Abstract
This report used high-performance affinity microcolumns to examine the changes in binding by sulfonylurea drugs to in vivo glycated HSA that had been isolated from individual patients with diabetes. An immunoextraction approach was developed to isolate HSA and glycated HSA from clinical samples, using only 20 μL of plasma or serum and 6–12 nmol of protein to prepare each affinity microcolumn. It was found that the affinity microcolumns could be used in either frontal analysis or zonal elution studies, which typically required only 4–8 min per run. The microcolumns had good stability and allowed data to be obtained for multiple drugs and experimental conditions over hundreds of sample application cycles. Both the overall binding, as measured by frontal analysis, and site-specific interactions, as examined by zonal elution, showed good agreement with previous data that had been obtained for in vitro glycated HSA with similar levels of modification. It was also possible to directly compare the changes in site-specific binding that occurred between sulfonylurea drugs or as the level of HSA glycation was varied. This method is not limited to clinical samples of glycated HSA but could be adapted for work with other modified proteins of interest in personalized medicine.
INTRODUCTION
Personalized medicine holds great promise for improving disease diagnosis and therapy through the customization of treatment based on a patient’s specific needs.1–3 Diabetes is one disease for which this approach is of potential interest.4–7 The number of diabetic patients has increased by 90% over the last 20 years, with an estimated 26 million individuals in the U.S. suffering from this disease.4,5
Some of the complications of diabetes are associated with the non-enzymatic glycation of proteins.8 Glycation begins when a free amine group on a protein reacts with an aldehyde group on a reducing sugar (e.g., glucose) to form a reversible Schiff base, which can later rearrange to produce a more stable fructosamine residue, or Amadori product.9–11 Numerous studies have indicated that glycation can produce structural and functional modifications in proteins with long half-lives (e.g., collagen, lens crystalline, and hemoglobin).12,13 Recent research has also examined the effects of glycation on human serum albumin (HSA), the main protein in serum and an important binding agent for transporting many drugs and endogenous compounds in the circulatory system.14–19
The binding of drugs to HSA usually occurs at Sudlow sites I and II, which are located in subdomains IIA and IIIA of this protein.20 It has been suggested that the binding of HSA to some drugs can be altered by glycation-related modifications that occur in the vicinity of Sudlow sites I and II.16,18 A group of drugs that may exhibit such changes are the sulfonylureas, which have been used to treat type II diabetes for over 50 years.21 These drugs are highly bound (i.e., 90–99%) to serum proteins and particularly to HSA.22 Displacement or release of these drugs from serum proteins is known to cause hypoglycemia,23 while the presence of an effective dose that is too low for the same drugs may result in elevated glucose levels.
In previous studies, high-performance affinity chromatography (HPAC) has been used to examine the interactions between several first- and second-generation sulfonylurea drugs with normal and in vitro glycated HSA.24–27 The use of HPAC over conventional methods for monitoring drug-protein binding (e.g., ultrafiltration and equilibrium dialysis) has several benefits, such as the high reproducibility of this method plus its good precision and ease of automation. Other advantages include the small sample requirements of HPAC and its ability to reuse the same protein preparation for hundreds of experiments.28,29
In this report, HPAC will be explored for use in studying drug interactions with in vivo glycated HSA that has been isolated from individuals known to have diabetes. Initial studies will examine the use of immunoextraction columns for isolating in vivo glycated HSA from small amounts of samples obtained from patients and will combine this approach with recent developments in the preparation of affinity microcolumns for drug-protein binding studies.28,30 The resulting affinity microcolumns will then be used to look at the overall binding of several sulfonylurea drugs to in vivo glycated HSA, as will be studied by frontal analysis. The same columns will be employed with zonal elution to detect any changes in binding that might occur for the same drugs at Sudlow sites I and II upon the in vivo glycation of HSA in individual patients. The results should provide information that can be used to better understand these interactions and should lead to new analytical tools that can be employed in personalized medicine for patients with diabetes or other diseases.
EXPERIMENTAL SECTION
Materials and Reagents
The following solutes were purchased from Sigma-Aldrich (St. Louis, MO): acetohexamide (99% pure), tolbutamide (99.8%), gliclazide (99.5%), R-warfarin (> 99%) and L-tryptophan (> 99%). A commercial sample of serum made from male AB plasma was also obtained from Sigma-Aldrich (product H4522, lot 039K0728; pre-screened and found to be negative for HIV-1/HIV-2, hepatitis B and hepatitis C). The Nucleosil Si-300 (particle size, 7 μm; pore size, 300 Å) was from Macherey-Nagel (Düren, Germany). Reagents for the bicinchoninic acid (BCA) protein assay, as used for determining the protein content of the isolated glycated HSA samples and chromatographic supports, were from Pierce (Rockford, IL). The measurement of glycation levels was conducted by using a fructosamine assay kit from Diazyme Laboratories (San Diego, CA), as described previously.25 The Vivapure anti-HSA resin was from Sartorius Stedim Biotech (Goettingen, Germany). All buffers and aqueous solutions were prepared using water from a Nanopure system (Barnstead, Dubuque, IA) and were filtered using 0.2 μm GNWP nylon filters from Millipore (Billerica, MA).
Apparatus
The isolation of HSA and glycated HSA from serum or plasma was performed using a 5702RH temperature-controlled centrifuge from Eppendorf (New York, NY) and a fixed-angle centrifuge rotor from VWR (West Chester, PA). The chromatographic experiments were carried out by using a Jasco 2000 HPLC system (Easton, MD) that contained a DG-2080-53 three solvent degasser, two PU-2080 isocratic pumps, an AS-2057 autosampler equipped with a 100 μL sample loop (operated in the partial loop injection mode), and a UV-2075 absorbance detector. The columns were kept at 37°C by using a Jasco CO-2060 column oven. A Rheodyne Advantage PF six-port switching valve (Cotati, CA) was used for alternating the passage of drug and buffer solutions through the columns during the frontal analysis studies. The system components were controlled by a Jasco LC-Net II/ADC system and EZChrom Elite software v3.2.1 (Scientific Software, Pleasanton, CA). The breakthrough times for the frontal analysis data and central peak moments for the zonal elution data were determined by using PeakFit 4.12 (SeaSolve Software, San Jose, CA). Linear and non-linear regression were conducted by utilizing DataFit 8.1.69 (Oakdale Engineering, PA).
