Abstract
B. dermatitidis belongs to a group of thermally dimorphic fungi that grow as sporulating mold in the soil and convert to pathogenic yeast in the lung following inhalation of spores. Knowledge about the molecular events important for fungal adaptation and survival in the host remains limited. The development of high-throughput analytic tools such as RNA sequencing (RNA-Seq) has potential to provide novel insight on fungal pathogenesis especially if applied in vivo during infection. However, in vivo transcriptional profiling is hindered by the low abundance of fungal cells relative to mammalian tissue and difficulty in isolating fungal cells from the tissues they infect. For the purpose of obtaining B. dermatitidis RNA for in vivo transcriptional analysis by RNA-Seq, we developed a simple technique for isolating yeast from murine lung tissue. Using a two-step approach of filtration and centrifugation following lysis of murine lung cells, 91% of yeast cells causing infection were isolated from lung tissue. B. dermatitidis recovered from the lung yielded high-quality RNA with minimal murine contamination and was suitable for RNA-Seq. Approximately 87% of the sequencing reads obtained from the recovered yeast aligned with the B. dermatitidis genome. This was similar to 93% alignment for yeast grown in vitro. The use of near-freezing temperature along with short ex vivo time minimized transcriptional changes that would have otherwise occurred with higher temperature or longer processing time. In conclusion, we have developed a technique that recovers the majority of yeast causing pulmonary infection and yields high-quality fungal RNA with minimal contamination by mammalian RNA.
Keywords: Blastomyces dermatitidis, RNA-Seq, in vivo transcriptional profiling, experimental murine infection
1. Introduction
Blastomyces dermatitidis, the etiologic agent of blastomycosis, belongs to a group of human pathogenic fungi that exhibit thermal dimorphism (Gauthier and Klein, 2008). These pathogens include Histoplasma capsulatum, Coccidioides immitis, Coccidioides posadasii, Paracoccidioides brasiliensis, Sporothrix schenckii, and Penicillium marneffei (Gauthier and Klein, 2008). In the soil (22–25°C), these fungi grow as mold that produce infectious conidia. Following soil disruption, aerosolized conidia inhaled into the lungs of a mammalian host (37°C) convert into pathogenic yeast to cause pneumonia (Gauthier and Klein, 2008; Schwarz and Baum, 1951). Once pulmonary infection is established, these pathogens can disseminate to almost any organ in the body including the brain, skin, and bone (Gauthier et al., 2007)
At core body temperature, the ability of B. dermatitidis to grow as yeast is critical for pathogenesis and essential for virulence (Nemecek et al., 2006; Finkel-Jimenez et al., 2002; Finkel-Jimenez et al., 2001). Despite the importance of yeast growth in vivo, few genes have been identified that contribute to survival in lung tissue at 37°C. Upregulation of Blastomyces adhesion-1 (BAD1), which is expressed only in the yeast phase, induces down-regulation of tumor necrosis factor alpha (TNF-α) to facilitate evasion of host immune defenses (Finkel-Jimenez et al., 2002; Finkel-Jimenez et al., 2001). Blastomyces yeast-phase-specific 1 (BYS1) is highly expressed in the yeast phase and has a minor influence on B. dermatitidis yeast morphology in vitro at 37°C and during murine pulmonary infection (Krajaejun et al., 2010). With the exception of dimorphism-regulating kinase (DRK1) controlling the morphologic switch to yeast (Nemecek et al., 2006), knowledge about genes that promote survival and morphogenesis in vivo remains limited to BAD-1 and BYS1, respectively.
The development of high-throughput analytic tools such as gene expression microarrays and RNA sequencing (RNA-Seq) allow for transcript analysis on a genome-wide scale and has provided new insights on fungal pathogenesis (Ngamskulrungroj et al., 2011; Nguyen and Sil, 2008; Dhamgaye et al., 2012). The application of these techniques for human fungal pathogens has been primarily limited to in vitro growth conditions including co-cultivation with cell culture lines and ex vivo experiments using murine macrophages (Ngamskulrungroj et al., 2011; Nguyen and Sil, 2008; Jong et al., 2008; Monteiro et al., 2009; Tavares et al., 2007; Johannesson et al., 2006; Lin et al., 2012; Thewes et al., 2007). The ability to capture transcriptional events on a genome-wide scale for pathogenic yeast during infection has potential to provide novel insight on mechanisms used by fungi for adaptation and survival in tissue to cause disease. However, in vivo transcriptional profiling is hindered by the low ratio of fungal cells to mammalian cells and the difficulty of isolating fungi from tissue. Carryover of excessive mammalian RNA results in suboptimal hybridization of fluorescently labeled fungal cDNA for gene expression microarrays (Thewes et al., 2007). For RNA-Seq, there is potential for reduced sensitivity to detect low abundance transcripts and difficulty distinguishing fungal from mammalian transcripts if excessive mammalian nucleic acids are present.
Although several techniques have been described for isolation of fungi from the kidney, liver or gastrointestinal tract (Thewes et al., 2007; Zakikhany et al., 2007; Rosenbach et al., 2010; White et al., 2007; Andes et al., 2005; Walker et al., 2009), isolation of fungi from lung tissue has been limited to those cells obtained by bronchoalveolar lavage (Hu et al., 2008; McDonagh et al., 2008). None of these existing techniques could be efficiently applied to B. dermatitidis pulmonary infection because yeast recovered by bronchoalveolar lavage would be limited to cells in the alveolar and endobronchial spaces, but not within pyogranulomas. Herein, we describe a simple, new two-step technique to isolate B. dermatitidis yeast from lung tissue for the purpose of analyzing the transcriptional response of this pathogen during experimental murine infection. This technique eliminates the majority of murine RNA and yields high-quality, fungal RNA suitable for in vivo transcriptional analysis by RNA-Seq.
