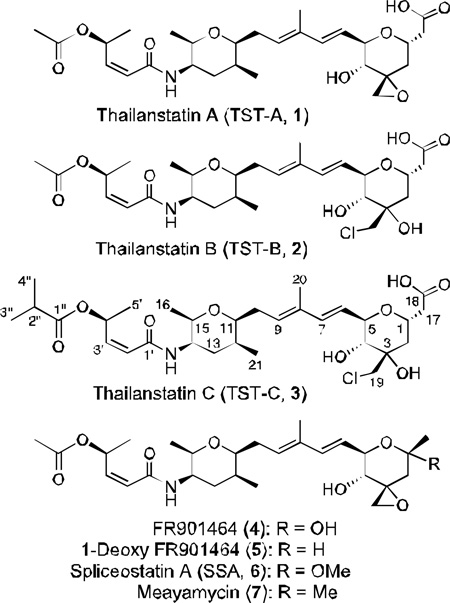
Abstract
Mining the genome sequence of Burkholderia thailandensis MSMB43 revealed a cryptic biosynthetic gene cluster resembling that of FR901464 (4), a prototype spliceosome inhibitor produced by Pseudomonas sp. No. 2663. Transcriptional analysis revealed a cultivation condition in which a regulatory gene of the cryptic gene cluster is adequately expressed. Consequently, three new compounds, named thailanstatins A (1), B (2) and C (3), were isolated from the fermentation broth of B. thailandensis MSMB43. Thailanstatins are proposed to be biosynthesized by a hybrid polyketide synthase-nonribosomal peptide synthetase pathway. They differ from 4 by lacking an unstable hydroxyl group and by having an extra carboxyl moiety; those differences endow thailanstatins with a significantly greater stability than 4 as tested in phosphate buffer at pH 7.4. In vitro assays showed that thailanstatins inhibit pre-mRNA splicing as potently as 4, with half-maximal inhibitory concentrations in the single to sub µM range. Cell culture assays indicated that thailanstatins also possess potent antiproliferative activities in representative human cancer cell lines, with half-maximal growth inhibitory concentrations in the single nM range. This work provides new chemical entities for research and development, and new structure-activity information for chemical optimization of related spliceosome inhibitors.
Keywords: Burkholderia thailandensis MSMB43, genomics-guided discovery, natural product, pre-mRNA splicing inhibitor, thailanstatin
The complexity of the eukaryotic genome is manifest not only in the number of genes it contains, but also by how those genes are structured and regulated. The protein coding sequences (exons) of most eukaryotic genes lie between non-coding regions (introns) which are excised during pre-mRNA splicing by the multi-component spliceosome complex, with the alternative splicing of more than 90% of human genes generating protein variants far greater than the number of the encoding genes.1, 2 While this processing inherently provides for transcriptome diversity, aberrant alternative splicing has been implicated in numerous disease conditions such as cancer and neurodegeneration.3 As such, dozens of small molecule effectors targeting the alternative splicing process have been identified and evaluated as drug candidates;4 these include FR901464 (4), a natural product of Pseudomonas sp. No. 2663,5–7 1-deoxy 4 (5),8 spliceostatin A (6)9 and meayamycin (7),10 all derivatives of 4, and pladienolides, a structurally different group of natural products from Streptomyces platensis Mer-11107,11–13 and sudemycins, synthetic compounds that possess the pharmacophore common to 4 and pladienolides.14 Many of those compounds demonstrated potent antiproliferative properties in human cancer cell lines, and were found to be generally much less toxic to normal human cells.14 In particular, E7107, a derivative of pladienolide D, exhibited promising preclinical activities and progressed to clinical trials.15 Mechanistically, both 6 and pladienolide B were found to bind non-covalently to the splicing factor 3b (SF3b) subcomplex of the U2 small nuclear ribonucleoprotein (snRNP) particle of spliceosome, thus inhibiting pre-mRNA splicing and causing pre-mRNA leakage into the cytoplasm.16–18 Therefore, pre-mRNA splicing inhibition represents a new mechanism of anti-neoplastic action, warranting further exploration of more and diverse chemical entities with this activity.
Natural products have been traditionally sought from actinomycetes, filamentous fungi and medicinal plants. Gram-negative bacterial species, such as Burkholderia, Chromobacterium, Lysobacter, Pseudomonas and Xenorhabdus, however, have emerged as new sources of diverse bioactive natural products (see recent reviews19–22 and references cited herein). For example, the 6.7-Mb combined genome of B. thailandensis E264 strain contains a total of 226.5 kb of thiotemplate modular system (TMS) genes that encode at least 11 nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS) or hybrid biosynthetic pathways.23 This vast pool of metabolic potentiality has been confirmed by recent discovery of diverse natural products from cultures of this bacterium, including thailandamides,24,25 bactobolins A–D,26 burkholdacs A and B,27 spiruchostatin C,28 thailandepsins A–F,29, 30 and burkholderic acid.31
Genomics-guided approach for the discovery of microbial natural products, which utilizes biosynthetic gene cluster information extracted from microbial genomes for the prediction of product structures or structural features, has gained significant ground since its inception in 2005.32–34 Our recent discovery of a series of six thailandepsin compounds from the B. thailandensis E264 strain, shown to be potent histone deacetylase inhibitors and broad-spectrum antiproliferative agents,29, 30 reinforced the approach and spurred our exploratory efforts to include a broader range of Gram-negative bacteria. To this end, we have re-discovered FK228 (Romidepsin), an FDA approved anticancer drug for the treatment of refractory T-cell lymphomas, from B. thailandensis MSMB43 strain, after having found that the genome of this bacterium contains a gene cluster highly homologous to the FK228 biosynthetic gene cluster in C. violaceum No. 968.35
We report here the discovery of three new natural products, named thailanstatin A (TST-A, 1), thailanstatin B (TST-B, 2) and thailanstatin C (TST-C, 3), from the culture broth of B. thailandensis MSMB43 with a genomics-guided approach. Thailanstatins have an overall structural similarity with 4 but differ by lacking an unstable hydroxyl group and by having an extra carboxyl moiety; these differences make thailanstatins significantly more stable than 4. Thailanstatins possess equally potent pre-mRNA splicing inhibitory activity in vitro and antiproliferivative activities in human cancer cell lines. This work provides new chemical entities with pre-mRNA inhibitory activity for research and development, and new structure-activity information for chemical optimization of related spliceosome inhibitors.
