordance rate 52%, mother-child transmission rate 26%, and unrelated concordance rate 4%. The highly consistent results imply that the findings from our study are not dataset specific and may be reasonably generalizable to other studies, at least for this marker set.
Table 1-2.
Mean overlap quantities (± SEM) in the preterm delivery dataset.
| Hap550 |
Human660W-Quad |
|
|---|---|---|
| GC | GC | |
| Num of dup samples (avg. CNV/sample) | 40 (21) | 40 (383) |
| Num of non-dup samples(avg. CNV/sample) | 3564 (48.8) | 3564 (438.6) |
| Duplicate concordance rate | 0.52 (±0.06) | 0.64(±0.02) |
| Unrelated pair concordance rate* | 0.04(±0.002) | 0.21(±0.002) |
| Mother-child transmission rate | 0.26 (±0.004) | 0.38(±0.002) |
The unrelated pair concordance rate was derived from mothers. The rate derived from children was very similar.
1.4.2 Addition of CNV Markers from the HumanHap610 Chip and the Human660W-Quad Chip
The HumanHap610 chip (dental caries study) and the Human660W Quad chip (pre-term delivery study) each consist of the HumanHap550 SNP set augmented by different sets of CNV probes. The second columns of Table I and Table II give results for each study using the full chip for that study. For the HumanHap610 chip, the parent-child transmission rate and inheritance rate are similar to those from the HumanHap550 SNP set, but the average unrelated pair concordance rate is higher: 13% vs. 5%. One of the likely explanations is that the 60K additional CNV probes on the HumanHap610 chip contain more probes for common CNVs, and this is supported by evidence from later analyses (common vs. rare CNVs). Another noticeable difference is that the average duplicate concordance rate for HumanHap610 is much lower than for HumanHap550 (45% vs. 55%). This suggests quite poor performance of the CNV probes on this chip.
The full Human660W-Quad chip performs very differently than the HumanHap610, finding about seven times as many CNVs per sample. It also has much higher concordance and transmission rates, suggesting that the CNV probes have much better performance. The average duplicate concordance rate for the Human660W-Quad is 64%, as compared to 45% for the HumanHap610 and 55% for the HumanHap550. The average unrelated pair concordance rate for the Human660W-Quad is also much higher, 21%, suggesting that the CNV markers on the Human660W-Quad find many common CNVs. This is likely to also be the reason that the mother-child transmission rate is much higher than that on the HumanHap550 (38% vs. 26%).
1.4.3 GC Model Adjustment
PennCNV includes a GC model adjustment feature that adjusts the CNV calls to account for varying GC-content of the chromosome in different locations. Our analyses above included that adjustment, but the third column of Table 2-1 shows an analysis without the GC adjustment. Removing the adjustment increased the number of called CNVs and decreased the reliability measures (compare column 3 to column 2), but only very slightly. We conclude that the GC adjustment probably does improve quality, but does not make a major difference.
1.4.4 Sample Filtering
Column four of Table 2-1 shows an analysis in which we omitted the samples (about 13%) that had the PennCNV variability measure lrrsd (log R ratio standard deviation) greater than 0.3. As with the GC model adjustment, this improved reliability, but only very slightly. It is clearly a good idea in CNV analyses to omit poor-quality samples, but it appears that lrrsd might not be the most useful quality measure.
1.4.5 Deletion vs. Amplification CNVs
We used the dental caries dataset with the GC adjustment and the full HumanHap610 marker set to ask whether concordance rates differed for deletion and amplification CNVs. Table 2-3 shows the results. The number of deletion CNVs is 1.5∼2 times that of amplification CNVs, but this does not necessarily reflect frequency in the human genome, since any given CNV calling algorithm may have higher sensitivity to either deletions or amplifications. It is interesting to note that while duplicate concordance and parent-child transmission rates are higher for deletions, the inheritance rate (percent of the child’s CNVs that are inherited from parents) is higher for amplifications. It is possible that this means that de novo deletions are more common in viable offspring than de novo amplifications, but that would clearly merit further investigation.
Table 1-3.