2. Methods
2.1 Strains and Media
Blastomyces dermatitidis American Type Culture Collection (ATCC) strain 26199 was used for in vivo and in vitro experiments. This strain was isolated from a patient from South Carolina and is highly virulent in a murine model of infection (Brandhorst et al., 1999; Wüthrich et al., 2002). B. dermatitidis cultures were maintained as yeast on Histoplasma macrophage medium (HMM) at 37°C (Worsham and Goldman, 1988).
2.2 Murine Infection
B. dermatitidis ATCC 26199 yeast were used to infect C57BL/6 male mice (7 weeks of age). Each mouse received 2 × 103 B. dermatitidis yeast intratracheally using the protocol described by Wüthrich and colleagues (Wüthrich et al, 2002). Following inoculation, mice were clinically monitored for signs and symptoms of infection. At 17 days post infection, mice were euthanized by carbon dioxide and the lungs were immediately excised for isolation of yeast.
2.3 Isolation of B. dermatitidis yeast from murine lung tissue
Excised lungs were placed in individual 50 ml polypropylene conicals containing 20 ml of double-distilled, sterile water (ddH2O) prechilled (4°C) and supplemented with 100 μl DNase I (10 ug/ml; Roche). Twenty milliliters was selected because smaller volumes such as 5 ml resulted in coagulation of blood at near-freezing temperature, which in turn, impaired isolation of yeast from lung tissue. Water was used to facilitate lysis of murine cells without altering integrity of B. dermatitidis yeast. Excised lungs were homogenized for 15 seconds using an Omni TH tissue homogenizer at full speed (Omni International, Kennesaw, GA).
Following homogenization, a two-step process consisting of filtration and centrifugation was used to isolate B. dermatitidis yeast. Homogenized samples were passed through a 40 μm nylon mesh cell strainer (ThermoFisher Scientific, Waltham, MA). The rubberized end of a syringe plunger was used to help press the homogenate through the strainer. Tissues larger than 40 μm were retained, whereas B. dermatitidis yeast (8–15 μm in size) and finely homogenized tissues were collected. To separate yeast from the remaining lung tissue fragments, samples were centrifuged at 770 × g for 5 minutes at 4°C. B. dermatitidis – yeast formed a pellet, while the murine tissue was located in the supernatant and the interface (between the pellet and supernatant). The supernatant and interface were removed using a serologic pipette (referred to as supernatant-interface fraction or SIF). The yeast pellet was resuspended in 500 μl of ice-cold sterile ddH2O and transferred to a new polypropylene conical tube. This was followed by a 20 ml wash using ice-cold sterile ddH2O and centrifugation at 770 × g for 5 minutes at 4°C. The wash step facilitated removal of residual murine tissue. The supernatant was removed using a serologic pipette, and the yeast pellet (referred to as recovered yeast fraction or RYF) was suspended in 1 ml of ice-cold sterile ddH2O and rapidly frozen in liquid nitrogen in a drop-by-drop manner. Samples were kept on ice at all times. To minimize time spent ex vivo, mice were euthanized in pairs and lungs were processed immediately. Time ex vivo was less than 30 minutes and except for lung harvest, samples were kept at near-freezing temperature during all processing steps. For quality control, the SIF and 40 μl of the RYF were saved for microscopic analysis and quantitation of yeast enrichment. Yeast in the RYF and SIF were enumerated using a hemocytometer.
2.4 Analysis of the effect of temperature, time, and water treatment
Cultures of B. dermatitidis yeast were grown while shaking (220 rpm) in liquid HMM overnight at 37°C and collected by centrifugation (770 × g for 2 minutes). Parallel culture pellets were treated in several different ways: 1) immediately frozen in liquid nitrogen; 2) resuspended in 20 ml of ddH2O either pre-chilled to 4°C or pre-warmed to 37°C, recentrifuged, and the pellets frozen in liquid nitrogen (no-incubation sample), 3) resuspended in 20 ml of ddH2O either pre-chilled to 4°C or pre-warmed to 37°C and held without shaking for 30 minutes, 6 hours, and 24 hours at either 4°C or 37°C in 50 ml polypropylene conicals prior to centrifugation and freezing in liquid nitrogen. All frozen yeast samples were stored at −80°C prior to total RNA extraction.
2.5 Isolation of total RNA
Total RNA was extracted from yeast using the phenol-guanidinium thiocyanate-1-bromo-3-chloropropane extraction method (Sambrook and Russell, 2001). Frozen yeast cells were ground into a fine powder by a mortar and pestle and resuspended in TRI Regent (Molecular Research Center, Inc., Cincinnati, OH). Following separation by 1-bromo-3-chloropropane (Molecular Research Center, Inc., Cincinnati, OH), RNA was precipitated using isopropanol and high-salt solution (1:1 ratio; Molecular Research Center, Inc.), washed with 75% ethanol, and resuspended in RNase-free water. To remove residual DNA for qRT-PCR or RNA-Seq, samples were treated in solution with Turbo DNase (BioRad, Hercules, CA) followed by additional purification using an RNeasy kit (Qiagen). Nanodrop spectrophotometry (Nanodrop Products, Wilmington, DE), 0.8% agarose gel electrophoresis, and an Agilent Bioanalyzer (Agilent Technologies; Santa Clara, CA) were used to assess RNA integrity. Removal of genomic DNA was confirmed by PCR on cDNA using primers spanning the intron of alpha-1,3-glucan synthase.