RESULTS AND DISCUSSION
Identification of the Thailanstatin Biosynthetic Gene Cluster
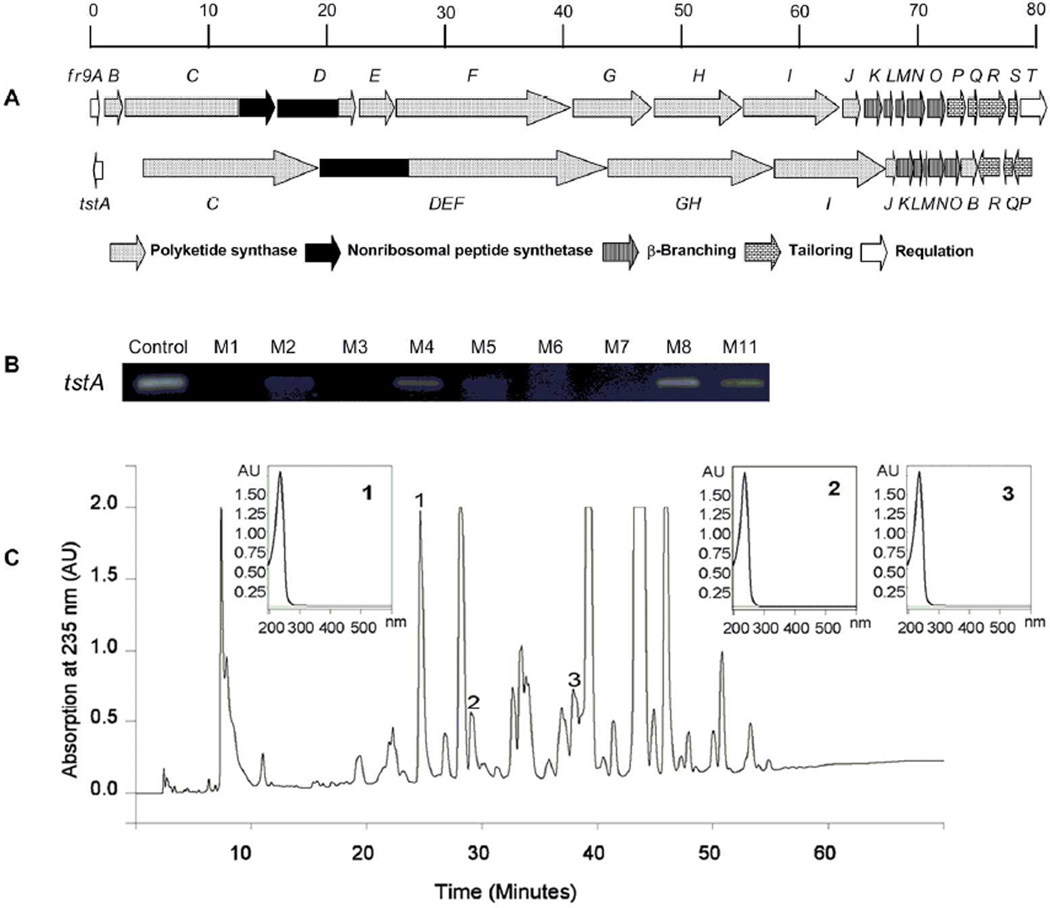
Mining the improved genome sequence of B. thailandensis MSMB4336 revealed a cryptic gene cluster highly homologous to the fr9 gene cluster responsible for the biosynthesis of 4 in Pseudomonas sp. No. 2663.37 This gene cluster (named tst, referring to the thailanstatin compounds it produces; see below) spans a 78.1-kb DNA region that contains 15 opening reading frames (ORFs, designated tstA through tstR), whereas two transposon-like segments are present between tstA and tstC, and tstR and tstQ (Figure 1A). Comparing the physical arrangement of the two gene clusters, tstDEF and tstGH are the apparent equivalents of the fused fr9D-fr9E-fr9F or fr9G-fr9H genes, respectively, tstB is translocated, and the orientation of tstA and tstP-tstQ-tstR is inverted. The most conspicuous difference with the fr9 gene cluster is the absence of the equivalent fr9S and fr9T genes from the tst gene cluster. All putative functions of the deduced gene products were assigned by sequence comparisons with the FR9 proteins and with other bacterial homologs (Table 1).
Figure 1.
Genomics-guided discovery of thailanstatins (1–3). (A) A graphical comparison of the FR901464 (4) biosynthetic gene cluster (fr9) and the thailanstatin biosynthetic gene cluster (tst). (B) Identification with RT-PCR analysis of three growth media in which the regulatory tstA gene is expressed. (C) HPLC profiling of a crude extract of B. thailandensis MSMB43 fermentation broth and identification of three compound peaks with a conjugated diene-like characteristic UV absorbance (UVmax at 235 nm).
Table 1.
Comparison of genes and deduced proteins between the thailanstatin biosynthetic gene cluster (tst) from Burkholderia thailandensis MSMB43 (GenBank accession no. JX307851) and the FR901464 biosynthetic gene cluster (fr9) from Pseudomonas sp. No. 2663 (GenBank accession no. HM047288).
| The tst gene cluster | The fr9 gene cluster | DNA sequence identity / gap |
Protein sequence similarity / identity |
Confirmed or deduced protein function |
||
|---|---|---|---|---|---|---|
| Gene | Deduced protein (# of aa) |
Gene | Deduced protein (# of aa) |
|||
| tstA | TstA (276) | fr9A | FR9A (294) | 71% / 1% | 71% / 57% | LuxR-type transcription regulator |
| tstC | TstC (4,997) | fr9C | FR9C (5,061) | 78% / 2% | 81% / 75% | PKS / Hybrid PKS-NRPS |
| tstDEF | TstDEF (8,036) | fr9D | FR9D (1,170) | 81% / 3% | 85% / 79% | Hybrid PKS-NRPS |
| fr9E | FR9E (1,188) | 79% / 3% | 81% / 76% | PKS | ||
| fr9F | FR9F (4,904) | 81% / 2% | 81% / 75% | PKS | ||
| tstGH | TstGH (4,855) | fr9G | FR9G (3,017) | 66% / 3% | 80% / 74% | PKS |
| fr9H | FR9H (2,662) | 69% / 4% | 83% / 77% | PKS | ||
| tstI | TstI (3,061) | fr9I | FR9I (3,052) | 67% / 3% | 75% / 69% | PKS |
| tstJ | TstJ (294) | fr9J | FR9J (296) | 81% / 1% | 86% / 80% | Acyltransferase |
| tstK | TstK (492) | fr9K | FR9K (429) | 86% / <1% | 88% / 82% | HMG-CoA synthase |
| tstL | TstL (261) | fr9L | FR9L (251) | 80% / <1% | 88% / 80% | Enoyl-CoA hydratase |
| tstM | TstM (79) | fr9M | FR9M (79) | 78% / <1% | 92% / 78% | Acyl carrier protein |
| tstN | TstN (422) | fr9N | FR9N (421) | 79% / <1% | 85% / 79% | KetosynthetaseS |
| tstO | TstO (392) | fr9O | FR9O (377) | 75% / 4% | 92% / 74% | Acyltransferase |
| tstB | TstB (322) | fr9B | FR9B (360) | NSHa | 53% / 35% | Acyltransferase |
| tstR | TstR (482) | fr9R | FR9R (482) | 85% / <1% | 93% / 87% | Cytochrome P450 |
| tstQ | TstQ (145) | fr9Q | FR9Q (145) | 73% / <1% | 88% / 69% | Cyclase |
| tstP | TstP (341) | fr9P | FR9P (341) | 81% / <1% | 91% / 85% | Dioxygenase |
| -- | -- | fr9S | FR9S (152) | --b / -- | -- / -- | Acetyltransferase |
| -- | -- | fr9T | FR9T (467) | -- / -- | -- / -- | Diguanylate cyclase |
NSHa, no significant homology; --b, not available.