Sample pretreatment and column preparation
Pre-existing EDTA plasma samples, acquired through venipuncture and generated according to standard procedures for work with whole blood (see Supporting Information), were obtained from de-identified patients known to have diabetes, as provided by W. Clarke at the Johns Hopkins University School of Medicine. The collection and provision of these samples was performed with approval of the Johns Hopkins Institutional Review Board. All of these samples were handled with appropriate precautions for materials that may contain bloodborne pathogens. The level of hemoglobin A1c (HbA1c) was measured and provided for each plasma sample, as is used in clinical laboratories for the diagnosis and long-term monitoring of glucose control in diabetic patients.31–35
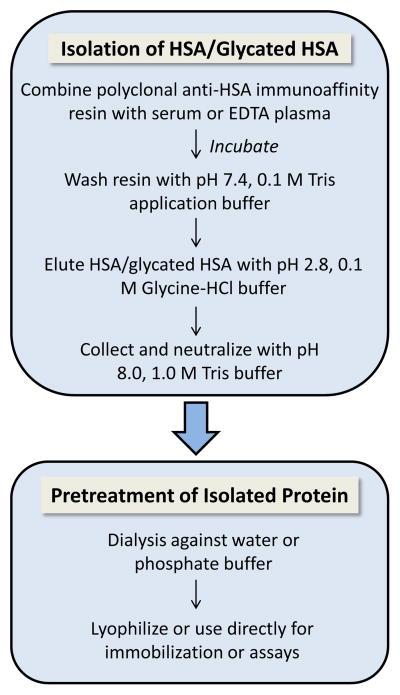
Figure 1 shows the general procedure that was used to isolate HSA and in vivo glycated HSA from serum or plasma samples. This procedure was conducted by using a 20 μL serum or plasma sample. In this study, such a sample was used as a representative portion of 1–2 mL remnant EDTA plasma that was acquired for an individual patient; in future work, finger stick samples might also be employed, as are commonly used in routine glucose monitoring. The 20 μL of serum or plasma was added to a Vivaspin 6 spin-filter column that contained a fresh 400 μL portion of a 50% packed resin slurry with polyclonal anti-HSA antibody fragments immobilized onto cross-linked agarose beads. Further details on the procedure for this isolation step, including the sample loading and elution conditions, are provided in the Supporting Information and Results and Discussion.
Figure 1.
Scheme for the purification of human serum albumin (HSA) and in vivo glycated HSA from serum or plasma, as used prior to the immobilization of these proteins in affinity microcolumns. More details are provided in the Supporting Information.
Nucleosil Si-300 silica was converted into a diol-bonded form.36 One portion of this support was used to immobilize HSA/glycated HSA by the Schiff base method,37 while a second portion was used to make a control support to which no protein was added during the immobilization step. Each HSA/glycated HSA support and control support was downward slurry-packed at 24 MPa (3500 psi) into a separate 2.0 cm × 2.1 mm I.D. stainless steel column using pH 7.4, 0.067 M potassium phosphate buffer as the packing solution and was stored in this buffer at 4°C when not in use. A BCA assay was performed to determine the protein content of each support, using normal HSA or in vitro glycated HSA as the standard and the control support as the blank.25,38
Chromatographic studies
All chromatographic experiments were performed at 37°C and using pH 7.4, 0.067 M potassium phosphate buffer as the mobile phase. All drug and solute solutions were prepared in this mobile phase and filtered through a 0.2 μm nylon filter, followed by a 15 min degassing step, prior to use. The solutions and samples of the sulfonylurea drugs and R-warfarin were used within two weeks of preparation.24–27,39–40 The L-tryptophan solutions were prepared fresh daily in the pH 7.4 buffer.40 The samples were applied or injected at a typical flow rate of 0.5 mL/min, as shown in prior work to give reproducible retention factors and binding capacities for these drugs and solutes on similar HSA columns.24–27,39,40
In the frontal analysis studies, each column was first equilibrated with the pH 7.4 mobile phase buffer. The mobile phase was then switched to a solution that contained a known concentration of the desired drug in the same buffer. These solutions included up to sixteen concentrations of acetohexamide that ranged from 1 to 1000 μM and ten concentrations of gliclazide or tolbutamide that ranged from 1 to 200 μM. These conditions included the typical therapeutic levels for these drugs and provided a sufficiently broad range of conditions to allow the observation of both weak and moderate-to-high affinity interactions of these solutes with glycated HSA.24–27,41 Each drug solution was continuously applied to the column until a breakthrough curve with a level plateau was produced. The system was later switched back to the pH 7.4, 0.067 M phosphate buffer to elute the retained analyte from the column under isocratic conditions. This process was repeated three times at each concentration for all drugs on the test columns and control column.
To keep the absorbance values within the linear range of the detector, the breakthrough curves for acetohexamide were monitored at 248 nm for concentrations of 1–20 μM and at 315 nm for concentrations of 30–1000 μM. Gliclazide and tolbutamide were monitored at 248 nm for all of the concentrations used in frontal analysis. The central location of each breakthrough curve was determined by using a Savitzky-Golay first derivative algorithm for smoothing, followed by fitting of the first derivative to an exponentially-modified Gaussian curve. A correction for any non-specific binding of a drug to the support was made by subtracting the breakthrough times of the control column from those obtained on the test column at each drug concentration, as described previously for the same analytes on columns based on normal HSA or in vitro glycated HSA.24–27,39,40
Zonal elution studies were performed by using R-warfarin and L-tryptophan as site-specific probes for Sudlow sites I and II of HSA.24–27,39,40 The pH 7.4 mobile phase was used to prepare all of the injected samples and competing agents for these experiments. The acetohexamide, gliclazide, or tolbutamide solutions ranged in concentration from 0 to 20 μM. The samples consisted of 20 μL injections of 5 μM R-warfarin or 5 μM L-tryptophan that were prepared by diluting each probe with a mobile phase that contained the final desired concentration of the competing agent. Elution of the injected solutes was monitored at 308 nm for R-warfarin and at 280 nm for L-tryptophan. Non-specific binding was evaluated and corrected by injecting all of the samples onto a control column. To determine the void time of each column, 20 μL injections of 20 μM sodium nitrate were made (i.e., a non-retained solute on HSA columns), and the resulting peak was monitored at 205 nm. The central moment of each peak was determined by using a fit to an exponentially-modified Gaussian curve.39
RESULTS AND DISCUSSION
Isolation of HSA and glycated HSA