2.6 Quantitative Real-Time PCR (qRT-PCR)
Total RNA extracted from RYFs and in vitro cultures was treated in solution with TurboDNase (BioRad) and cleaned using RNeasy column (Qiagen) to remove genomic DNA and other contaminants. Following this, 1 μg of RNA was converted to cDNA using an iScript cDNA synthesis kit (Bio-Rad) per manufacturer’s instructions. Total RNA extracted from infected lungs that were frozen in liquid nitrogen immediately after excision (referred to as whole lung RNA) was treated in similar fashion. Quantitative real-time PCR was performed with SsoFast EvaGreen Supermix (Bio-Rad) using a MyiQ real-time PCR detection system (Bio-Rad). Cycle conditions were as follows: 95°C 30 seconds, 1 cycle; 95°C 5 seconds, 60°C 10 seconds, 40 cycles. Following completion of qRT-PCR, melt curve analysis was performed. Primers used for qRT-PCR were B. dermatitidis GAPDH forward ACCCCCGCTCCTCCATCTTC, B. dermatitidis GAPDH reverse GAGTAGCCCCACTCGTTGTCATACC, M. musculus FTH1 forward GCTGAATGCAATGGAGTGTG, M. musculus FTH1 reverse GTCACATAAGTGGGGATCATTCT, TPS1 forward GGCAGCATCATCGTCAATC, TPS1 reverse CTCTGTTCATCACTCATCGTCAC, TPS2 forward CAAGCGTGGAAGAGGAGAAG, TPS2 reverse GCCGTTGATAGTGGAAACAAG, YPD1 forward AAAGAGGCGTGCGAAAAG, YDP1 reverse CTTCAGCGAGACTTCCTTATTG, HOG1 forward TCCGCAGGAGTCGTCCATC, and HOG1 reverse GCAAGACCAAAGTCGCAAATC. Relative expression (RE) was calculated by the method of RE = 2−ΔCt or 2−ΔΔCt (Livak and Schmittgen, 2001).
2.7 RNA sequencing (RNA-Seq)
Total RNA isolated from yeast during in vivo infection (two pools of 5 mice each) and yeast grown in liquid HMM (two replicates) was analyzed by RNA-Seq. The amount of RNA submitted for sequencing was 8.5 μg for pool #1, 8.2 μg for pool #2, 8.5 μg for in vitro biological replicate #1, and 8.8 μg for in vitro biological replicate #2. Strand-specific libraries were constructed for all RNA samples using the dUTP second strand marking method (Parkhomchuk et al., 2009; Levin et al., 2010) as previously described (Cuomo et al., 2012). Briefly, polyA RNA was purified using the Dynabeads mRNA Purification kit (Life Technologies, Grand Island, NY), and the resulting mRNA was treated with the Turbo DNA-free kit. Fragmentation buffer (Affymetrix, Santa Clara, CA) was added to samples on ice; samples were incubated at 80°C for 4 minutes, and then on ice for 5 minutes. RNAClean XP beads (Beckman Coulter Genomics) were used to purify the RNA. To convert the mRNA to cDNA, SuperScript Double-Stranded cDNA synthesis kit (Life Technologies) was used. For second strand synthesis, dTTP was replaced by dUTP. The cDNA samples were used to make Illumina-adapted libraries, starting at the end-repair step (Fisher et al., 2011). Samples were pooled and sequenced on the Illumina HiSeq to generate 101 base reads.
RNA-Seq quality was first evaluated by examining the percent called pass filter (PF) by the Illumina pipeline; all samples contained at least 93% PF reads. Base quality measured by Q20 (1 error every 100 bases) was also very high, with at least 90% of bases showing at least Q20 for all samples. Reads were aligned to the B. dermatitidis 26199 genome (Genbank accession AEII01000000) using the Burrows-Wheeler alignment tool (Li and Durbin, 2009). To estimate the proportion of reads with non-specific mapping to both the Blastomyces and mouse (mm9, http://genome.ucsc.edu/cgi-bin/hgGateway?db=mm9) genomes, reads were re-aligned to each genome using bowtie and the proportion of reads with alignments to both genomes was calculated (Langmead et al., 2009).
3. Results
3.1 Efficient isolation of B. dermatitidis yeast from murine lung tissue
At 17 days post infection, lungs from 11 mice were enlarged with multiple granulomas as expected (data not shown). The lungs were excised, homogenized in sterile water at near-freezing temperature, passed through a nylon-mesh filter, and yeast cells were separated from lung tissue by centrifugation. B. dermatitidis yeast, but not pseudohyphae or hyphae, were found in the recovered yeast fraction (RYF) and the fraction containing the supernatant and interface fraction (SIF) (Figure 1A, B).
Figure 1. Recovered yeast fraction and supernatant-interface fractions.
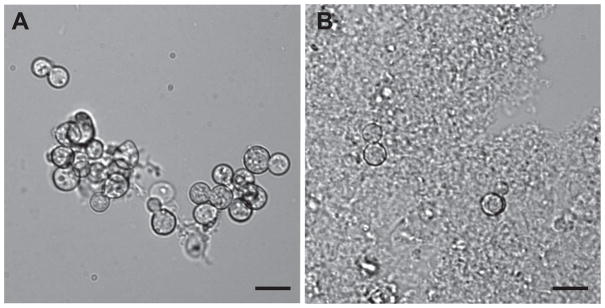
(A) B. dermatitidis yeast in the recovered yeast fraction (RYF). There was minimal residual murine lung tissue. (B) Supernatant-interface fraction (SIF) contained abundant murine tissue and comparatively little yeast. Scale bar represents 10 μm. Images are representative of RYF and SIF samples and are from a single mouse. Similar findings were observed for the RYFs and SIFs from the other 10 mice.
To quantify recovery of B. dermatitidis yeast from infected lung tissue at the time of lung harvest, yeast in the RYF and SIF were enumerated using a hemocytometer (Table 1). The burden of murine lung infection, calculated by addition of SIF and RYF, ranged between 8.2 × 106 to 3.81 × 107 (Table 1). Nearly 91% of yeast cells in infected lungs were found in the RYF (Table 1).
Table 1.