Isolation and Characterization of Thailanstatins
Semi-quantitative RT-PCR analysis indicated that the regulatory tstA gene of the tst gene cluster was expressed in three of the nine media (M4, M8 and M11) tested in flask fermentation conditions, with the highest expression found in M8 medium (glucose, 5 g/L; peptone, 5 g/L; NaCl, 3 g/L; Na2HPO4, 1.2 g/L; KH2PO4, 0.5 g/L; pH 7.0) (Figure 1B). Compound 4 has a linear polyketide-peptide framework with three discrete chiral segments joined by conjugated linkages including a diene; this characteristic diene linkage gives the compound a maximal UV absorption at 235 nm.7 B. thailandensis MSMB43 was thus grown in a large volume in M8 medium and the HPLC profile of the ethyl acetate extract of the fermentation broth was inspected for compound peaks with a conjugated diene-like characteristic UV absorption (UVmax at 235 nm). This led to the identification of three such peaks (Figure 1C), and subsequent compound isolation and purification steps were guided by monitoring this wavelength.
Compounds 1, 2 and 3 (20.0 mg, 37.2 mg and 10.7 mg, respectively, all with a greater than 98% purity) were purified from the ethyl acetate extract of 160 L of B. thailandensis MSMB43 fermentation broth by UV-guided silica gel chromatography, ODS C18 chromatography and semi-preparative HPLC, and their structures and assignments were determined with a combination of high-resolution mass spectrometry (HR-MS), 1H, 13C and 15N NMR spectrometry, UV and infrared (IR) spectrometry (Supporting Materials, Method and Results, Supporting Tables S1–S3 and Supporting Figures S1–S35). Their physico-chemical properties are summarized in Supporting Table S4. Thailanstatins share the same linear polyketide-peptide framework with 4 but differ from 4 by lacking a hydroxyl group at the C1 position and by having an extra carboxyl moiety at the C17 position, 2 and 3 differ from 1 and 4 by having a chloride substituent instead of an epoxide functionality at the C3 position, and 3 differs from 2 by having a dimethyl acetyl group instead of an acetyl group at the distal end. The terminal carboxyl moiety imparts thailanstatins with a positive bromocresol green (BCG) visualization reaction (yellow spot on dark blue background) on thin layer chromatography (TLC)38 (Supporting Figure S38).
Proposed Biosynthetic Pathway of Thailanstatin A
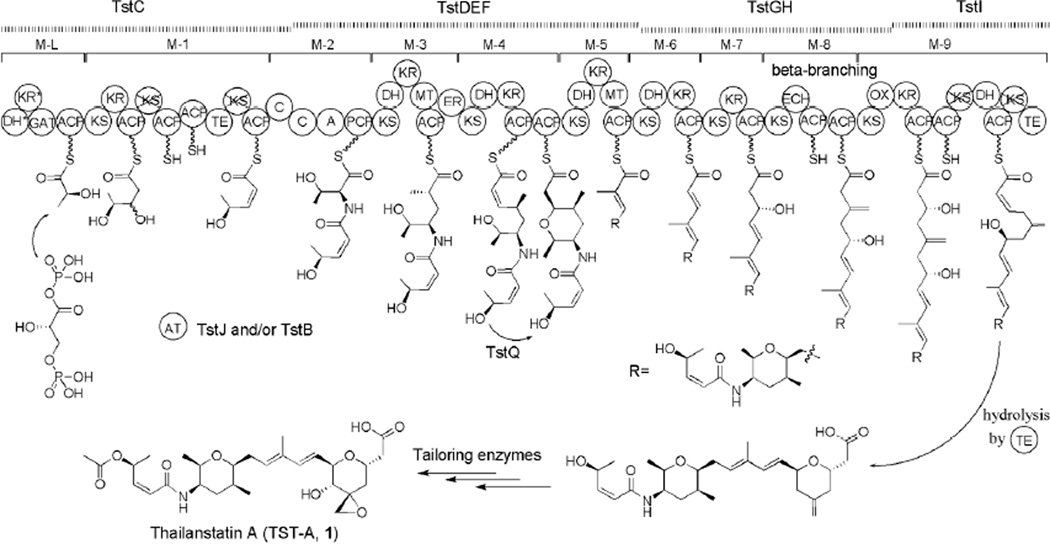
As the first identified natural product that inhibits eukaryotic pre-mRNA splicing, 4 is regarded as a prototype spliceosome inhibitor. The genetic basis for the biosynthesis of 4 in Pseudomonas sp. No. 2663 has been decoded recently by us.37 The biosynthetic (fr9) gene cluster spans an 80.6-kb DNA region that contains 20 genes (fr9A through fr9T) (Figure 1A and Table 1). Compound 4 is biosynthesized by a hybrid PKS-NRPS pathway which features with four acyltransferases (AT)-less PKSs complemented by three discrete ATs acting in trans, and with unusual PKS/NRPS organizations such as domain redundancy and domain inactivation. In addition, a 3-hydroxy-3-methylglutaryl-CoA synthase (HCS) is involved in polyketide β-branching, a glyceryl transferase/phosphatase (GAT) and an acyl carrier protein (ACP) in the start-module are proposed to incorporate D-1,3-bisphosphoglycerate, and an oxidative Baeyer-Villiger reaction is followed by chain release to form a pyran moiety.
The elucidated structure of thailanstatins and the detailed bioinformatic analysis of the tst gene cluster (Table 1 and Supporting Figure S39) allowed us to propose a model for the biosynthesis of 1 (Figure 2), which is similar to the biosynthetic route of 4.37 Overall, the tst gene cluster contains four genes that encode a total of 54 domains arranged into one loading module and nine extension modules of a proposed PKS-NRPS pathway, with a trans-acting AT activity supplemented by TstJ and/or TstB. The loading module of the hybrid PKS-NRPS assembly line has an unusual dehydratase (DH)*-ketoreductase (KR)*-GAT-ACP domain architecture and may be responsible for the incorporation of a glyceryl starter unit. During the chain elongation, a proposed cyclase (TstQ) may act on the intermediate tethered to the ACP domain of module 4, results in the formation of a tetrahydropyran moiety via intramolecular Michael addition. Five discrete proteins (TstK, TstL, TstM, TstN and TstO; omitted from Figure 2 for clarity), in addition to the enoyl CoA hydratase (ECH)-ACP-ACP domains located on TstGH, are predicted to be essential for β-branch incorporation. When the full-length product is achieved, the terminal thioesterase domain (TE) may catalyze the chain release via hydrolysis to generate a terminal carboxyl moiety and the formation of the second pyran moiety by a mechanism that may be similar to the formation of the first tetrahydropyran moiety. Additional tailoring steps, including hydroxylation, epoxidation and acetylation, are needed to mature the final product to form 1. The cytochrome P450 TstR and dioxygenase TstP are candidates for catalyzing the tailoring hydroxylation and epoxidation. However, an fr9S-equivalent gene encoding an acetyltransferase necessary for the tailoring acetylation is not present within the tst gene cluster, and therefore an as-yet unknown acetyltransferase may be involved in the acetylation step.