Mean overlap quantities (±SEM) in deletion vs. amplification CNVs.
| Deletion | Amplification | |
|---|---|---|
| Avg CNVs* in dup samples/person | 36.1 | 24.9 |
| Avg CNVs in non-dup samples/person | 46.6 | 21.9 |
| Duplicate concordance rate | 0.51 (±0.02) | 0.40 (±0.02) |
| Unrelated pair concordance rate | 0.11 (±0.004) | 0.14 (±0.007) |
| Father-child transmission rate | 0.32 (±0.006) | 0.25 (±0.007) |
| Mother-child transmission rate | 0.30 (±0.006) | 0.27 (±0.007) |
| Child inheritance rate | 0.41 (±0.009) | 0.46 (±0.009) |
Avg CNVs: average number of CNVs
1.4.6 Common vs. Rare CNVs
Again using the dental caries dataset with the GC adjustment and the full HumanHap610 marker set, we asked whether concordance rates differed for rare and common CNVs. Parent-child and unrelated-pair concordance rates are clearly expected to be higher for common CNVs because of chance matching, but duplicate concordance rates should not be different for rare and common CNVs if the algorithm is equally good at finding both. However, one of the concerns in CNV calling is that common CNVs can be hard to detect, since the deviation of the log R ratio between case and reference is small after normalization.
Results are given in Table 2-4. For the purposes of this analysis we arbitrarily considered a CNV to be common if it occurred in 2% or more of the sample. The concordance rates in unrelated pairs are 3% for rare CNVs and 19% for common CNVs, which confirms that most of the concordance between unrelated individuals is due to CNVs that are common in the population. This may also explain the higher transmission rate in common CNVs than rare ones (32% vs. 20%). The finding that the average duplicate concordance rate in common CNVs is higher than in rare ones (51% vs. 44%) suggests that PennCNV does not in fact have more difficulty detecting common CNVs.
Table 1-4.
Mean overlap quantities (± SEM) in common vs. rare CNVs.
| Common | Rare | |
|---|---|---|
| Avg CNVs in dup samples/person * | 41.7 | 19.4 |
| Avg CNVs in non-dup samples/person ** | 34 | 23.9 |
| Duplicate concordance rate | 0.51 (±0.03) | 0.44 (±0.04) |
| Unrelated pair concordance rate | 0.19 (±0.006) | 0.03 (±0.003) |
| Father-child transmission rate | 0.32 (±0.006) | 0.20 (±0.007) |
| Mother-child transmission rate | 0.31 (±0.006) | 0.21 (±0.007) |