Immunoextraction based on polyclonal anti-HSA antibodies was used to isolate HSA and glycated HSA from serum or EDTA plasma. This was conducted by using a commercial anti-HSA support with a binding capacity of at least 2 mg HSA/mL resin. The extraction efficiency of this approach was tested by combining a fresh 400 μL portion of the immunoaffinity support with a 20 μL standard sample that contained 800 μg HSA, or 40 g/L HSA (i.e., a representative concentration for this protein in serum or plasma).41 This sample was applied to the support in pH 7.4, 0.10 M Tris buffer, which was also added in a 200 μL portion to wash away non-retained sample components. Two 200 μL portions of pH 2.8, 0.10 M glycine-HCl buffer were then added to elute the retained protein, as recommended by the manufacturer. A fresh portion of the immunoaffinity resin was then used for the next sample application, thus avoiding the need for column regeneration and providing stable antibodies with a high and consistent level of binding. However, regeneration and reuse of such a resin could also have been employed, as has been described for other anti-HSA columns.42,43
Only 54% of the applied HSA was recovered by the initial immunoextraction approach, in agreement with prior observations made under similar conditions with the same type of anti-HSA resin.44 As the number of elution steps was increased from two to six, the recovery of HSA increased to 90%. A recovery of at least 90% and approaching 100% was also noted when this study was repeated for samples that contained highly glycated HSA that was previously prepared in vitro with a level of modification of 3.35 (± 0.14) mol hexose/mol protein.25 A high level of recovery was not essential to the binding studies that were later carried out with the final affinity microcolumns, as long as a representative portion of the original protein population had been captured; however, a high recovery did reduce the sample volume that was needed to prepare the affinity microcolumns. The specificity of the modified purification approach was examined by using it with a 20 μL commercial sample of human serum known to contain 1020 μg protein and 840 μg HSA. The retained and isolated protein fraction showed only one detectable band by SDS-PAGE which matched that of an HSA standard, in agreement with previous work using this support.44–46
A number of buffer combinations were tested with the immunoextraction method to obtain a high recovery for HSA and glycated HSA while avoiding long-term changes in the activities of these proteins. In these experiments, 20 μL of 40 mg/mL HSA or highly glycated HSA was incubated at room temperature for 20 min with 200 μL of 0.10 M glycine-HCl buffer that had a pH of 2.8 to 4.0. This incubation step was followed by neutralization of the solution to a pH of 7.0–7.4 by adding 10 μL of pH 8.0, 1.0 M Tris buffer. A binding study based on ultrafiltration, as described in the Supporting Information and using tolbutamide, was then carried out with 1) the treated protein samples and 2) control samples that had the same final concentrations of drug and HSA/glycated HSA but were placed directly into pH 7.4, 0.10 M Tris buffer. In each case, the controls and treated samples had no significant difference in their binding at the 95% confidence level. The effect of the elution pH on protein recovery was also considered. A decrease in the recovery of HSA from over 90% to 25% was observed when increasing the elution pH from 2.8 to 3.5. Based on these results, an approach using an elution pH of 2.8 was used in all later experiments involving immunoextraction for the isolation of in vivo glycated HSA.
Preparation of affinity microcolumns using clinical samples
The scheme optimized in the previous section was next used to isolate glycated HSA from individual clinical samples. This was of interest because recent studies have found that there are differences in the binding of sulfonylurea drugs to samples of normal HSA, as prepared from pooled serum, versus in vitro glycated HSA that has various levels of glycation.24–27 However, it is not yet known whether changes with glycation also occur for these drugs with HSA that has been glycated in vivo. Also, no known previous studies have examined the creation of affinity columns for binding studies that have used HSA or other proteins from specific individuals.28,29 Work in this section sought to develop tools for obtaining such information by creating affinity microcolumns that were prepared by using in vivo glycated HSA from individual patients.
To explore this approach, microcolumns were prepared using two different preparations of in vivo glycated HSA. The first clinical sample, glycated HSA-CS1, was from a diabetic patient who had an HbA1c level of 10.0%, which represented an estimated long-term glucose level of 15.5 mM in blood47 (Note: an HbA1c value of ≥6.5% is often used to indicate the presence of diabetes is a patient).33–35 The second sample, glycated HSA-CS2, came from a patient who had an HbA1c level of 13.7%, which represented a case of uncontrolled and/or advanced diabetes and an estimated long-term glucose level of 22.8 mM.47 The measured levels of glycation were 1.19 (± 0.15) and 1.51 (± 0.20) mol hexose/mol HSA for the isolated samples of HSA-CS1 and HSA-CS2. The results obtained for these columns were also compared to those obtained previously for normal HSA that was obtained from pooled human serum.24
The column size that was used with these samples was 2.0 cm × 2.1 mm I.D. It was known from previous work with normal HSA that this size was suitable for use in drug binding studies and offered good precision along with reasonably fast analysis times. It was also known from prior work that a column of this size could contain roughly 1 mg of immobilized HSA when using the same type of silica as employed in this report.24 This amount of HSA corresponded to what is found in approximately 20–28 μL of human serum or plasma.41 When the isolated in vivo glycated HSA was immobilized by the Schiff base method, the final supports contained 26 (± 1) mg protein/g silica for glycated HSA-CS1 and 14 (± 1) mg protein/g silica for glycated HSA-CS2. These results represented a total protein content of 0.4–0.8 mg or 6–12 nmol (3.6–7.2 × 1015 molecules) in the 2.0 cm × 2.1 mm I.D. columns. Previous work with in vitro glycated HSA and the same column size has found good agreement with binding parameters that have been obtained using much larger amounts of protein in ultrafiltration studies,25,27 indicating that the amount of protein in these microcolumns was sufficient to provide a good representation of the glycated HSA populations and their binding properties in the original clinical samples.