Recovery of Blastomyces dermatitidis yeast from infected murine lungs
| Mouse | Yeast in the Supernatant-Interface fraction (SIF) | Recovered yeast fraction(RYF) from lung tissue | Total Yeast*(SIF + RYF) | % Recovery of yeast from lung tissue† | Total RNA(μg) |
|---|---|---|---|---|---|
| 1 | 2.44 × 106 | 3.57 × 107 | 3.81 × 107 | 93.60% | 55.28 |
| 2 | 1.39 × 106 | 3.38 × 107 | 3.52 × 107 | 96.05% | 91.04 |
| 3 | 6.84 × 106 | 3.10 × 107 | 3.78 × 107 | 81.92% | 26.71 |
| 4 | 7.60 × 105 | 2.20 × 107 | 2.28 × 107 | 96.66% | 9.67 |
| 5 | 3.55 × 106 | 3.10 × 107 | 3.46 × 107 | 89.73% | 115.64 |
| 6 | 4.37 × 106 | 3.05 × 107 | 3.49 × 107 | 87.47% | 37.90 |
| 7 | 4.20 × 105 | 8.14 × 106 | 8.56 × 106 | 95.09% | 20.35 |
| 8 | 6.65 × 105 | 1.24 × 107 | 1.31 × 107 | 94.91% | 36.13 |
| 9 | 2.30 × 106 | 1.63 × 107 | 1.86 × 107 | 87.36% | 70.27 |
| 10 | 1.70 × 106 | 9.98 × 106 | 1.17 × 107 | 85.45% | 41.30 |
| 11 | 9.00 × 105 | 7.30 × 106 | 8.20 × 106 | 89.02% | 26.36 |
|
| |||||
| Average (SD) | 2.30 × 106 (1.95 × 106) | 2.16 × 107 (1.11 × 107) | 2.40 × 107 (1.24 × 107) | 90.69% (4.88%) | 48.24 (32.38) |
|
| |||||
| Median | 1.70 × 106 | 2.20 × 107 | 2.28 × 107 | 89.73% | 37.90 |
SD refers to standard deviation
Total yeast after the lungs were excised was calculated by addition of SIF and RYF.
Percent recovery was calculated as follows: % recovery = (RYF / total yeast) × 100.
3.2 Recovered yeast RNA was of high quality and suitable for RNA-Seq
The filtration-centrifugation technique for isolation of B. dermatitidis yeast facilitated recovery of total RNA with yields greater than 20 μg in 91% of mice (Table 1). For mouse #4, total RNA yield was low (9.67 μg) despite recovery of 2.20 × 107 yeast from lung tissue (Table 1) and may have been related to suboptimal RNA extraction. Analysis of ribosomal RNA by agarose gel electrophoresis suggested the majority of RNA from the recovered yeast fraction was from B. dermatitidis rather than from mouse (Figure 2). RNA extracted from infected mouse lungs immediately frozen in liquid nitrogen (referred to as whole lung RNA) demonstrated an electrophoretic pattern distinct from RNA isolated from in vitro culture or RYF (Figure 2). The murine ribosomal large subunit (LSU, 28S) band had a higher molecular weight than the B. dermatitidis ribosomal LSU band, whereas the ribosomal small subunit (SSU, 18S) bands for Mus musculus and B. dermatitidis were similar in size (Figure 2). For whole lung RNA, the murine ribosomal LSU band was more prominent than the B. dermatitidis LSU band. In contrast, the murine ribosomal LSU band was not visible for RNA harvested from in vitro culture or RYF; the B. dermatitidis LSU band was predominant (Figure 2).
Figure 2. Agarose gel electrophoresis of RNA.
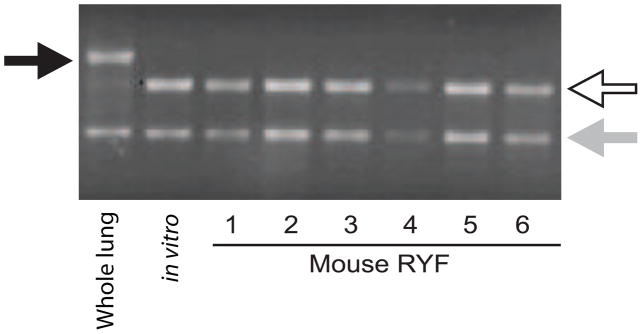
RNA extracted from infected whole lungs, in vitro culture of B. dermatitidis yeast, and recovered yeast fraction (RYF) from mice 1 – 6. For the whole lung sample, infected lungs from 2 mice were immediately placed into liquid nitrogen following excision and RNA was extracted. The black arrow marks the murine ribosomal large subunit band (LSU, 28S) band. The white arrow indicates B. dermatitidis LSU ribosomal band. The light gray arrow denotes the small subunit (SSU, 18S) ribosomal band, which was similar in size for B. dermatitidis yeast and murine lung tissue. For the whole lung sample, the murine ribosomal LSU band (black arrow) was bright, whereas the B. dermatitidis ribosomal LSU band was faint (white arrow). In contrast, the predominant ribosomal LSU band for RYF (mice 1–6) was the same size as RNA obtained from in vitro cultures. The murine ribosomal LSU band was not visible. The agarose gel is a representative example and similar results were observed for RYFs from mice 7–11. Each lane was loaded with 300 ng of total RNA based on spectrophotometric quantification.
To confirm that RYF was enriched for B. dermatitidis RNA and depleted of murine RNA, transcript abundance of B. dermatitidis GAPDH and M. musculus FTH1 (ferritin heavy chain 1) was analyzed using qRT-PCR (Figure 3). GAPDH transcript is abundant in yeast, whereas FTH1 is not encoded in the B. dermatitidis genome (fungi use siderophores rather than ferritin to store iron). Equal amounts of DNase-treated RNA from RYFs for mice 1, 3, 5, 7, 9 (pool #1) and 2, 6, 8, 10, 11 (pool #2) along with controls (in vitro culture and whole lung) were converted to cDNA. Mean GAPDH Ct values (± standard deviation) were 21.56 (± 0.22), 25.79 (± 0.27), 22.15 (± 0.01), and 22.23 (± 0.06) for in vitro culture, whole lung, RYF pool #1, and RYF pool #2, respectively. To quantify enrichment for B. dermatitidis RNA, transcript values for whole lung, RYF pool #1, and RYF pool #2 were normalized to GAPDH transcript from in vitro cultures and relative expression was calculated. GAPDH transcript for pools #1 and #2 were 12.4 and 11.8-fold more abundant when compared to whole lung (Figure 3A). To assess for reduction in murine RNA, M. musculus FTH1 transcript abundance was analyzed. Mean FTH1 values (± standard deviation) were 25.32 (± 0.80), 34.07 (± 0.62), and 33.30 (±0.50) for whole lung, RYF pool #1, and RYF pool #2, respectively. As expected, M. musculus FTH1 was not detected for B. dermatitidis yeast grown in liquid culture. FTH1 from RYF pools #1 and #2 were normalized to FTH1 from whole lung and relative expression was calculated. For RYF pools #1 and #2, transcript abundance for M. musculus FTH1 was > 250-fold lower compared to whole lung RNA (Figure 3B). Collectively, these data indicated that separation of B. dermatitidis yeast from murine lung tissue and reduction of murine RNA resulted in enrichment and enhanced detection of B. dermatitidis RNA.