Only CNVs from unrelated subjects were used to infer the common vs. rare CNVs.
Total sample number of duplicates was 66.
Total sample number of non-duplicate subjects was 984.
1.4.7 Samples With High CNV Number
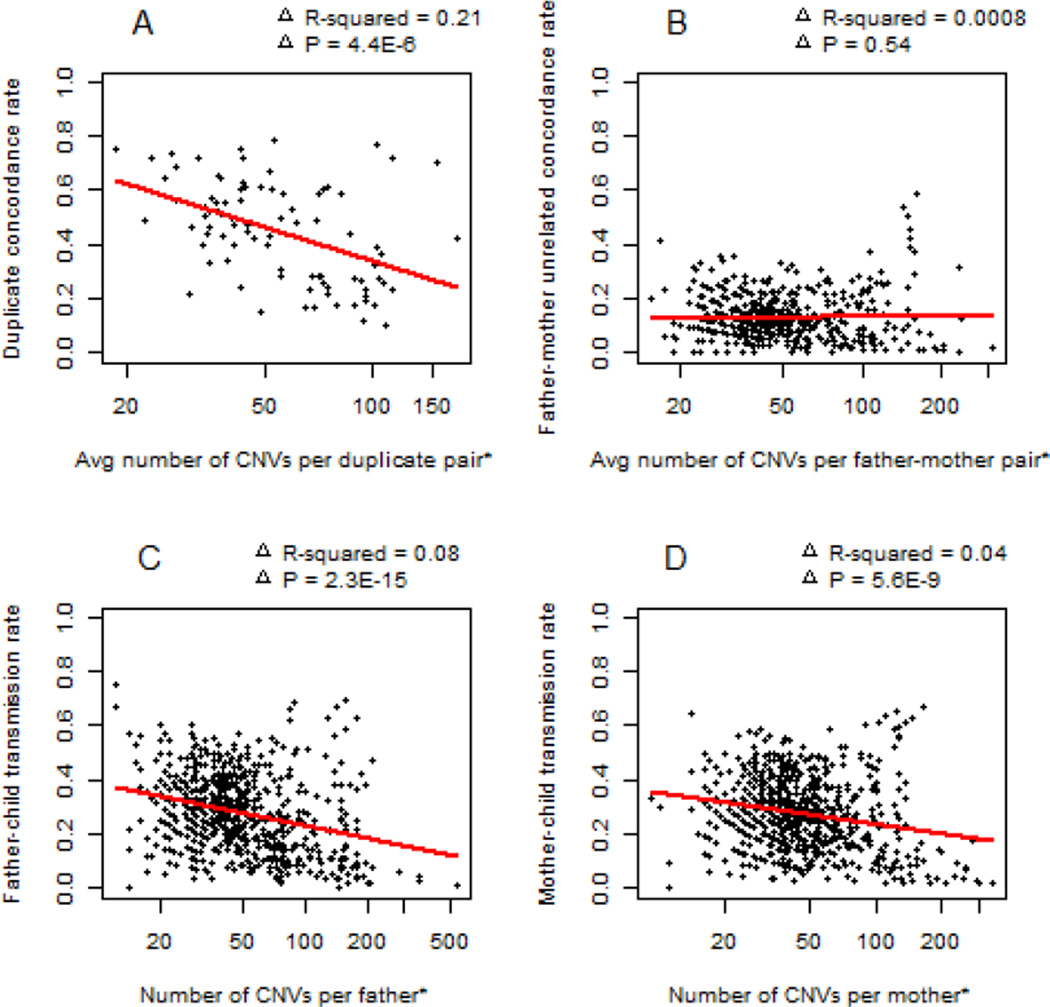
It might be logical to presume that samples with very high CNV numbers are of low quality and that the CNVs called in those samples are not real. To investigate this, we plotted CNV number vs. concordance rate using the dental caries dataset (HumanHap 610 marker set) in Figure 2-2. In general, the concordance/transmission rates tend to decrease with the number of CNV calls, but there are clearly some pairs that have high concordance and/or transmission rates even with more than 100 CNVs. This suggests that while it might be advisable to filter samples with very high numbers of CNV calls out of association studies, there are in fact some individuals who do carry high numbers of real CNVs.
Figure 2-2.
Relationship between number of CNV calls per sample and concordance rate in the dental caries dataset.
*All x–axes are log scale.
(A) shows relation between average number of CNV calls per pair of duplicate samples and duplicate concordance rate. (B) shows relation between average number of CNV calls per pair of unrelated samples and unrelated concordance rate. (C) shows relation between number of CNV calls in each father and father-child transmission rate. (D) shows relation between number of CNV calls in each mother and mother-child transmission rate.
1.4.8 CNV Size
It is often assumed that calls of larger CNVs are more likely to be accurate, and our results using the dental caries dataset (HumanHap 610 marker set) (Table 2-5) support that. We measured the “size” of the CNV by the number of markers rather than the physical length, and found that the shortest CNVs (3 – 5 markers) had only an 18% mean parent-child transmission rate, while the longest (> 54 markers) had a 42% mean parent-child transmission rate. This is a substantial difference, but it is not substantial enough to justify filtering out the smallest CNVs or to justify assuming that the largest ones are necessarily correct. Thus while our results support the common wisdom, they do not suggest a workable filtering strategy for association studies.
Table 1-5.