Figure 2.
Proposed biosynthetic pathway of thailanstatin A (1) in B. thailandensis MSMB43. Crossed domains are inactive or nonfunctional, star-labeled domains are atypical. See text for explanations. A, adenylation; ACP, acyl carrier protein; AT, acyltransferase; C, condensation; DH, dehydratase; ECH, enoyl-CoA hydratase; ER, enoyl reductase; GAT, glyceryl transferase/phosphatase; KR, ketoreductase; KS, ketosynthetase; MT, methyltransferase; OX, FAD-dependent monooxygenase; PCP, peptidyl carrier protein; TE, thioesterase.
Compound 2 is presumably derived from 1 through an epoxide ring opening reaction. This chemical SN2 reaction by a chlorine atom may be spontaneous or can be catalyzed by an unknown enzyme, such as one of the halohydrin dehalogenases which are known to be highly promiscuous enzymes that can catalyze enantioselective epoxide ring opening with different anionic nucleophiles.39 The route to 3 appears to be more complicated due to the presence of an unusual dimethyl acetyl group, which is probably derived from valine.
Reasoning that the tst gene cluster and the fr9 gene cluster are highly homologous and their encoded PKS-NRPS pathways possess a very similar module and domain organization, one would expect that the products biosynthesized by the two pathways should be the same. However, 1 differs from 4 by having an extra carboxyl moiety. We proposed that in the biosynthetic pathway leading to 4, an oxidative chain release mechanism is mediated by a Baeyer-Villiger monooxygenase (BVMO; OX) domain on FR9H, which results in the formation of the backbone of 4.37 In contrast, we propose that, in the biosynthetic pathway leading to 1 (Figure 2), the homologous OX domain on TstGH in the termination module skips its function, although detailed sequence analysis indicates that it would be an active enzyme (Supporting Figure S39C); consequently, a hydrolysis release of the product chain takes place to generate a terminal carboxyl moiety. Identifying other factors that dictate the differential chain release between the two homologous biosynthetic pathways requires further investigation.
Thailanstatins Are More Stable Than FR901464
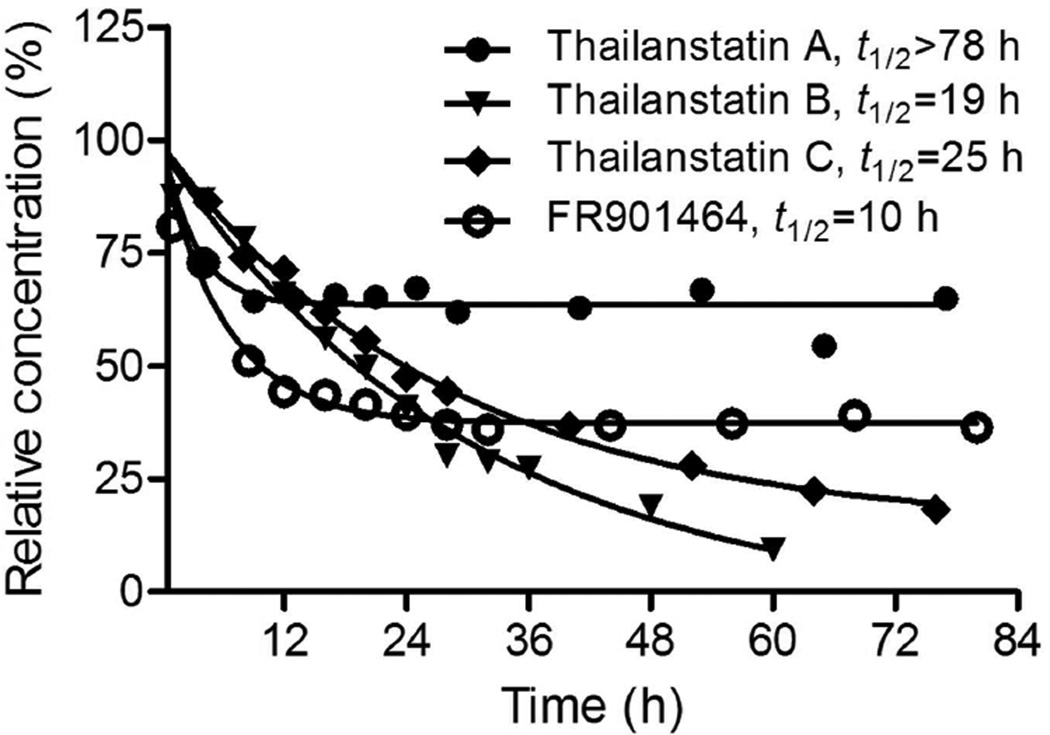
Compound 4 was reported to be unstable under physiologically relevant conditions in phosphate buffer at 37 °C, with a half-life (t1/2) of 4 h (pH 7) to 8 h (pH 7.4).40 To find out whether thailanstatins are more stable than 4, all four compounds were subjected to the same stability test in phosphate buffer (pH 7.4) at 37 °C. While the half-life of 4 was found to be 10 h, largely consistent with the previously assessed 8 h,40 the half-lives of 1–3 were found to be >78 h, 19 h and 25 h, respectively (Figure 3). A longer half-life is generally a more desirable property for cancer drug development. Interestingly, after a quick decay within the first 8 h, 1 remained at 65% of its starting level for at least 78 h as tested; 4 exhibited a similar trend of decay but remained at a much lower (38%) level. In contrast, 2 and 3 decayed continuously at slower paces but to very low levels over time. Each compound appeared to decay into a specific product whose chemical identity requires further investigation (Supporting Figure S40).
Figure 3.
Thailanstatins (1–3) are more stable than FR901464 (4). Compounds were placed in phosphate buffer (pH 7.4) at 37 °C and examined by HPLC at multiple time points.