Frontal analysis studies for clinical samples
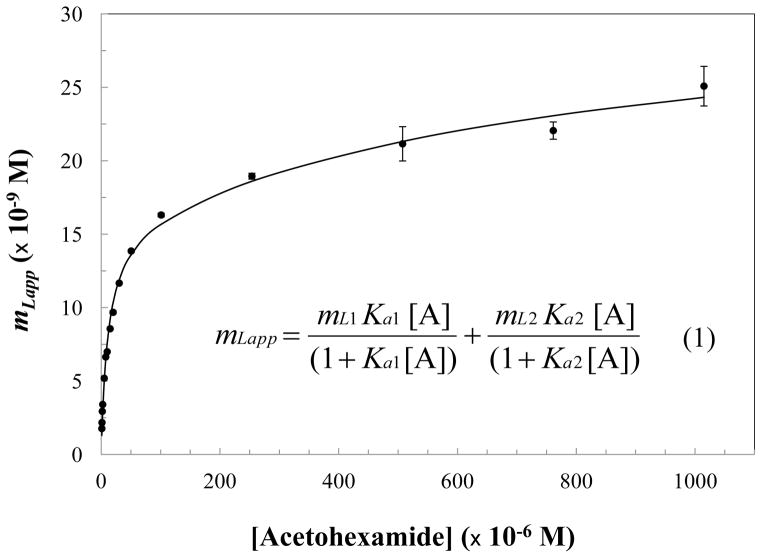
The columns containing in vivo glycated HSA were examined by frontal analysis to study their overall binding to various sulfonylurea drugs. Each frontal analysis experiment took roughly 6–8 min to complete at 0.5 mL/min on a 2.0 cm × 2.1 mm I.D. microcolumn (see Supporting Information for examples). This meant that even triplicate experiments over a wide range of concentrations could be carried out in a reasonable amount of time. Each of the in vivo glycated HSA columns was used over the course of approximately 9–12 months and at least 250 sample application cycles for the work described in this report. The stability of these columns was monitored by examining their retention for R-warfarin and L-tryptophan, as determined through the site-specific competitive studies that are described later for these same columns. During the entire period of this study, the maximum variation in the retention factor for R-warfarin was 13–31%, and the maximum variation in the retention factor for L-tryptophan was 3–22%; changes of less than 5–11% and 1–3%, respectively, were noted in these same parameters over the first 2–3 months of column operation. This level of variation did not create any problems during the analysis of data from the frontal analysis experiments, as illustrated by Figure 2 and eq (1), which provided association equilibrium constants that were determined independently from the column binding capacity.28 This feature also meant it was not necessary to capture all of the glycated HSA/HSA from a sample or to have the same amount of protein in each microcolumn for these measurements, as long as a representative portion of the original protein population was present.
Figure 2.
Fit of frontal analysis data obtained for acetohexamide on the glycated HSA-CS1 column when using a two-site binding model, as described by eq (1).25 Each data point represents the average of three measurements for mLapp, the moles of analyte required to reach the mean point of the breakthrough curve at a given molar concentration of the applied analyte. The error bars represent ± 1 S.D. for this value and ranged in size from ± 0.01–5.5% (average, ± 1.1%). Terms: molar concentration of applied analyte A, [A]; Ka1 and Ka2, association equilibrium constants for the analyte at binding sites 1 and 2; mL1 and mL2, total moles of binding sites 1 and 2 in the column.
The frontal analysis data were analyzed by preparing a graph of the moles of applied analyte that were required to reach the central point of the breakthrough curve versus the concentration of analyte that was applied to the column. An example of such a plot is shown in Figure 2. These data were fit to various binding models. The results for acetohexamide in Figure 3 had the best-fit to the two-site model in eq (1), with a correlation coefficient of 0.993 (n = 16) and only random variations in the corresponding residual plot. Fitting the same data to the one-site model (see Supporting Information) gave a lower correlation coefficient of 0.967 and non-random variations in a residual plot. No further improvement in the fit was obtained when using a three-site model. Similar results were obtained for acetohexamide on the glycated HSA-CS2 column and for tolbutamide and gliclazide with both microcolumns containing in vivo glycated HSA (i.e., correlation coefficients of 0.999 for the two-site model versus 0.949–0.997 for the one-site model at n = 10–16). The best fit noted to a two-site binding model is consistent with previous results reported for the same drugs on columns that contained normal HSA or in vitro glycated HSA.24–27 Table 1 summarizes the equilibrium constants and moles of binding sites that were estimated with the two-site model for the sulfonylurea drugs on the in vivo glycated HSA columns. Previous values obtained for normal HSA are also provided for reference.24,27
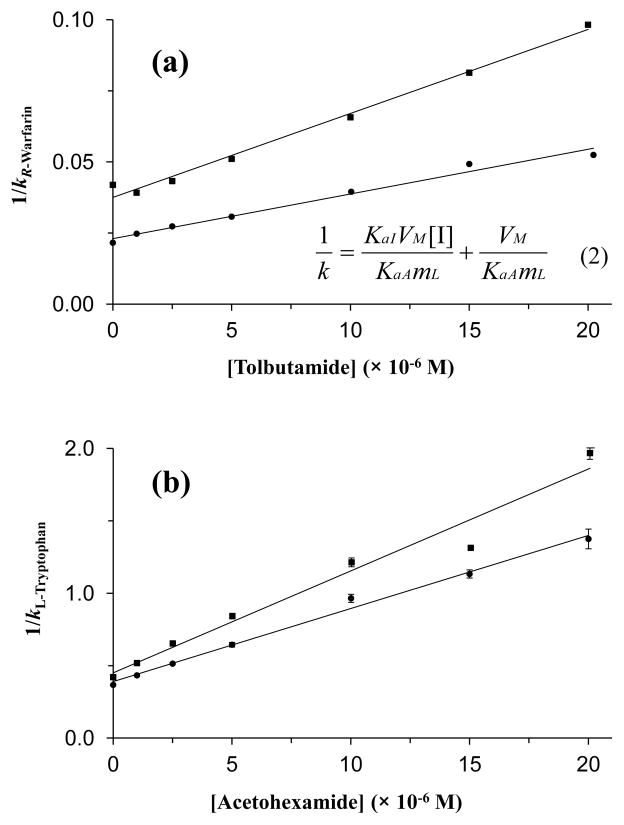
Figure 3.
Change in the retention factor (k) at (a) Sudlow site I for R-warfarin in the presence of tolbutamide as a competing agent and (b) at Sudlow site II for L-tryptophan in the presence of acetohexamide, as obtained for columns containing glycated HSA-CS1 (●) or glycated HSA-CS2 (■). The error bars represent ± 1 S.D. for three replicate measurements of 1/k and are often comparable in size to the markers used in these plots, especially in (a). The size of these error bars ranged from (a) ± 0.08–0.93% (average, ± 0.24%) to (b) ± 0.60–5.0% (average, ± 1.9%). Eq (2) represents the relationship that is expected for a system with direct competition at a single type of binding site between the retention factor for the injected probe (A) and the molar concentration of the competing agent in the mobile phase, [I]. Terms: KaA and KaI, association equilibrium constants for the injected probe and competing agent, respectively, at their site of competition; VM, column void volume; mL, moles of common binding sites for A and I. Based on eq (2), a plot of 1/k versus [I] should produce a linear relationship for a system with single-site competition, where the value of KaI can be found by taking the ratio of the slope over the intercept.25
Table 1.