Mean overlap quantities (± SEM) by size of CNV call.
| Size of CNV call |
||||||
|---|---|---|---|---|---|---|
| 3-5 SNPs | 6-10 SNPs | 11-22 SNPs | 23-54 SNPs | > 54 SNPs | All | |
| Avg num of CNVs/person* | 13 | 19.2 | 18.6 | 13.4 | 3.5 | 67.6 |
| Duplicate concordance rate | 0.31 (±0.02) | 0.49 (±0.02) | 0.48 (±0.02) | 0.55 (±0.03) | 0.64 (±0.04) | 0.45 (±0.02) |
| Unrelated pair concordance rate | 0.07 (±0.004) | 0.11 (±0.004) | 0.12 (±0.005) | 0.23 (±0.008) | 0.16 (±0.01) | 0.13 (±0.004) |
| Father-child transmission rate | 0.18 (±0.007) | 0.29 (±0.006) | 0.30 (±0.008) | 0.41 (±0.01) | 0.42 (±0.02) | 0.28 (±0.005) |
| Mother-child transmission rate | 0.19 (±0.007) | 0.27 (±0.006) | 0.30 (±0.008) | 0.40 (±0.01) | 0.43 (±0.02) | 0.27 (±0.005) |
In all 1736 non-duplicate samples.
1.4.9 DNA Source
Next, we compared the reliability of CNV calls for different DNA sample types. The dental caries study includes samples from blood, saliva, mouthwash and buccal swabs. To investigate the effect of sample type, we compared transmission rates in pairs with different combinations of sample types using the HumanHap 610 marker set. The resulting transmission rates in the dental caries dataset are shown in Table 2-6. There is no detectable difference in reliability between the blood and saliva samples. The comparative reliability of mouthwash is not conclusive due to the small sample size. A limitation of the results shown in Table 2-6 is that because all parental samples are either blood or saliva, it is difficult to tell from transmission rates if the other sample types are more error-prone. That is, child samples with more false CNVs due to poor DNA quality may not necessarily show lower transmission rates if the true CNVs were also called. Thus in Table 2-7 we also show the number of CNVs called per sample by sample type. The mouthwash, buccal and WGA samples do have significantly more CNVs called per person, and we conclude that it is likely that they have higher false-positive rates than the blood and saliva samples. It is also intriguing that there is a higher number of CNVs per person in the children's saliva and mouthwash samples than in their parents. It appears that children may in general be producing lower-quality saliva samples than adults. By contrast, we saw comparable CNV numbers from blood samples in children and parents in both the dental caries (Table 2-7) and preterm delivery (Table 2-8) datasets.
Table 1-6.
Mean parent-child transmission rate (± SEM) by sample type in the dental caries dataset.
| Sample sources |
Num of sample pairs |
Father-child transmission |
Mother- child transmission |
|
|---|---|---|---|---|
| Parent | Child | |||
| Mouthwash | Mouthwash | 9 | 0.22 (±0.05) | 0.34 (±0.05) |
| Saliva | Saliva | 98 | 0.27 (±0.01) | 0.29 (±0.01) |
| Blood | Blood | 349 | 0.28 (±0.008) |
0.27 (±0.007) |
| Blood | Buccal | 89 | 0.24 (±0.01) | 0.23(±0.01) |
| Blood | Saliva | 51 | 0.30 (±0.02) | 0.26 (±0.02) |
| Blood | Mouthwash | 40 | 0.32 (±0.02) | 0.32 (±0.02) |
| Blood | WGA | 10 | 0.32(±0.06) | 0.32(±0.06) |
Table 1-7.
Mean number of CNVs called per sample (± SEM) by sample type in the dental caries dataset.
| Child |
Father |
Mother |
||||
|---|---|---|---|---|---|---|
| sample num | CNV num/sample | sample num | CNV num/sample | sample num | CNV num/sample | |
| Blood | 349 | 61.5±3.3 | 539 | 55.9±2.1 | 539 | 55.7±1.9 |
| Saliva | 150 | 82.0±6.7 | 98 | 58.4±3.8 | 98 | 51.1±3.8 |
| Mouthwash | 49 | 120.7±23.1 | 9 | 71.8±19.4 | 9 | 49±11.0 |
| Buccal | 89 | 109.8±9.3 | ||||
| WGA | 10 | 153.6±38.4 | ||||
Table 1-8.