The absence of a hydroxyl group at the C1 position and the presence of a carboxyl moiety at the C17 position on thailanstatins are bound to contribute to the compounds’ stability because these are the only structural differences between 1 and 4, but the relative contribution of these two moieties remain unclear. On the other hand, it seems likely that the absence of an epoxide group and presence of chlorine at the C3 position contributes to the more uniform decay pattern of 2 and 3. It was reported that a combination of the epoxide at the C3 position and the hydroxyl group at the C1 position causes a rapid decay of 4 under physiologically relevant conditions, and replacing the hydroxyl group with a methyl group has led to 7, a more stable analogue of 4.40 It was reasoned that the methylketal group in 7 has a greater chemical stability than the hemiketal group in 4.8, 40 Complete elimination of the hydroxyl group of 4 resulted in 5, a even more stable analogue of 4.8 Theoretically, the carboxyl moiety on thailanstatins may provide a buffering capacity under weak alkaline conditions, thus increases the stability of thailanstatins.
Thailanstatins Are Potent Pre-mRNA Splicing Inhibitors
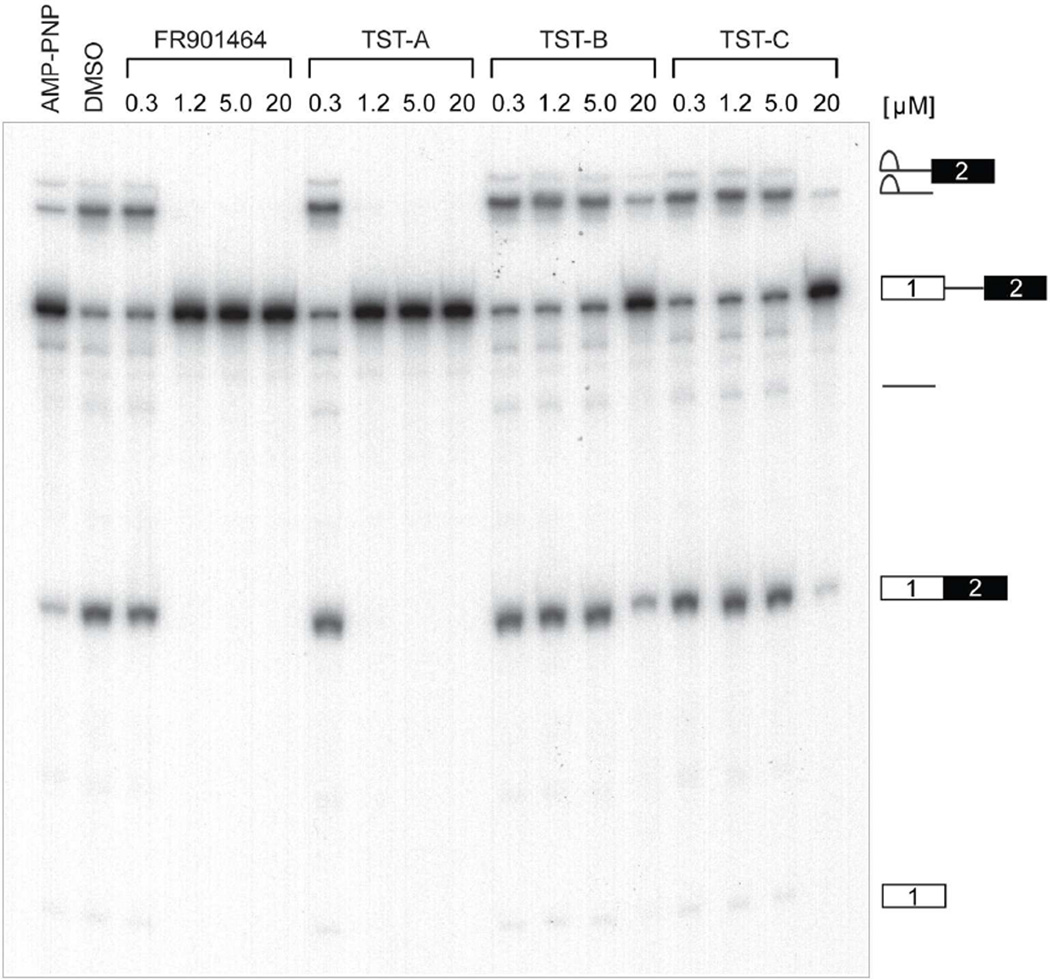
Compound 4, and a methylated derivative of this, 6, have been shown to be potent eukaryotic pre-mRNA splicing inhibitors that binds noncovalently to the SF3b subcomplex in the U2 snRNP particle of spliceosome, to prevent base-pairing interactions with sequences 5’ of the branch point.16, 41 Here, we measured the in vitro pre-mRNA splicing inhibitory activities of thailanstatins together with 4, using our recently described high-throughput assay termed EJIPT for exon-junction complex (EJC) immunoprecipitation.42 Splicing leaves in its wake a unique molecular signature in the form of an EJC that assembles ~20–24 nt upstream of splice junctions and whose core is comprised of four proteins (eIF4AIII, Y14, Magoh, and MLN51) which remain bound at their original position after export to the cytoplasm.43–45 Briefly, biotin tagged Ad2 pre-mRNAs were spliced in cell extracts for 90 minutes in the presence of increasing amounts of drug or the appropriate controls. Following 1 hr incubation with Y14 antibody immobilized on magnetic protein A beads, EJC-containing mRNAs were detected with a horseradish peroxidase (HRP)-conjugated secondary antibody, followed by the addition of an HRP substrate for enzymatic amplification and detection of chemiluminesence signals with a plate reader. Compound 4 dose-dependently decreased the EJIPT signal, with a half-maximal inhibitory concentration (IC50) of 580 nM. Compound 1 exhibited a similar activity in this assay, with an IC50 of 650 nM, whereas both 2 and 3 were nearly 10-fold less potent, each with IC50s of around 6 µM (Table 2). Taken together, the data suggests that 1 is as potent as 4 and that the lack of an epoxide group in 2 and 3 greatly influences this activity.
Table 2.
In vitro pre-mRNA splicing inhibitory activities. The final half-maximal inhibitory concentration (IC50 in µM) is the average of two independently performed triplicate assay results (in nM) with standard deviation provided.
| Compound | IC50-1 (nM) | IC50-2 (nM) | Final IC50 (µM) |
|---|---|---|---|
| Thailanstatin A (1) | 399.03 | 902.01 | 0.65±0.36 |
| Thailanstatin B (2) | 4432.58 | 7928.34 | 6.18±2.47 |
| Thailanstatin C (3) | 4786.66 | 8883.36 | 6.84±2.90 |
| FR901464 (4) | 535.56 | 633.09 | 0.58±0.07 |
To confirm these conclusions, conventional splicing gels were performed utilizing a different pre-mRNA substrate (CDC14–15) labeled with 32P-UTP. As expected, 1 inhibited splicing with the same potency as 4, while much greater concentrations of 2 and 3 were required to produce the same effect (Figure 4). These compounds caused pre-mRNA to accumulate and prevented the production of mRNA and splicing intermediates (e.g. lariat, lariat-exon 2) in the same manner as 4. This suggests they also target an early step in spliceosome dynamics presumably by inhibiting U2 snRNP’s recognition of the branch A sequence during assembly.