Binding parameters obtained by frontal analysis and using a two-site model to describe the interactions of various sulfonylurea drugs with columns containing in vivo glycated HSAa
| Column/Drug | High affinity sites | Lower affinity site | ||||
|---|---|---|---|---|---|---|
| Ka1 (× 105 M−1) | ML1 (× 10−8 M−1) | Relative activity (mol/mol HSA) | Ka2 (× 103 M−1) | ML2 (× 10−8 M−1) | Relative activity (mol/mol HSA) | |
| Normal HSAb | ||||||
| Tolbutamide | 0.87 (± 0.06) | 2.0 (± 0.1) | 1.12 (± 0.08) | 8.1 (± 1.7) | 1.8 (± 0.1) | 1.01 (± 0.08) |
| Gliclazide | 0.71 (± 0.19) | 0.71 (± 0.22) | 0.50 (± 0.16) | 8.9 (± 1.5) | 2.7 (± 0.1) | 1.9 (± 0.2) |
| Acetohexamide | 1.3 (± 0.2) | 2.4 (± 0.1) | 1.3 (± 0.1) | 0.35 (± 0.30) | 9.3 (± 5.5) | 5.2 (± 3.1) |
| Glycated HSA-CS1 | ||||||
| Tolbutamidec | 0.97 (± 0.12) | 1.2 (± 0.2) | 0.96 (± 0.22) | 5.7 (± 2.2) | 1.2 (± 0.3) | 1.0 (± 0.2) |
| Gliclazide | 0.65 (± 0.25) | 0.55 (± 0.27) | 0.45 (± 0.22) | 11 (± 3.0) | 1.6 (± 0.2) | 1.3 (± 0.2) |
| Acetohexamide | 1.0 (± 0.1) | 1.6 (± 0.1) | 1.3 (± 0.1) | 1.2 (± 0.9) | 1.7 (± 0.5) | 1.4 (± 0.4) |
| Glycated HSA-CS2 | ||||||
| Tolbutamide | 0.91 (± 0.19) | 0.80 (± 0.17) | 1.2 (± 0.25) | 10 (± 5) | 0.71 (± 0.16) | 1.04 (± 0.17) |
| Gliclazide | 0.77 (± 0.13) | 0.38 (± 0.06) | 0.56 (± 0.09) | 5.0 (± 0.9) | 1.4 (± 0.1) | 2.08 (± 0.09) |
| Acetohexamide | 1.4 (± 0.1) | 0.86 (± 0.03) | 1.3 (± 0.1) | 1.0 (± 0.3) | 2.2 (± 0.3) | 3.2 (± 0.5) |
All of these data were obtained at pH 7.4 and 37°C. The values in parentheses represent a range of ± 1 S.D., as obtained through non-linear regression for the best-fit parameters to eq (1) to the group of results (n = 10–16, each measured in triplicate) or through error propagation based on the standard deviations for these parameters.
The values provided for acetohexamide or tolbutamide and gliclazide with normal HSA are provided for reference and were also measured by HPAC. These results were obtained from Refs. [24] and [27].
The Ka1 and mL1 values for tolbutamide with glycated HSA-CS1 were estimated from the linear region of a double reciprocal plot made according to eq. (1).
The two classes of sites taking part in these interactions consisted of relatively high affinity interactions involving 1–2 binding regions (i.e., as estimated based on the specific activities in Table 1) and a group of lower affinity sites involving up to 3–5 regions on HSA. These results were consistent with those reported for the same drugs with normal HSA or in vitro glycated HSA.24–27 The apparent association equilibrium constants that were estimated for the group of high affinity regions had relative standard deviations of ±7.1 to ±38% (average, ±18%) and were in the general range of 0.65–1.4 × 105 M−1. This range was consistent with earlier values reported for the same drugs with in vitro glycated HSA that had similar or slightly lower levels of glycation (i.e., binding constants of 0.84–1.2 × 105 M−1 for tolbutamide, 1.0 × 105 M−1 for gliclazide and 1.2–2.0 × 105 M−1 for acetohexamide).25–27
Site-specific competition studies for clinical samples
The next set of studies used zonal elution and competition studies with site-specific probe compounds to explore the high affinity regions on in vivo glycated HSA that were interacting with the sulfonylurea drugs. A representative set of chromatograms for these studies is provided in the Supporting Information. These experiments typically required 4–7 min per run, which again made it easy to employ the microcolumns for replicate studies with various drugs and experimental conditions. As noted earlier, the in vivo glycated HSA columns had good stability and showed only relatively small changes in their retention factors for R-warfarin and L-tryptophan during the course of these studies. In addition, the approach that was used to analyze the zonal elution data, as illustrated in Figure 3 and eq (2), provided association equilibrium constants that were determined in a manner that was independent of any modest variations that occurred in the column binding capacity.28 This fact again meant that only a representative portion of the glycated HSA/HSA from a sample was needed for this type of study and that this method could be used even when there were moderate differences in the amounts of protein in the affinity microcolumns that were prepared from different samples.
Competition studies at Sudlow site I were conducted by using R-warfarin as a site-specific probe.20,24,28,29 Figure 3(a) gives examples of the resulting plots of 1/k versus the mobile phase concentration of the tested drug, as prepared according to eq (2). These plots gave linear relationships for all of the sulfonylurea drugs and in vivo glycated HSA columns, with correlation coefficients of 0.951–0.990 (n = 6–7) and only random variations in the data points about the best-fit lines. These results were consistent with a model in which the sulfonylurea drugs had binding and direct competition with R-warfarin at Sudlow site I. Similar behavior has been noted for columns containing normal HSA or in vitro glycated HSA.24–27
The best-fit lines for plots like those in Figure 3(a) were used to obtain the association equilibrium constant for each sulfonylurea drug at Sudlow site I on the samples of in vivo glycated HSA (see Table 2), giving values that had relative standard deviations ranging from ± 5.1 to ± 10.0% (average, ± 7.4%). These equilibrium constants were in the range expected for one of the high affinity regions of these drugs on HSA and showed good agreement with previous values of 6.5–6.9 × 104 M−1 for tolbutamide, 1.8–3.6 × 104 M−1 for gliclazide and 3.8–5.9 × 104 M−1 for acetohexamide that have been measured at the same site when using in vitro glycated HSA with similar or slightly lower levels of glycation.25–27
Table 2.