Mean number of CNVs called per sample (± SEM) in buffy coat blood samples in the preterm delivery dataset.
| Child |
Mother |
|||
|---|---|---|---|---|
| sample num |
CNV num/sample |
sample num |
CNV num/sample |
|
| Buffy coat |
1257 | 379.2±2.9 | 1257 | 380.8±2.3 |
In the pre-term delivery study most maternal samples were from buffy coat, but a substantial proportion of the infant samples were from dried blood spots. Some of the buffy coat samples and some of the blood spot samples were whole-genome amplified (WGA). Mother-child transmission rates calculated using the Human660W-Quad marker set are listed in Table 2-9 according to the sample type for both mother and child. When both mother and child are buffy coat (no WGA), the transmission rate is 40%. If the child is WGA, it only drops to 31%, so we can infer that most of the real CNVs are still being found in buffy coat WGA. However, when the mother is WGA transmission percentages drop substantially, from which we can infer that the WGA samples are giving us many spurious CNV calls in addition to the real ones. These findings suggest that the WGA samples give us reasonable sensitivity, but very poor specificity.
Table 1-9.
Mean mother-child transmission rate (± SEM) by sample type in the preterm delivery dataset.
| Sample source |
Num of sample pairs |
Mother-child transmission |
|
|---|---|---|---|
| mother | child | ||
| Buffy coat | Buffy coat | 1347 | 0.40 (±0.003) |
| Buffy coat | Blood spot | 346 | 0.35 (±0.004) |
| Buffy coat | Buffy coat WGA | 52 | 0.31 (±0.008) |
| Buffy coat WGA | Buffy coat | 13 | 0.06 (±0.006) |
| Buffy coat WGA | Blood spot | 18 | 0.14 (±0.01) |
| All | 1782 | 0.38 (±0.002) | |
1.4.10 Age
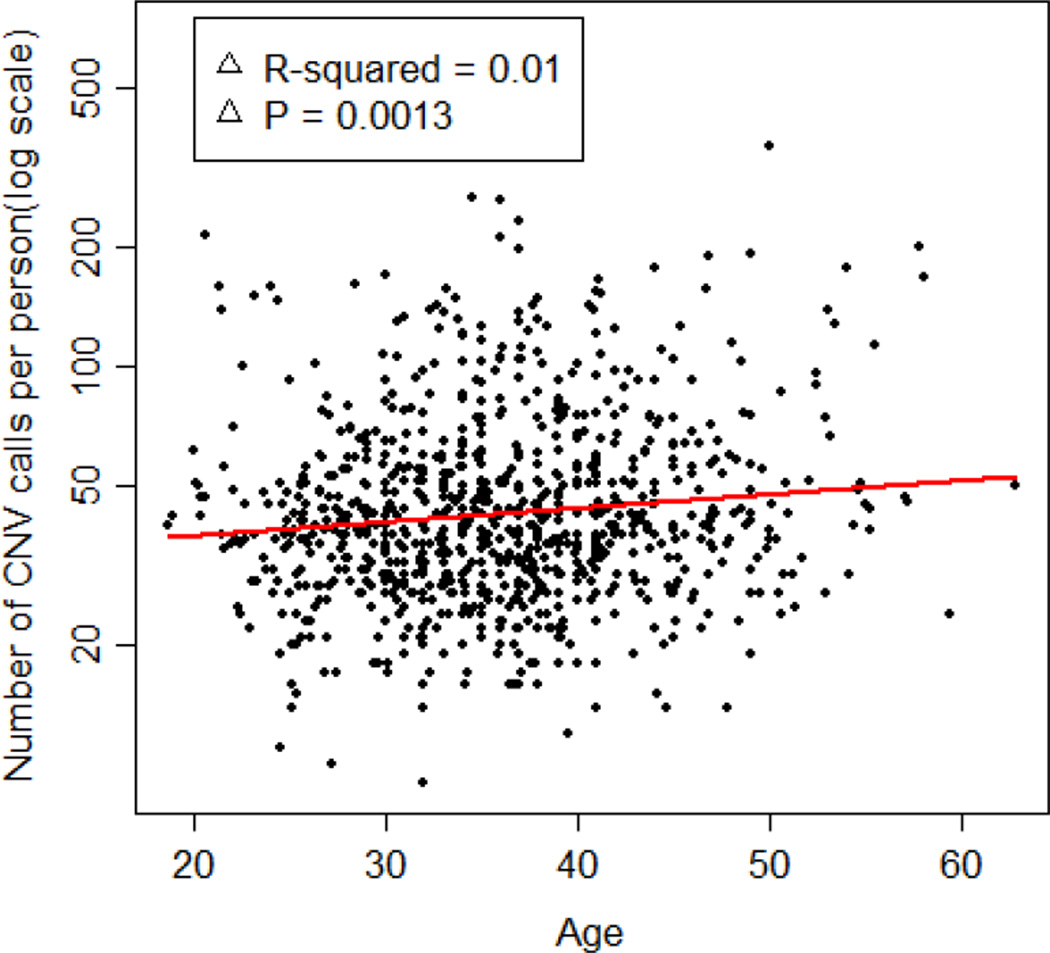
Another interesting question is that of somatic mutations with age. Using parents only from the two studies separately, we regressed the log of the number of CNVs on the age of the individual. After excluding a few extreme outliers, we found a very small but statistically significant increase in the number of CNVs with age (Figure 2-3). This finding is consistent with suggestions made in previous studies [Martin et al., 1996; Maslov and Vijg, 2009].