Figure 4.
Thailanstatins (1–3) inhibit pre-mRNA splicing in vitro. FR901464 (4) and 1–3 were added to splicing reactions containing 32P-labeled CDC14–15 pre-mRNA at the final concentrations indicated. AMP-PNP (5 mM) and DMSO were used as positive and negative controls, respectively. RNA was purified and resolved on denaturing PAGE and gels were autoradiographed at −80 °C. Identities of splicing intermediates are depicted to the right of gels.
Thailanstatins Possess Potent In Vitro Antiproliferative Activities
Standard MTT assays were performed to assess the antiproliferative activities of thailanstatins and 4 in four representative human cancer cell lines: DU-145 (prostate cancer), NCI-H232A (non-small cell lung cancer), MDA-MB-231 (triple-negative breast cancer) and SKOV-3 (ovarian cancer). All thailanstatins exhibited potent antiproliferative activities with half-maximal growth inhibitory concentrations (GI50s) in the single nM range and with an overall potency ranking of 1 > 2 > 3. Reference compound 4 appeared to be slightly more potent than 1 (Table 3). Amongst the four cancer cell lines, DU-145 appeared to be most sensitive to the 4-class of natural products. This overall ranking of antiproliferative activities correlates well with the in vitro pre-mRNA splicing trends.
Table 3.
Antiproliferative activities in human cancer cell lines. The half-maximal growth inhibition concentration (GI50 in nM) is the average of triplicate well results with standard deviation provided.
| GI50 (nM) | ||||
|---|---|---|---|---|
| Compound | DU-145 | NCI-H232A | MDA-MB-231 | SKOV-3 |
| Thailanstatin A (1) | 1.11±0.02 | 2.26±0.17 | 2.58±0.11 | 2.69±0.37 |
| Thailanstatin B (2) | 3.00±0.92 | 2.50±0.06 | 6.22±1.67 | 4.94±1.76 |
| Thailanstatin C (3) | 2.98±0.90 | 3.67±0.53 | 8.82±2.20 | 5.57±2.01 |
| FR901464 (4) | 1.05±0.02 | 1.94±0.24 | 2.10±0.19 | 1.06±0.01 |
Summary and Perspective
Collectively from a structure-activity relationship (SAR) point of view, the presence of a carboxyl moiety at the C17 position and the absence of a hydroxyl group at the C1 position of thailanstatins are advantageous to the compounds’ stability, and the highly reactive epoxide at the C3 position of 1 and 4 is critical for high potency in inhibiting pre-mRNA splicing as well as cancer cell proliferation. The epoxide ring opening through halogenation modification on 2 and 3 diminishes compounds’ bioactivities despite the fact that such a modification on many other natural products generally enhances bioactivities.46 Additional methyl groups on 3 (compared to 2) have no significant effect on compound bioactivity. While the highly reactive epoxide functionality that allows 1 to exhibit as potent in vitro pre-mRNA splicing inhibitory activity and antiproliferative activity as 4, the carboxyl moiety endows 1 a much greater degree of stability under physiologically relevant conditions than 4. Therefore, 1 is a better natural product candidate for potential anticancer drug development, and the new structure-activity information resulting from this work will facilitate chemical optimization of related spliceosome inhibitors.
EXPERIMENTAL SECTION
General Experimental Procedures
Melting point was measured with a Stuart® melting point apparatus SMP3. Optical rotation was measured with a JASCO DIP370 polarimeter. UV spectrum was obtained with a Cary 50 UV-Vis Spectrophotometer. IR spectrum was recorded on an AgCl silver slide on a Thermo Scientific NICOLET 380 FT-IR Spectrometer. 13C NMR spectrum was acquired on a Bruker DRX 500 MHz NMR spectrometer with a 5-mm BBO probe at 25 °C; all other NMR spectra were obtained on the same spectrometer with a 5-mm BBI probe at 25 °C. The proton and carbon NMR chemical shifts were calibrated using solvent signals (CDCl3: δH 7.26/δC 76.7, CD2Cl2: δH 5.32/δC 53.8) as references. For 1H-15N HSQC spectrum, chemical shift was referenced to liquid NH3 (0 ppm) at 25 °C. NMR data analyses were performed with a Bruker TopSpin 3.1.b.11 software package. HR-MS analyses were carried out on an LC/MSD-TOF mass spectrometer (Agilent, Palo Alto, CA) in positive mode. The samples were directly injected with a 5-µL aliquot at 30 µL mL−1 from the 4.6 mm diameter auto-syringe delivery system (Harvard Apparatus, Holliston, Massachusetts) in 100% Methanol. The following instrumental parameters were used to generate the optimal protonated ions [M + H]+ in positive mode: capillary voltage 3400 V; drying Gas 6.0 L min−1; nebulizer 20 psi; gas temperature 325 °C; oct DC1 39.5 V; fragmentor 180 V; oct RF 250 V; skimmer 60V. Internal calibration was achieved with assisted spray of two reference masses, 922.0098 m/z and 121.0509 m/z. Acquired data was processed using Analyst QS 1.1 build: 9865 software (Agilent, Palo Alto, CA) to extract parent masses observed in the range from 100 to 3200 AMU (atomic mass units), formulas were generated with integrated molecular mass calculator. Analytical HPLC was performed on the Varian ProStar HPLC system equipped with an Agilent Eclipse XDB-C18 column (4.6 × 250 mm, 5 µm), UV absorbance was monitored at 235 nm, according to a described protocol.37 Briefly, analyte was centrifuged at 10,000 g for 3 min and 10 µL of the supernatant was injected into the column. The column was first eluted by a linear gradient from 15% to 55% acetronitrile (+ 0.1% FA) in 35 min, then by a linear gradient from 55% to 100% acetronitrile (+ 0.1% FA) in 20 min, and then isocratically in 100% acetronitrile (+ 0.1% FA) for 8 min. Standard curves were generated with the peak areas of purified compounds, and applied back to calculated the titer and final yield of each compound.
Bacterial Strains
Burkholderia thailandensis MSMB43 strain, originally isolated by Dr. Bart Currie and associates at Darwin University, Australia,47 was obtained under a Material Transfer Agreement from the US Centers for Diseases Control (CDC). Pseudomonas sp. No. 2663 strain, the producer of 4,7 was purchased from the National Institute of Bioscience and Human-Technology, Agency of Industrial Science and Technology, Japan.