Association equilibrium constants (KaI) measured at Sudlow sites I and II for various sulfonylurea drug with normal HSA and in vivo glycated HSA.
| Binding site and column | KaI (× 104 M−1)a | ||
|---|---|---|---|
| Tolbutamide | Gliclazide | Acetohexamide | |
| Sudlow site I | |||
| Normal HSAb | 5.5 (± 0.2) | 1.9 (± 0.04) | 4.2 (± 0.4) |
| Glycated HSA-CS1 | 6.8 (± 0.5) | 2.3 (± 0.2) | 4.7 (± 0.3) |
| Glycated HSA-CS2 | 7.9 (± 0.4) | 1.4 (± 0.1) | 4.0 (± 0.4) |
| Sudlow site II | |||
| Normal HSAb | 5.3 (± 0.2) | 6.0 (± 0.5) | 13 (± 1.0) |
| Glycated HSA-CS1 | 7.3 (± 0.5) | 11 (± 1.0) | 13 (± 0.2) |
| Glycated HSA-CS2 | 7.9 (± 0.3) | 9.2 (± 0.7) | 17 (± 0.4) |
These binding parameters were calculated from data obtained at pH 7.4 and 37°C using the best-fit lines generated according eq (3). The values in parentheses represent a range of ± 1 S.D. for the population of results (n = 6–7, each measured in triplicate) and were determined by using error propagation with the standard deviations of the slopes and intercepts of the best-fit lines that were obtained when using eq (2).
As has been noted in prior work with in vitro glycated HSA,25–27 the strength of these interactions, and the change in these interactions with the extent of glycation, differed between the drugs that were tested. For instance, the association equilibrium constant for tolbutamide at Sudlow site I increased by 1.2- to 1.4-fold in going from normal HSA to both samples of in vivo glycated HSA. These shifts in affinity, as well as the 1.2-fold increase in binding strength seen for tolbutamide in going from glycated HSA-CS1 to glycated HSA-CS2, were all significant at the 95% confidence level (note: the same confidence level was used in all further comparisons in this study). For gliclazide, an increase of 1.2-fold or a decrease of 0.74-fold in affinity occurred when comparing normal HSA with glycated HSA-CS1 or glycated HSA-CS2. A small increase of 1.1-fold or a decrease of 0.95-fold in affinity may have been present for acetohexamide in going from normal HSA to glycated HSA-CS1 or glycated HSA-CS2, but these differences were not statistically significant. In addition, a 0.61-fold decrease in affinity was noted for gliclazide between glycated HSA-CS1 and glycated HSA-CS2.
It has been suggested that changes in binding like those seen in Table 2 may be produced by variations in the extent and types of modifications that occur at various regions on HSA during the glycation process.16,18,25–27 This is supported by the fact that such modifications have been previously noted to occur at or near Sudlow sites I and II for both in vivo glycated HSA and in vitro glycated HSA,48–50 with the latter being prepared under the same or similar conditions to those used for the binding studies in Refs. 25–27. The shifts in binding seen in Table 2 are consistent with the fact that glycation-related modifications at Sudlow sites I and II have also been found for samples of the in vivo glycated HSA that was used in this current study (see Supporting Information). In addition, it has been observed with in vitro glycated HSA that the pattern of these modifications can vary with the level of glycation.48,49 This explains why the affinity for a drug at individual binding sites may change between normal HSA and glycated HSA or between two different preparations of glycated HSA,25–27 as is illustrated in Table 2.
Additional competition studies were carried out by using L-tryptophan as a probe for Sudlow site II,20,24,28,29 as illustrated in Figure 3(b). Plots prepared according to eq (2) gave linear behavior for all the sulfonylureas tested, indicating that these drugs had binding and direct competition with L-tryptophan at Sudlow site II. The correlation coefficients for these graphs ranged from 0.975–0.999 (n = 6–7) and gave only random variations in the data points about the best-fit lines. These results agreed with data that have been obtained for the same drugs using columns that contained normal HSA or in vitro glycated HSA.24–27
Table 2 shows the association equilibrium constants at Sudlow site II that were calculated from plots like those in Figure 3(b). These values had relative standard deviations ranging from ± 1.5 to ± 9.1% (average, ± 5.2%). These equilibrium constants were in the range expected for one of the high affinity regions on glycated HSA and were similar to values of 5.9–7.2 × 104 M−1 for tolbutamide, 3.8–7.6 × 104 M−1 for gliclazide and 7.9–12 × 104 M−1 for acetohexamide that have been measured previously at Sudlow site II with in vitro glycated HSA that had similar or lower levels of glycation.25–27 In this current study, tolbutamide had a 1.4- to 1.5-fold increase in affinity at Sudlow site II in going from normal HSA to glycated HSA-CS1 or glycated HSA-CS2. For gliclazide, an increase of 1.8- or 1.5-fold was seen at the same binding region in going from normal HSA to glycated HSA-CS1 or glycated HSA-CS2, respectively. No apparent change was seen when comparing the binding of acetohexamide at this site on normal HSA versus glycated HSA-CS1; however, a 1.3-fold increase in affinity was noted when the binding of acetohexamide at Sudlow site II on either of these columns was compared with the affinity noted for glycated HSA-CS2.
CONCLUSION
It was found that affinity microcolumns could be used as analytical tools to examine the binding of drugs to modified proteins from individual clinical samples. This approach was demonstrated by using it to investigate the binding of several sulfonylurea drugs to in vivo glycated HSA. Only 20 SL of serum or plasma was needed to prepare each affinity microcolumn, which contained 6–12 nmol protein. These microcolumns could be used to conduct binding studies with typical run times of 4–8 min and were quite stable, making it possible to obtain good reproducibility and to reuse the same protein preparation for many studies. For instance, over the course of more than 250 sample application cycles on each microcolumn, the equivalent of only 25–50 pmol protein was needed per experiment.