Figure 2-3.
Relationship between age and number of CNV calls in each adult (log scale) in the dental caries dataset.
1.4.11 Comparison to genoCN
Finally, in order to compare with the performance of PennCNV, we conducted a limited study using another algorithm - genoCN [Sun et al., 2009]. We chose two pairs of duplicates and two pairs of unrelated samples at random (from samples with lrrsd<0.3) and tested them using genoCN (Table 2-10 and Table 2-11). genoCN detected 4∼20 fold more CNVs than PennCNV (with GCmodel and HumanHap610 markers). Most of the CNVs called by PennCNV were also called by genoCN (table 2-9), but the duplicate concordance rates for the genoCN calls were much lower than those for PennCNV. From this we can infer that genoCN may give a lot of spurious CNV calls in addition to the real ones (reasonable sensitivity but poor specificity). We also observed that the unrelated pair concordance rates in genoCN were lower than in PennCNV, presumably also due to low specificity.
Table 1-10.
Comparison of duplicate concordance rate among two pairs of duplicate samples from the dental caries dataset using PennCNV and genoCN.
| Subject ids |
Num of CNVs |
Num of concordant CNV | Duplicate concordance rate | |||
|---|---|---|---|---|---|---|
| dup1 | dup2 | dup1 | dup2 | |||
| PennCNV | 175040850 | 9942 | 88 | 23 | 16 | 0.28 |
| 175043297 | 9950 | 37 | 37 | 24 | 0.65 | |
| genoCN | 175040850 | 9942 | 540 | 616 | 67 | 0.12 |
| 175043297 | 9950 | 129 | 101 | 52 | 0.45 | |
Table 1-11.
Unrelated concordance rate among two pairs of father-mother samples from the dental caries dataset using PennCNV and genoCN.
| Subjects id |
Num of CNVs |
Num of concordant CNV |
Unrelated concordance rate |
|||
|---|---|---|---|---|---|---|
| father | mother | father | mother | |||
| PennCNV | 175192256 | 175049618 | 64 | 63 | 7 | 0.11 |
| 175133191 | 175097605 | 58 | 51 | 10 | 0.18 | |
| genoCN | 175192256 | 175049618 | 195 | 507 | 21 | 0.06 |
| 175133191 | 175097605 | 322 | 304 | 44 | 0.14 | |
An additional problem that we observed in both algorithms was that the largest CNVs were not “called” as single units, but were broken into several reported smaller CNVs. This problem was worse in genoCN than in PennCNV.
1.5 CONCLUSIONS
In summary, CNV association studies have been of great interest lately, but a key problem is how to identify a set of reliable CNVs. Molecular validation is not feasible for GWAS-sized datasets, and without gold-standard data it has been quite difficult to compare CNV calling algorithms to make recommendations for the best ones to use in association studies. We have taken advantage of two large family-based GWAS studies to use inheritance as a substitute for molecular validation and ask questions about what kind of sample, SNP, and CNV filtering leads to the most reliable CNV calls. While many authors have previously reported concordance rates for CNV calls in duplicate samples, we hope that by also looking at very large samples of parent-child pairs we have added depth to that picture.