Gene Cluster Analysis
The first draft genome sequence of B. thailandensis MSMB43 was reported by T. D. Read and associates,48 but the quality of was found to be suboptimal for our natural product discovery purpose due to insufficient coverage. We thus re-sequenced the bacterial genome and obtained a significantly improved draft genome sequence, which contains at least 13 secondary metabolite biosynthetic gene clusters.36 Searches for homologous gene and protein sequences were performed with Blast algorithms available at the NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi/). Sequence alignments were performed by BioEdit sequence alignment editor (Ibis Biosciences). Analysis of the domain and module organization of PKSs and NRPS was carried out with a search engine available at the NRPS-PKS database49 The nucleotide sequence of thailanstatin biosynthetic (tst) gene cluster has been deposited into the GenBank under accession no. JX307851.
Reverse Transcription (RT)-PCR
Cultivation of B. thailandensis MSMB43 in nine growth media (see details in ref 29) and RT-PCR detection of the expression conditions of a key regulatory gene, tstA, was performed according to previously described protocols.29 Primers used in RT-PCR are as follows: tstA-RT-PCR-FP (5’-CCAGCATTGGCGCCGAGCGC-3’) and tstA-RT-PCR-RP (5’-GGTCGGAGCACGTTCATGTGG-3’). This primer set was designed to generate a 277-bp product. RNA samples that had not been subjected to reverse transcription were used as negative controls; a positive control amplified the tstA sequence from bacterial genomic DNA template.
Isolation and Identification of Thailanstatins and FR901464, and General Physical Chemical Analyses
Details are provided in Supporting Information. Compound 4 was freshly purified from the fermentation culture of Pseudomonas sp. No. 2663 and was used as a reference compound in this study. Compounds were dissolved in DMSO as 10 mM stock solutions for all subsequent experiments.
Stability Test
Compound stability was tested according to a published method.50 Briefly, a 15-mL Corning tube was filled with 10 mL of phosphate buffer (pH 7.4), 20 µL of benzoic acid (10 mM in DMSO), and 70 µL of DMSO under an open atmosphere and warmed to 37 °C. Ten µL of each compound (10 mM in DMSO) was added to the reaction mixture and the resulting mixture was sealed with a cap, vortexed for 15 s, and then placed in a 37 °C incubator. HPLC analysis for compound stability was done with an Agilent Eclipse Plus C18 column (4.6 × 100 mm, 3.5 µm). A 20 µL sample was injected into the HPLC system. Flow rate was 0.5 mL min−1, UV absorbance was monitored at 235 nm. For 1, the sample was eluted isocratically with 40% acetonitrile in water (containing 0.1% formic acid, FA). For other compounds, a linear gradient elution from 10% acetonitrile in water (containing 0.1% FA) to 100% acetonitrile was achieved in 14 min. The resulting data were normalized by the ratio factor of compound/benzoic acid at time zero, and were analyzed with GraphPad Prism 5.01 software. The half-life t1/2 was generated by a nonlinear fit analysis of these data.
In Vitro Pre-mRNA Splicing Inhibition Assay
32P-UTP-labeled CDC14–15 pre-mRNA was transcribed in vitro according to the Riboprobe Combination SP6/T7 system (Promega). The Megascript T7 kit (Ambion) was used to generate Ad2ΔIVS biotin-labeled pre-mRNAs with a biotin-16-UTP (Roche) to cold UTP ratio of 1:14 (350 µM final). Pre-mRNAs were gel purified, phenol:chloroform extracted, ethanol precipitated and resuspended in nuclease free water. Whole cell splicing extracts from HEK 293T cells were prepared and in vitro splicing reactions of 32P-labeled CDC14–15 pre-mRNAs were performed as described.51 Samples were run on 6% urea-PAGE and autoradiographed at −80 °C. Splicing-dependent exon junction complex immunoprecipitation (EJIPT) assay was performed as recently described.42 Briefly, 10 µL splicing reactions consisting of 1X SP buffer, 40 µg extract, 20 nM biotin Ad2 pre-mRNA, and 1 U µL−1 of RNasin were assembled on ice in 96-well plates with or without drug in serial dilutions and incubated for 1.5 hr. All incubations were performed at 30 °C in an Eppendorf tabletop shaker set at 750 rpm. Y14 antibody (4C4; 0.7 µL per well) immobilized on Protein A magnetic beads (Invitrogen; 8.6 µL per well) for >1 hr at 4 °C in 1X PBS/0.1% NP-40 prior to splicing was then washed in HNT buffer (20 mM HEPES-KOH pH 7.9, 150 mM NaCl, 0.5% Triton X-100) and 100 µL suspensions were added to each well. Immunoprecipitation was carried out for 1 hr, beads were washed five times (200 µL of HNT) on a KingFisher apparatus, then biotinylated RNA/protein complexes bound to 4C4 were reacted with an avidin-HRP conjugate (Pierce; 1:25,000 dilution) for another hour. Following five additional washes, Super Signal ELISA Fempto chemiluminescent substrate (150 µL) was added to beads and luminescence was read on a Wallac Victor multi-label counter. The log values of compound concentrations were plotted against the relative luminescence units with Sigma plot 12.0. The IC50 values were generated by a 4-parameter logistic nonlinear regression fit analysis of two independent sets of experiment results.
Antiproliferative Assay
Cancer cell lines (DU-145, NCI-H232A, MDA-MB-231 and SKOV-3) were obtained as gift from Professor Kit Lam at the University of California-Davis Medical Center. Cell proliferation was evaluated using the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) dye reduction assay according to a detailed protocol available at the NCI-DTP server (http://www.dtp.nci.nih.gov/branches/btb/ivclsp.html), with slight modifications and performed in triplicate. Briefly, cells were seeded in 96 well plates at 10,000 or 15,000 cells per well and incubated overnight at 37 °C. Cells were then exposed to various concentrations (serial three-fold dilutions) of a drug for 48 h at 37 °C. After MTT solution was added, and cells were incubated at 37 °C for 4 h. The absorption signals were measured at 570 and 690 nm on an Infinite M200 TECAN plate reader. Growth inhibition was calculated as defined by the National Cancer Institute [viability (%) =100 × (T−T0)/(C−T0); T0 = cell density at time zero; T = cell density of the test well after period of exposure to test compound; C = cell density of the vehicle treated]. The log values of compound concentrations were plotted against normalized cell viability (%) with GraphPad Prism 5.01. The GI50 value was generated by a nonlinear fit analysis of the data.