These affinity microcolumns were utilized in several formats to examine the binding of sulfonylurea drugs with in vivo glycated HSA that was obtained from individual patients. Frontal analysis was employed to investigate the overall binding of each sulfonylurea drug and indicated that these drugs gave the best-fit to a model involving a group of 1–2 high affinity sites and a group of up to 3–5 lower affinity binding regions. The apparent association equilibrium constants measured for the high affinity sites of the in vivo glycated HSA were in the general range of 0.65–1.4 × 105 M−1, which agreed with values that have been reported for the same drugs with comparable preparations of in vitro glycated HSA.25–27
Zonal elution was utilized to study the interactions that occurred at specific binding sites on HSA. These experiments indicated that both Sudlow sites I and II on the in vivo glycated HSA were binding to all of the tested drugs. The association equilibrium constants for these interactions were consistent with values expected for the high affinity sites of these drugs. These values also agreed with previous data obtained for these drugs at the same sites on in vitro glycated HSA with similar levels of modification.25–27 All of these results confirmed that the in vivo glycated HSA columns could be used to study the binding of drugs to modified proteins in a disease such as diabetes and in comparing samples for separate patients. It was further possible to use the data from these columns to see how these interactions changed with the extent of glycation. In some cases, the change in affinity at a given site was as large as 1.8-fold in going from normal HSA to in vivo glycated HSA. This observation also agrees with previous results for the same drugs and similar preparations of in vitro glycated HSA, for which changes in affinity of up to 1.4- to 1.9-fold have been reported.25–27 It is possible that even larger shifts in binding may be observed in future work when this approach is applied to a larger number of clinical samples. These changes are of possible clinical importance in that they would be expected to alter the displacement of these drugs by other agents and the amount of such a drug that is present in the non-bound protein form in serum or plasma. These effects, in turn, could change the effective dose of the drug in a patient and lead to undesirable effects such as hypoglycaemia or poor control of glucose levels.22,23,25–27
The methods that were described in this study for preparing and using affinity microcolumns in binding studies are not limited to individual clinical samples of glycated HSA but could be adapted for work with alternative proteins or modified proteins. Examples of other serum carrier proteins that should be amenable to this approach include α1-acid glycoprotein and lipoproteins, both of which have been used in prior drug-binding studies by HPAC involving proteins prepared from pooled serum51–55 and which are known to vary in their composition or structure with disease state.56–58 It is expected that the application of this method could also be expanded to include proteins that have been modified through processes other than glycation (e.g., glycosylation or alterations due to oxidative stress).56–58 In the future, this method could be extended to systems such as cytosolic proteins that bind to lipids, hormones or drugs.59–61 The main requirement in each of these cases is the use of an appropriate immunoaffinity support, or comparable protein isolation system, and a sufficient sample volume to isolate the desired protein for placement in an affinity microcolumn. Continued improvements in the design of affinity microcolumns, along with the development of better isolation and immobilization schemes for work with small amounts of proteins, should further assist in this research. The expected result is a set of powerful tools for binding studies that will be applicable for use in personalized medicine and in the study of various disease states, ranging from diabetes to cancer and autoimmune disorders.56–61
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under grants R01 DK069629 and R01 GM044931 and was conducted in facilities that were renovated under grant RR015468. The authors also thank C. Bi for her assistance with the fructosamine assay.
References
- 1.Hood L, Heath J, Phelps M, Lin B. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 2.Weston A, Hood L. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 3.Lim D, Dickherber A, Compton C. Anal Chem. 2011;83:8–13. doi: 10.1021/ac1018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unwin N, Whiling D, Gan D, Jacqmain O, Ghyoot G, editors. IDF Diabetes Atlas. 5. International Diabetes Federation; Brussels: 2011. [Google Scholar]
- 5.National Diabetes Fact Sheet: General Information and National Estimates on Diabetes in the United States, 2007. U.S. Centers for Disease Control and Prevention; Atlanta, GA: 2008. [Google Scholar]
- 6.Rendell M. Drugs. 2004;64:1339–1358. doi: 10.2165/00003495-200464120-00006. [DOI] [PubMed] [Google Scholar]
- 7.Lapolla A, Fedele D, Seraglia R, Traldi P. Mass Spectrom Rev. 2006;25:775–797. doi: 10.1002/mas.20090. [DOI] [PubMed] [Google Scholar]
- 8.Jones RL, Cerami A. Rec Adv Diabetes. 1984;1:173–180. [Google Scholar]
- 9.Shaklai N, Garlick RL, Bunn HF. J Biol Chem. 1984;259:3812–3817. [PubMed] [Google Scholar]
- 10.Rohovec J, Maschmeyer T, Aime S, Peters JA. Chem Eur J. 2003;9:2193–2199. doi: 10.1002/chem.200204632. [DOI] [PubMed] [Google Scholar]
- 11.Thornalley PJ, Langborg A, Minhas HS. Biochem J. 1999;344:109–116. [PMC free article] [PubMed] [Google Scholar]
- 12.Khan MWA, Rasheed Z, Khan WA, Ali R. Biochemistry (Moscow) 2007;72:146–152. doi: 10.1134/s0006297907020034. [DOI] [PubMed] [Google Scholar]
- 13.Murtiashaw MH, Winterhalter KH. Diabetologia. 1986;29:366–370. doi: 10.1007/BF00903346. [DOI] [PubMed] [Google Scholar]
- 14.Seedher N, Kanojia M. Chem Biol Drug Design. 2008;72:290–296. doi: 10.1111/j.1747-0285.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 15.Hervé F, Urien S, Albengres E, Duché JC, Tillement JP. Clin Pharmacokin. 1994;26:44–58. doi: 10.2165/00003088-199426010-00004. [DOI] [PubMed] [Google Scholar]
- 16.Barzegar A, Moosavi-Movahedi AA, Sattarahmady N, Hosseinpour-Faizi MA. Prot Pept Lett. 2007;14:13–18. doi: 10.2174/092986607779117191. [DOI] [PubMed] [Google Scholar]