Figure 3. Quality control analyses for enrichment of B. dermatitidis RNA and reduction of murine RNA.
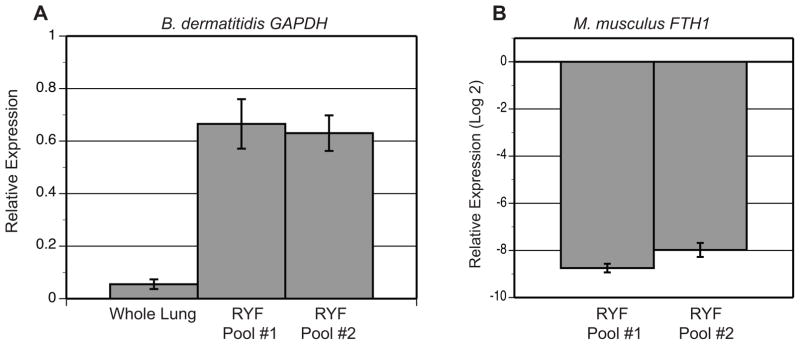
RNA extracted from in vitro liquid culture, infected whole lungs, and RYFs were analyzed by qRT-PCR for B. dermatitidis GAPDH and M. musculus FTH1. (A) Transcript abundance for B. dermatitidis GAPDH was 12.4 and 11.8-fold higher for RYF pool #1 and RYF pool #2 compared to the whole lung sample, respectively. The results were averaged from 2 experiments. Relative expression (RE) was calculated by RE = 2−ΔCt = 2− ((GAPDH whole lung, pool #1, or pool #2) − (GAPDH in vitro culture)). (B) M. musculus FTH1 was 250-fold lower in RYF from pools #1 and #2 when normalized to FTH1 transcript abundance in infected whole lungs. Relative expression was calculated by RE = 2− ((FTH1 pool #1 or #2) − (FTH1 whole lung)) and converted to log base 2. The results were averaged from 2 experiments.
Quality control analysis for RNA-Seq demonstrated the RNA sequence reads from RYF pools #1 and #2 were predominately B. dermatitidis in origin (Table 2). Ninety-three percent of RNA sequences from the pooled RYFs and in vitro cultures passed initial quality control (Table 2). This data along with measured RNA integrity numbers using an Agilent Bioanalyzer (7.6 for pool #1 and 7.8 for pool #2) indicated RYF RNA was of high quality. Importantly, 86% and 88% of sequencing reads for RYF pool #1 and pool #2 aligned to the B. dermatitidis 26199 genome, respectively (Table 2). In comparison, 93.15% and 92.8% of sequencing reads for in vitro cultures could be aligned to the B. dermatitidis 26199 genome. Moreover, less than 0.27% of the RNA-Seq reads from the RYFs that aligned to the B. dermatitidis genome also cross-mapped to the M. musculus genome. These data indicate the majority of contaminating murine RNA was removed from the RYFs and that the RYF RNA-Seq reads are highly specific for B. dermatitidis.
Table 2.
RYF versus in vitro RNA for RNA-Seq analysis
| Sample | Reads that passed filter (PF)a | PF Reads aligned to the 26199 genomeb |
|---|---|---|
| RYF for pool #1 | 61,752,858 (93.14%) | 54,451,587 (88.18%) |
| RYF for pool #2 | 61,541,994 (93.12%) | 52,961,006 (86.06%) |
| In vitro culture #1 | 63,803,410 (93.14%) | 59,429,719 (93.15%) |
| In vitro culture #2 | 58,667,296 (93.04%) | 54,443,098 (92.80%) |
RYF refers to recovered yeast fraction from murine lungs.
Pool #1 contains RYF RNA from mice 1, 3, 5, 7, 9
Pool #2 contains RYF RNA from mice 2, 6, 8, 10, 11
Total and percent of reads that passed filter (PF) of Illumina pipeline.
Total and percent of PF reads aligned to the B. dermatitidis 26199 genome
Techniques that did not enrich for B. dermatitidis yeast failed to yield RNA of suitable quality for genome-wide expression analysis. Treatment of lung homogenates with a cocktail of RNase A, DNase I and collagenase degraded murine RNA, but did not completely remove it. Agarose electrophoresis demonstrated a large smear of degraded murine RNA, which precluded accurate determination of the quantity and quality of B. dermatitidis RNA (data not shown). Homogenization of infected lungs in a RNA stabilization solution (RNAlater, Life Technologies) followed by filtration and centrifugation did not adequately separate B. dermatitidis yeast from lung tissue because the pellet consisted of both yeast cells and murine tissue.