We found several classes of samples that clearly have low reliability and should be filtered out of CNV association studies, including any with whole-genome amplification and any with excessive numbers of called CNVs. These results are quite concordant with conclusions of previous authors. But filtering out these samples did not result in high reliability rates in the remaining samples, an issue that we believe has not received adequate attention previously. The most prognostic variable we looked at was CNV size, but even that did not guarantee high reliability for large CNVs nor low reliability for small CNVs. Thus we suggest that the common strategy of using only the largest CNV calls and assuming they are correct is excessively crude and probably quite detrimental to statistical power.
Overall, we conclude from our data that it is probably not possible to find a CNV calling strategy that will give us a set of “reliable” CNV calls using current chip technologies. For now, CNV calls will need to be understood as having high error rates. But if we understand and model the features of that error process, we can still use them appropriately in genetic association studies. In particular, the most critical issue will be to make sure that cases and controls are well matched on any features that we know affect CNV call reliability rates, such as DNA sample type.
We also made some contributions to the growing picture of what “normal” variability in copy number means for the human genome. In particular, we found a subset of individuals who carry a fairly high load of rare CNVs (100 or more) that appear from inheritance rates to be real. We also found a modest increase in the number of CNVs with age, suggesting a non-trivial rate of somatic mutation, although this clearly bears further study. Finally, we found some intriguing results related to the relative inheritance rates of deletions vs. amplifications, which would be interesting to follow up further.
1.6 ACKNOWLEDGMENTS
The work of XZ was supported by T32MH015169. The work of JRS, MLM, and EF was supported by U01DE018903 and U01HG004423. The work of BF, MM, and JCM was supported by U01HG004423. The work of CPM and CCL was supported by U01HG004446. Genotyping was performed by the Johns Hopkins University (JHU) Center for Inherited Disease Research (CIDR) through contract HHSN268200782096C. Dental caries subjects were collected by the Center for Oral Health Research in Appalachia (PI M. Marazita, a collaboration of the University of Pittsburgh and West Virginia University funded by NIDCR R01-DE 014899) and the Iowa Fluoride Study and the Iowa Bone Development Study (PI S. Levy), funded by NIDCR R01-DE09551 and R01-DE12101, respectively). Pre-term birth subjects were a part of the Danish National Birth Cohort (DNBC), which was established with the support of a major grant from the Danish National Research Foundation. Additional support for the DNBC has been provided by the Danish Pharmacist's Fund, the Egmont Foundation, the March of Dimes Birth Defects Foundation, The Augustinus Foundation, and the Health Fund of the Danish Health Insurance Societies.
1.7 REFERENCES
- Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger AE, Saw SM, Goh LK, Seielstad M, Young TL, Li YJ. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res. 2010;38:e105. doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Ogburn CE, Colgin LM, Gown AM, Edland SD, Monnat RJ., Jr. Somatic Mutations Are Frequent and Increase with Age in Human Kidney Epithelial Cells. Hum Mol Genet. 1996;5:215–221. doi: 10.1093/hmg/5.2.215. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J, Melbye M, Olsen SF, Sørensen TI, Aaby P, Andersen AM, Taxbøl D, Hansen KD, Juhl M, Schow TB, Sørensen HT, Andresen J, Mortensen EL, Olesen AW, Søndergaard C. The Danish National Birth Cohort. Its background, structure and aim. Scand J Public Health. 2001;29:300–307. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald JR, Mills R, Prasad A, Noonan K, Gribble S, Prigmore E, Donahoe PK, Smith RS, Park JH, Hurles ME, Carter NP, Lee C, Scherer SW, Feuk L. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nature Biotechnology. 2011;29:512–521. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Sun W, Wright FA, Tang Z, Nordgard SH, Van Loo P, Yu T, Kristensen VN, Perou CM. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wineinger NE, Kennedy RE, Erickson SW, Wojczynski MK, Bruder CE, Tiwari HK. Statistical issues in the analysis of DNA Copy Number Variations. Int J Comput Biol Drug Des. 2008;1:368–395. doi: 10.1504/IJCBDD.2008.022208. [DOI] [PMC free article] [PubMed] [Google Scholar]