- 17.Lindup WE. Prog Drug Metab. 1987;10:141–185. [Google Scholar]
- 18.Mohamadi-Nejad A, Moosavi-Movahedi AA, Hakimelahi GH, Sheibani N. Intl J Biochem Cell Biol. 2002;34:1115–1124. doi: 10.1016/s1357-2725(02)00031-6. [DOI] [PubMed] [Google Scholar]
- 19.Vidal P, Nielsen E, Welinder BS. J Chromatogr. 1992;573:201–206. doi: 10.1016/0378-4347(92)80120-f. [DOI] [PubMed] [Google Scholar]
- 20.Sudlow G, Birkett DJ, Wade DN. Mol Pharmacol. 1975;11:824–832. [PubMed] [Google Scholar]
- 21.Skillman TG, Feldman JM. Am J Med. 1981;70:361–372. doi: 10.1016/0002-9343(81)90773-7. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya S, Sakurai T, Sekiguchi SI. Biochem Pharmacol. 1984;33:13542–13545. doi: 10.1016/0006-2952(84)90595-1. [DOI] [PubMed] [Google Scholar]
- 23.Harrower A. Drug Safety. 2000;22:312–320. doi: 10.2165/00002018-200022040-00004. [DOI] [PubMed] [Google Scholar]
- 24.Joseph KS, Hage DS. J Chromatogr B. 2010;878:1590–1598. doi: 10.1016/j.jchromb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph KS, Anguizola J, Hage DS. J Chromatogr B. 2010;878:2775–2781. doi: 10.1016/j.jchromb.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph KS, Anguizola J, Hage DS. J Pharm Biomed Anal. 2011;54:426–432. doi: 10.1016/j.jpba.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuda R, Anguizola J, Joseph KS, Hage DS. Anal Bioanal Chem. 2011;401:2811–2819. doi: 10.1007/s00216-011-5382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiel JE, Joseph KS, Hage DS. In: Advances in Chromatography. Grinsberg N, Grushka E, editors. Vol. 48. Vol. 4 Taylor & Francis; New York: 2009. [Google Scholar]
- 29.Hage DS. J Chromatogr B. 2002;768:3–30. doi: 10.1016/s0378-4347(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 30.Yoo MJ, Schiel JS, Hage DS. J Chromatogr B. 2010;878:1707–1713. doi: 10.1016/j.jchromb.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacks D. Clin Chem. 2009;55:1612–1614. doi: 10.1373/clinchem.2009.132704. [DOI] [PubMed] [Google Scholar]
- 32.Bennett C, Guo M, Dharmage S. Diabetes UK Diabetic Med. 2007;24:333–343. doi: 10.1111/j.1464-5491.2007.02106.x. [DOI] [PubMed] [Google Scholar]
- 33.Weykamp C, Garry W, Mosca A, Hoshino T, Little R, Jeppsson JO, Goodall I, Miedema K, Myers G, Reinauer H, Sacks D, Slingerland R, Siebelder C. Clin Chem. 2008;54:240–248. doi: 10.1373/clinchem.2007.097402. [DOI] [PubMed] [Google Scholar]
- 34.Miedema K. Clin Chem Lab Med. 2003;41:1259–1265. doi: 10.1515/CCLM.2003.193. [DOI] [PubMed] [Google Scholar]
- 35.Saudek C, Herman W, Sacks D, Bergenstal R, Edelman D, Davidson M. J Clin Endocrinol Metab. 2008;93:2447–2453. doi: 10.1210/jc.2007-2174. [DOI] [PubMed] [Google Scholar]
- 36.Ruhn P, Garver S, Hage DS. J Chromatogr A. 1994;669:9–19. doi: 10.1016/0021-9673(94)80332-3. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Hage D. In: Handbook of Affinity Chromatography. Hage DS, editor. Vol. 3 Taylor and Francis; New York: 2006. [Google Scholar]
- 38.Yang J, Hage DS. J Chromatogr A. 1997;766:15–25. doi: 10.1016/s0021-9673(96)01040-0. [DOI] [PubMed] [Google Scholar]
- 39.Joseph KS, Moser AC, Hage DS. J Chromatogr A. 2009;1216:3492–3500. doi: 10.1016/j.chroma.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad M, Moser A, Hage D. J Sep Sci. 2009;32:1145–1155. doi: 10.1002/jssc.200800567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burtis CA, Ashwood ER, Bruns DE. Tietz Textbook of Clinical Chemistry and Molecular Diagnosis. 5. Saunders; St Louis: 2006. [Google Scholar]
- 42.Hage DS, Walters RR. J Chromatogr. 1987;386:37–49. doi: 10.1016/s0021-9673(01)94582-0. [DOI] [PubMed] [Google Scholar]
- 43.Hage DS, Thomas DH, Beck MS. Anal Chem. 1993;65:1622–1630. doi: 10.1021/ac00059a023. [DOI] [PubMed] [Google Scholar]
- 44.Gundry RL, White MY, Nogee J, Tchernyshyov I, Van Eyk JE. Proteomics. 2009;9:2021–2028. doi: 10.1002/pmic.200800686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Proteomics Clin Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polaskova V, Kapur A, Khan A, Molloy MP, Baker MS. Electrophoresis. 2010;31:471–482. doi: 10.1002/elps.200900286. [DOI] [PubMed] [Google Scholar]
- 47.Rohlfing CL, Wiedmeyer HM, Little RR, England JD, Tennill A, Goldstein DE. Diabetes Care. 2002;25:275–278. doi: 10.2337/diacare.25.2.275. [DOI] [PubMed] [Google Scholar]
- 48.Barnaby OS, Cerny RL, Clarke W, Hage DS. Clin Chim Acta. 2011;412:277–285. doi: 10.1016/j.cca.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnaby OS, Cerny RL, Clarke W, Hage DS. Clin Chim Acta. 2011;412:1606–1615. doi: 10.1016/j.cca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frolov A, Hoffman R. Anal Bioanal Chem. 2010;397:2349–2356. doi: 10.1007/s00216-010-3810-9. [DOI] [PubMed] [Google Scholar]
- 51.Xuan H, Hage DS. Anal Biochem. 2005;346:300–310. doi: 10.1016/j.ab.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 52.Mallik R, Xuan H, Guiochon G, Hage DS. Anal Biochem. 2008;376:154–156. doi: 10.1016/j.ab.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xuan H, Joseph KS, Wa C, Hage DS. J Sep Sci. 2010;33:2294–2301. doi: 10.1002/jssc.201000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, Sobansky MR, Hage DS. Anal Biochem. 2010;397:107–114. doi: 10.1016/j.ab.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sobansky MR, Hage DS. Anal Biochem Chem. 2012;403:563–571. doi: 10.1007/s00216-012-5816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Israili ZH, Dayton PG. Drug Metab Rev. 2001;33:161–235. doi: 10.1081/dmr-100104402. [DOI] [PubMed] [Google Scholar]
- 57.Mohamed NA, Kuroda Y, Shibukawa A, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. J Chromatogr A. 2000;875:447–453. doi: 10.1016/s0021-9673(99)01288-1. [DOI] [PubMed] [Google Scholar]
- 58.Skipski VR. In: Blood Lipids and Lipoproteins: Quantitation, Composition, and Metabolism. Nelson GJ, editor. Wiley; New York: 1972. pp. 471–583. [Google Scholar]
- 59.Sugiyama Y, Takikawa H. Tanpakushitsu Kakusan Koso. 1990;35:941–956. [PubMed] [Google Scholar]
- 60.Ichikawa K, Hashizume K. Life Sci. 1991;49:1513–1522. doi: 10.1016/0024-3205(91)90323-4. [DOI] [PubMed] [Google Scholar]
- 61.Matarese V, Stone RL, Waggoner DW, Bernlohr DA. Progr Lipid Res. 1989;28:245–272. doi: 10.1016/0163-7827(89)90001-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.