3.3 Impact of temperature, time, and water on transcription
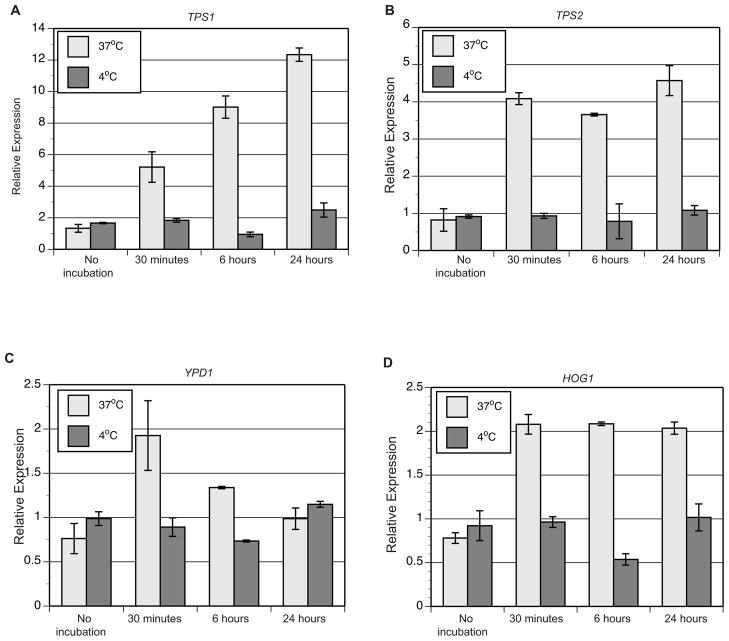
To minimize changes in transcript abundance, infected murine lungs were kept at near-freezing temperature during all processing steps and ex vivo time was limited to less than 30 minutes. Despite these efforts, there was potential for transcriptional changes, including those involved with adaptation to near-freezing temperature (Krandror et al., 2004), exposure to water to lyse murine cells, and processing time. The near-freezing response has been described in Saccharomyces cerevisiae and includes genes involved with the cold shock response and trehalose biosynthesis (Murata et al., 2006). This adaptive response occurs within 6 hour of exposure to 4°C (Murata et al., 2006). In addition, alterations in the hyperosmolarity glycerol (HOG) pathway have been described under cold stress (Panadero et al., 2006). The effect of a short exposure to near-freezing temperature on transcription has not been established in S. cerevisiae or the dimorphic fungi.
To investigate if transcription is altered during a short exposure to water and near-freezing temperature, qRT-PCR was used to analyze changes in transcript levels for genes involved with trehalose biosynthesis (TPS1, TPS2) and the HOG pathway (YPD1, HOG1). Homologs of S. cerevisiae cold shock response genes such as TIP1, TIR1-4, PAU1-7 were not identified in the B. dermatitidis genome by tBLASTn (data not shown). RNA was extracted from yeast grown in HMM at 37°C and yeast pellets resuspended at time 0 (no-incubation), 30 minutes, 6 hours, and 24 hours incubation in vitro at 37°C or 4°C (Figure 4). Except for a decline in transcript at 6 hours, TPS1, TPS2, YPD1, and HOG1 transcript levels were similar to the no-incubation time point at 30 minutes and 24 hours incubation at 4°C (Figure 4). In contrast, transcript levels for TPS1, TPS2, YPD1, and HOG1 were elevated at 37°C when compared to no-incubation at 30 minutes, 6 hours, and 24 hours (Figure 4). These data suggested that exposure to 4°C minimized changes in transcript abundance that would have otherwise occurred if the samples were processed at a higher temperature or for a longer duration.
Figure 4. Impact of temperature, time, and water on transcription.
To model the impact of temperature (4°C versus 37°C) during short and long exposures to incubation in water, qRT-PCR was used to analyze transcript abundance for genes involved in trehalose biosynthesis (TPS1, TPS2) and the HOG pathway (YPD1, HOG1) during in vitro culture. When compared to the no-incubation time point, transcript abundance increased for genes at 37°C at 30 minutes incubation (3.9-fold for TPS1, 4.9-fold for TPS2, 2.5-fold for YPD1, and 2.7-fold for HOG1). In contrast, cultures held for 30 minutes at 4°C prevented the increase in transcript abundance (≤ 0.1 fold change) when compared to the no-incubation time point. With the exception of a 1.8 and 1.7-fold reduction in transcript for TSP1 and HOG1 at 6 hours 4°C, changes in transcription were minimal at 4°C. In contrast, the transcriptional changes were more dynamic at 37°C. The results were averaged from 2 experiments. Relative expression was calculated using the 2−ΔΔCt method. ΔCt = (Ct of gene of interest) − (Ct of GAPDH) for each time point and cultures grown in HMM. ΔΔCt = (ΔCt of the time point) − (ΔCt of HMM).
4. Discussion
We developed a simple, new technique for isolating pathogenic yeast from murine lung tissue in order to obtain high-quality fungal RNA for genome-wide gene expression analysis. We applied the method to B. dermatitidis. Understanding the in vivo transcriptional profile of B. dermatitidis yeast during pulmonary infection has the potential to identify novel virulence genes and provide deeper insight into the regulatory pathways that contribute to pathogenesis. Knowledge of genes used by fungi to adapt to and survive in host tissue remains limited. DRK1 and BAD1 in B. dermatitidis facilitate the temperature-dependent conversion from mold to yeast and evasion of host immune defenses, respectively (Nemecek et al., 2006; Finkel-Jimenez et al., 2002; Finkel-Jimenez et al., 2001). H. capsulatum calcium binding protein-1 (CBP1) allows for survival in macrophages and yeast-phase specific-3 (YPS3) enables dissemination (Sebghati et al., 2000; Bohse and Woods, 2007). In contrast, required-for-yeast-phase genes (RYP1-3) encode transcriptional regulators that promote the conversion of H. capsulatum to yeast at 37°C (Nguyen and Sil, 2008; Webster and Sil, 2008). Deletion of spherule outer-wall glycoprotein (SOWgp), metalloproteinase (MEP1), and urease (URE) genes attenuates virulence of C. immitis and C. posadasii (Hung et al., 2005; Hung et al., 2002; Mirbod-Donovan et al., 2006). Although investigation of these genes has provided valuable insight on fungal virulence, a comprehensive picture of the regulatory genetic networks in which these genes participate during infection remains unknown.