Supplementary Material
ACKNOWLEDGEMENT
We thank Jay Gee (US Centers for Diseases Control, CDC) for facilitating the distribution of B. thailandensis MSMB43 strain from CDC to Y.-Q.C., Holger Foersterling (University of Wisconsin-Milwaukee NMR Facility) for assistance with NMR analysis, Grzegorz Sabat (University of Wisconsin-Madison Genetics-Biotechnology Center) for collecting HR-MS data, and Vyara Matson and Douglas Steeber for assistance with MTT assays. This work was supported in parts by a Catalyst Award from the University of Wisconsin-Milwaukee Research Foundation (to Y.-Q.C) and a grant from NIH/NCI (R01CA152212 to Y.-Q.C.). G.-L.T. and L.Z. are Awardees of the National Distinguished Young Scholar Program in China.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supporting materials, methods, results and references, and tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper TA, Wan L, Dreyfuss G. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaharieva E, Chipman JK, Soller M. Toxicology. 2012;296:1–12. doi: 10.1016/j.tox.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima H, Takase S, Terano H, Tanaka H. J. Antibiot. (Tokyo) 1997;50:96–99. doi: 10.7164/antibiotics.50.96. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K. J. Antibiot. (Tokyo) 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima H, Sato B, Fujita T, Takase S, Terano H, Okuhara M. J. Antibiot. (Tokyo) 1996;49:1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 8.Thompson CF, Jamison TF, Jacobsen EN. J. Am. Chem. Soc. 2001;123(41):9974–9983. doi: 10.1021/ja016615t. [DOI] [PubMed] [Google Scholar]
- 9.Motoyoshi H, Horigome M, Ishigami K, Yoshida T, Horinouchi S, Yoshida M, Watanabe H, Kitahara T. Biosci. Biotechnol. Biochem. 2004;68:2178–2182. doi: 10.1271/bbb.68.2178. [DOI] [PubMed] [Google Scholar]
- 10.Osman S, Albert BJ, Wang Y, Li M, Czaicki NL, Koide K. Chemistry. 2011;17:895–904. doi: 10.1002/chem.201002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizui Y, Sakai T, Iwata M, Uenaka T, Okamoto K, Shimizu H, Yamori T, Yoshimatsu K, Asada M. J. Antibiot. (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 12.Sakai T, Asai N, Okuda A, Kawamura N, Mizui Y. J. Antibiot. (Tokyo) 2004;57:180–187. doi: 10.7164/antibiotics.57.180. [DOI] [PubMed] [Google Scholar]
- 13.Sakai T, Sameshima T, Matsufuji M, Kawamura N, Dobashi K, Mizui Y. J. Antibiot. (Tokyo) 2004;57:173–179. doi: 10.7164/antibiotics.57.173. [DOI] [PubMed] [Google Scholar]
- 14.Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. ACS Chem. Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eskens FA, Ramos FJ, Burger H, de Jonge MJ, Wanders J, Lopez-Anaya A, Baselga J, Tabernero J. J. Clin. Oncol. (Meeting Abstracts) 2009;27:3508. [Google Scholar]
- 16.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 17.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 18.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, Mizui Y. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 19.Xie Y, Wright S, Shen Y, Du L. Nat. Prod. Rep. 2012;29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross H, Loper JE. Nat. Prod. Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 21.Bode HB. Curr. Opin. Chem. Biol. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Duran N, Menck CF. Crit. Rev. Microbiol. 2001;27:201–222. doi: 10.1080/20014091096747. [DOI] [PubMed] [Google Scholar]
- 23.Donadio S, Monciardini P, Sosio M. Nat. Prod. Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Nat. Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 25.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. J. Am. Chem. Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 26.Seyedsayamdost MR, Chandler JR, Blodgett JA, Lima PS, Duerkop BA, Oinuma K, Greenberg EP, Clardy J. Org. Lett. 2010;12:716–719. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biggins JB, Gleber CD, Brady SF. Org. Lett. 2011;13:1536–1539. doi: 10.1021/ol200225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klausmeyer P, Shipley SM, Zuck KM, McCloud TG. J. Nat. Prod. 2011;74:2039–2044. doi: 10.1021/np200532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Henkes LM, Doughty LB, He M, Wang D, Meyer-Almes FJ, Cheng Y-Q. J. Nat. Prod. 2011;74:2031–2038. doi: 10.1021/np200324x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Flemming CJ, Cheng Y-Q. Med. Chem. Comm. 2012;3:976–981. doi: 10.1039/C2MD20024D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke J, Ishida K, Hertweck C. Angew. Chem. Int. Ed. Engl. 2012;51:11611–11615. doi: 10.1002/anie.201205566. [DOI] [PubMed] [Google Scholar]
- 32.Challis GL. Microbiology. 2008;154(Pt 6):1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- 33.McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco AM, Zazopoulos E, Farnet CM. J. Nat. Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 34.Zotchev SB, Sekurova ON, Katz L. Curr. Opin. Biotechnol. 2012;23:941–947. doi: 10.1016/j.copbio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Liu XY, Wang C, Cheng Y-Q. Acta Crystallogr. Sect. E Struct. Rep. Online. 2012;68(Pt 9):o2757–o2758. doi: 10.1107/S160053681203601X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuo Y, Liu L, Wang Q, Liu X, Ren B, Liu M, Ni P, Cheng Y-Q, Zhang L. J. Bacteriol. 2012;194:4749–4750. doi: 10.1128/JB.00931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, He HY, Tang MC, Tang YM, Zhou Q, Tang G-L. J. Am. Chem. Soc. 2011;133:2452–2462. doi: 10.1021/ja105649g. [DOI] [PubMed] [Google Scholar]
- 38.Wardas W, Lipska I, Bober K. Acta Pol. Pharm. 2000;57:15–21. [PubMed] [Google Scholar]
- 39.Hasnaoui-Dijoux G, Majeric Elenkov M, Lutje Spelberg JH, Hauer B, Janssen DB. Chembiochem. 2008;9:1048–1051. doi: 10.1002/cbic.200700734. [DOI] [PubMed] [Google Scholar]
- 40.Albert BJ, Sivaramakrishnan A, Naka T, Czaicki NL, Koide K. J. Am. Chem. Soc. 2007;129:2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corrionero A, Minana B, Valcarcel J. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg MG, Wan L, Younis I, Diem MD, Soo M, Wang C, Dreyfuss G. Mol. Cell Biol. 2012;32:1271–1283. doi: 10.1128/MCB.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 44.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neumann CS, Fujimori DG, Walsh CT. Chem. Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Gee JE, Glass MB, Novak RT, Gal D, Mayo MJ, Steigerwalt AG, Wilkins PP, Currie BJ. BMC Microbiol. 2008;8:54. doi: 10.1186/1471-2180-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukhopadhyay S, Thomason MK, Lentz S, Nolan N, Willner K, Gee JE, Glass MB, Inglis TJ, Merritt A, Levy A, Sozhamannan S, Mateczun A, Read TD. J. Bacteriol. 2010;192:6313–6314. doi: 10.1128/JB.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. Nucleic Acids Res. 2004;32(Web Server issue):W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albert BJ, McPherson PA, O'Brien K, Czaicki NL, Destefino V, Osman S, Li M, Day BW, Grabowski PJ, Moore MJ, Vogt A, Koide K. Mol. Cancer Ther. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kataoka N, Dreyfuss G. Methods Mol. Biol. 2008;488:357–365. doi: 10.1007/978-1-60327-475-3_23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.