Genome-wide gene expression studies in the dimorphic fungi have been performed with RNA obtained from in vitro cultures (Nguyen and Sil, 2008; Monteiro et al., 2009; Tavares et al., 2007; Johannesson et al., 2006; Lin et al., 2012; Hwang et al., 2003). These findings may not accurately reflect the transcriptional changes required for yeast cell survival in vivo because the host environment is complex, dynamic, and difficult to mimic in vitro or ex vivo. Moreover, the type of tissue that is infected (i.e. oral cavity, esophagus, liver, kidney) can influence the transcriptional response (Kumamoto et al., 2008). Comparative analysis of C. albicans gene expression has demonstrated that the transcription profile for cells during in vivo oropharyngeal infection and in vitro growth were substantially different (Fanning et al., 2012; White et al., 2007). Similarly, differences in transcript abundance for in vivo and ex vivo infections were observed during invasion of hepatic tissue by C. albicans (Thewes et al., 2007). Moreover, C. albicans cells involved in gastrointestinal colonization have a unique transcriptional profile compared to cells grown in culture (Rosenbach et al., 2010; White et al., 2007). These studies illustrate the limitations of using in vitro methods for modeling in vivo events and highlight the importance of in vivo transcriptional profiling for genes associated with infection or colonization.
Enrichment of yeast RNA and reduction of mammalian RNA is an important technical hurdle to overcome for in vivo transcriptional profiling. C. albicans RNA has been successfully isolated from the oral cavity, peritoneal cavity, liver, kidney, and gastrointestinal tract for gene expression microarray analyses (Andes et al., 2005; Walker et al., 2009; Zakikhany et al., 2007; Thewes et al., 2007; Rosenbach et al., 2010; White et al., 2007). Similarly, C. neoformans RNA has been enriched from the cerebral spinal fluid and early in pulmonary infection for differential display RT-PCR and SAGE analyses (Rude et al., 2002; Steen et al., 2003; Hu et al., 2008). A variety of techniques have been used to extract fungal RNA including biopsy of infected hepatic tissue (Thewes et al., 2007), collection of intraperitoneal yeast by lavage (Thewes et al., 2007), scraping pseudomembranes from humans with oral candidiasis (Zakikhany et al., 2007), harvest of feces from the cecum (Rosenbach et al., 2010; White et al., 2007), enzymatic degradation of murine DNA and RNA from renal tissue (Andes et al., 2005), surgical excision of the renal cortex (Walker et al., 2009), collection of infected CSF (Rude et al., 2002; Steen et al., 2003), and bronchoalveolar lavage (Hu et al., 2008). However, none of these existing techniques were applicable for isolation of B. dermatitidis during experimental pulmonary infection. Surgical excision or selective biopsy of infected pulmonary tissue was impractical because an infected lung can have hundreds of pyogranulomas. RNA extracted from lungs immediately frozen in liquid nitrogen contained an overwhelming amount of murine RNA, which in turn, precluded accurate assessment of B. dermatitidis RNA for quality control and would limit detection of low abundant fungal transcripts. Excessive amounts of mammalian RNA have led to suboptimal results in genome-wide gene expression analysis (Thewes et al., 2007). Although bronchoalveolar lavage is feasible, the recovery of B. dermatitidis would be limited to yeast located in the alveolar spaces, but not within pyogranulomas. Treatment with RNase resulted in partial degradation, but not removal of murine RNA. Moreover, enzymatic digestion of nucleic acids requires prolonged incubation at 37°C, which would substantially alter transcript abundance. Thus, the technique described herein is simple, new, and tailored for isolation of yeast that cause invasive pulmonary infection.
The two-step procedure of filtration and centrifugation following lysis of infected lung tissue resulted in substantial enrichment of B. dermatitidis yeast cells, recovery of high-quality fungal RNA, and elimination of the majority of M. musculus RNA. The protocol was quick, straightforward, and did not require expensive reagents or equipment. Moreover, quality controls including visual inspection of RYF by light microscopy and use of agarose gel electrophoresis along with qRT-PCR accurately predicted reduction of M. musculus RNA, which was reflected by high percentage of RNA-Seq reads from the RYF that aligned to the B. dermatitidis genome. The effort to minimize ex vivo sample time and the use of near-freezing temperature appeared to minimize transcriptional changes that would have otherwise occurred with longer processing time or higher temperature.
Although the two-step protocol for isolation of B. dermatitidis yeast from lung tissue was simple and yielded high-quality fungal RNA suitable for genome-wide expression analysis, there were potential limitations. Recovery of yeast may be limited during early stages of infection (i.e. the first week post-inoculation) when the burden of infection in each animal is low. This may require protocol modification such as pooling of lungs for processing or pooling of RYFs prior to RNA extraction. There was potential for technical aspects of yeast recovery to cause subtle changes in transcription; however, the use of near-freezing temperature to slow transcription together with short processing time ex vivo appeared to minimize these changes. Finally, this protocol may be limited to fungi that grow as yeast in lung tissue. Homogenization of lungs infected with filamentous fungi such as Aspergillus fumigatus could destroy hyphal cells and preclude recovery of RNA, although this would need to be investigated.
In conclusion, we developed a simple, new technique for isolating B. dermatitidis yeast and RNA from lung tissue during experimental murine infection. The quality and quantity of RNA was suitable for RNA-Seq. This technique is likely applicable to other pathogenic fungi that cause pneumonia such as H. capsulatum, Coccidioides species, Paracoccidioides brasiliensis, and Cryptococcus species. The ability to effectively isolate yeast from lung tissue will provide new insight for understanding how pathogenic fungi adapt to and survive in the host to cause disease. This, in turn, will open novel avenues for exploring host-pathogen interactions.
Highlights.
A simple, new technique was developed to isolate yeast from murine lung tissue.
91% of B. dermatitidis yeast cells were recovered from murine lung tissue.
B. dermatitidis RNA was of high quality and had minimal contamination by murine RNA.
87% of RNA sequencing reads mapped back to the B. dermatitidis genome.
Acknowledgments
We thank the Broad Institute Genomics Platform for sequencing the RNA samples described herein.
6. Funding Sources
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract No.:HHSN272200900018C to CC; 5K08AI071004 to GG; and R37AI035681 to BK). The funding agency was not involved with the study design, collection of data, analysis or interpretation of data, or the decision to submit the article for publication.
Footnotes
7. Conflicts of Interest: G.M. Gauthier is an associate editor for Fungal Genetics and Biology. Otherwise, there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andes D, et al. A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J Infect Dis. 2005;192:893–900. doi: 10.1086/432104. [DOI] [PubMed] [Google Scholar]
- Bohse ML, Woods JP. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect Immun. 2007;75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst TT, et al. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012 doi: 10.1101/gr.142802.112. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamgaye S, et al. RNA sequencing revealed novel actors of the acquisition of drug resistance in Candida albicans. BMC Genomics. 2012;13:396. doi: 10.1186/1471-2164-13-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, et al. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell. 2012;11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel-Jimenez B, Wüthrich M, Klein B. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-α production through TGF-β–dependent and – independent mechanisms. J Immunol. 2002;168:5746–5755. doi: 10.4049/jimmunol.168.11.5746. [DOI] [PubMed] [Google Scholar]
- Finkel-Jimenez B, et al. The WI-1 adhesin blocks phagocyte TNF-α production imparting pathogenicity on Blastomyces dermatitidis. J Immunol. 2001;166:2665–2673. doi: 10.4049/jimmunol.166.4.2665. [DOI] [PubMed] [Google Scholar]
- Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier GM, et al. Blastomycosis in solid organ transplant recipients. Transpl Infect Dis. 2007;9:310–317. doi: 10.1111/j.1399-3062.2007.00227.x. [DOI] [PubMed] [Google Scholar]
- Gauthier GM, Klein BS. Insights into fungal morphogenesis and immune evasion. Microbe. 2008;3:416–423. doi: 10.1128/microbe.3.416.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, et al. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, et al. A parasitic phase-specific adhesion of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect Immun. 2002;70:3443–3456. doi: 10.1128/IAI.70.7.3443-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, et al. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immune. 2005;73:6689–6703. doi: 10.1128/IAI.73.10.6689-6703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L, et al. Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol Biol Cell. 2003;14:2314–2316. doi: 10.1091/mbc.E03-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H, et al. Phase-specific gene expression underlying morphological adaptations of the dimorphic human pathogenic fungus Coccidioides posadasii. Fungal Genet Biol. 2006;43:545–559. doi: 10.1016/j.fgb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Jong A, et al. Infectomic analysis of gene expression profiles of human brain microvascular endothelial cells infected with Cryptococcus neoformans. J Biomed Biotechnol. 2008;2008:375620. doi: 10.1155/2008/375620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajaejun T, et al. Discordant influence of Blastomyces dermatitidis yeast-phase-specific gene BYS1 on morphogenesis and virulence. Infect Immunol. 2010;78:2522–2528. doi: 10.1128/IAI.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krandror O, et al. Yeast adapt to near-freezing temperature by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA. Niche-specific gene expression during C. albicans infection. Curr Opin Microbiol. 2008;11:325–330. doi: 10.1016/j.mib.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, et al. Comprehensive transcription analysis of human pathogenic fungus Penicillium marneffei in mycelial and yeast cells. Med Mycol. 2012 doi: 10.3109/13693786.2012.678398. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;24:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McDonagh A, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbod-Donovan F, et al. Urease produced by Coccidioides posadasii contributes to virulence of this respiratory pathogen. Infect Immunol. 2006;74:504–515. doi: 10.1128/IAI.74.1.504-515.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JP, et al. Genomic DNA microarray comparison of gene expression patterns in Paracoccidioides brasiliensis mycelia and yeasts in vitro. Microbiology. 2009;155:2795–2808. doi: 10.1099/mic.0.027441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, et al. Genome-wide expression analysis of yeast response during exposure to 4°C. Extremophiles. 2006;10:117–128. doi: 10.1007/s00792-005-0480-1. [DOI] [PubMed] [Google Scholar]
- Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P, et al. Cryptococcus gattii virulence composite: candidate genes revealed by microarray analysis of high and less virulent Vancouver island outbreak strains. PLoS One. 2011;13:e16076. doi: 10.1371/journal.pone.0016076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panadero J, et al. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J Biol Chem. 2006;281:4638–4645. doi: 10.1074/jbc.M512736200. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acid Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbach A, et al. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell. 2010;9:1075–1086. doi: 10.1128/EC.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rude TH, et al. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun. 2002;70:5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 7.4–7.8. [Google Scholar]
- Schwarz J, Baum GL. Blastomycosis. Am J Clin Pathol. 1951;21:999–1029. doi: 10.1093/ajcp/21.11.999. [DOI] [PubMed] [Google Scholar]
- Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- Steen BR, et al. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell. 2003;2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares AH, et al. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect. 2007;9:583–590. doi: 10.1016/j.micinf.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Thewes S, et al. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- Walker LA, et al. Genome-wide analysis of Candida albicans gene expression patterns during infection of the mammalian kidney. Fungal Genet Biol. 2009;46:210–219. doi: 10.1016/j.fgb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RH, Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad, Sci, USA. 2008;105:14573–14578. doi: 10.1073/pnas.0806221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol 1988. 1988;26:137–143. [PubMed] [Google Scholar]
- Wüthrich M, et al. Requisite elements in vaccine immunity to Blastomyces dermatitidis: plasticity uncovers vaccine potential in immune-deficient hosts. J Immunol. 2002;169:6969–6976. doi: 10.4049/jimmunol.169.12.6969. [DOI] [PubMed] [Google Scholar]
- Zakikhany K, et al. In vivo transcript profiling of Candida albicans identified a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]