Summary
The availability of extensive genome information for many different microbes, including unculturable species in mixed communities from environmental samples, has enabled systems-biology interrogation by providing a means to access genomic, transcriptomic, and proteomic information. To this end, metaproteomics exploits the power of high performance mass spectrometry for extensive characterization of the complete suite of proteins expressed by a microbial community in an environmental sample.
Background and rationale for metaproteomics
The advent of genome-based science, which is founded primarily on high throughput DNA sequencing, enables an unprecedented view of the molecular machinery of life. In particular, the proliferation of large-scale genome sequencing centers, coupled with remarkable advancements in next-generation DNA sequencing, is revolutionizing molecular biology. While the completion of the human genome project over ten years ago1,2 was a monumental technical feat, it is important to keep in mind that this has spawned a concomitant increase in the number of complete genome sequences (> 3,000) for a plethora of lower organisms, such as bacteria, archaea, and viruses (see the Integrated Microbial Genomes with Microbiome Samples website; http://img.jgi.doe.gov/cgi-bin/m/main.cgi). More recent efforts have extended this experimental genomic technology to environmental field samples, thereby providing whole community (often termed metagenomic) sequence information on microbial members from various ecological communities.3 Interrogation of the protein complement of these microbial communities (termed either whole community proteomics4 or metaproteomics5) seeks to identify the functional expression of the metagenome and elucidate the metabolic activities occurring within a community at the moment of sampling.
While genomic information provides a wealth of important information about the potential molecular machinery that might be employed for life processes, it does not reveal the finer-level details of actual expression and function – that is the realm of RNA and proteins. The focus of systems-biology science hinges on four key “omics” technologies: genomics for DNA, transcriptomics for RNA, proteomics for proteins, and metabolomics for small molecules/metabolites. Clearly, a comprehensive view of molecular biology would involve an integration of all of these. However, these omics approaches represent the cutting-edge of experimental genome science, and each are in a state of rapid development. Integrating their outputs is an obvious and desirable goal, but the mechanics of how to best do this remain somewhat elusive at the present time.
This article will focus on proteomics, which can be broadly defined as the comprehensive characterization of the suite of proteins for an organism, tissue, community, etc. 6 In this regard, proteomics represents the identification of functional gene products, providing incredible information and insight into the molecular machinery produced and utilized by organisms to sustain the metabolic processes required for life. This is certainly not meant to imply that RNA, metabolites, lipids, etc. are unimportant, but rather that proteins are usually regarded as the key operational units in most metabolic pathways. From a systems-biology perspective, the starting point for all considerations and the key enabling data is the genome. The DNA sequence, and thus genes, for an organism defines the complete repertoire of all potential transcriptional and translational biomolecules that can be used for metabolic activities. RNA, or more specifically mRNA, as gleaned from transcriptome measurements, provides much more detailed information about genome expression and thus gene activity. It is feasible to measure RNA under a variety of experimental conditions in order to examine how genome transcription varies between each condition. However, the final product of mRNA are proteins, which constitute the basic molecular machinery that carry-out the majority of functional aspects of cellular metabolism. It is important to keep in mind that while measuring RNA is informative, there are additional levels of cellular localization and regulation at the protein level (such as post-translational modifications, controlled proteolysis / protein turnover for example) that are not captured in RNA measurements. Thus, one might argue that proteomic and metabolomic measurements provide the most informative details about the key players most responsible for cellular function.
The large-scale characterization of any given proteome is accomplished by comparing measured protein or peptide data with predicted protein or peptide data derived from genomic information. Thus, it is vital to have complete and relevant genome sequence information for the system being studied. This has led to the term proteogenomics to describe the strong linkage between genomics and proteomics.7 As implied, the quality of the proteomic measurements is inextricably linked to the quality of the genome or metagenomic sequence. As follows, sequencing/assembly/annotation errors in the genome propagate directly to proteome measurements, leading to complications with the proteome identifications.
The technological requirements for proteomic measurements include high throughput processing, sensitive protein/peptide detection, large dynamic range, ability to deal with very complex mixtures, accurate mass measurements, and ability to structurally characterize (and resolve) peptide sequences. In this regard, mass spectrometry (MS) has emerged as the unchallenged leader in the field, becoming the dominant technological platform for almost all proteomic measurements. Early work in proteomics was conducted with two-dimensional gel electrophoresis (2D-GE) 8,9, often accompanied by MS detection; this general approach has been supplemented by the use of multiple dyes in a single gel electrophoresis experiment (DIGE).10 More recently, the ability to interface multi-dimensional liquid chromatographic separations (either off-line or on-line) with MS (generally termed LC-MS) has enabled an unprecedented glimpse into very complex samples, such as those containing many thousands of proteolytic peptides.11,12 The advent of high performance MS platforms, such as the hybrid Quadrupole-time of flight-MS (Q-TOF-MS), linear trapping quadrupole – Fourier transform ion cyclotron resonance-MS (LTQ-FTICR-MS), and linear trapping quadrupole – Orbitrap- MS (LTQ-Orbitrap-MS), has provided much improved capabilities for rapid scanning and high performance (mass accuracy, mass resolution, dynamic range), thereby opening the door to more advanced and discriminatory proteomic measurements.
Single-celled organisms such as bacteria, archaea, and viruses, provide relatively straight-forward test-beds upon which to evaluate these advanced LC-MS proteome approaches. For example, much work has been directed at the proteomic characterization of bacteria grown in laboratory benchtop reactors, with specific inquiries into the proteomic response observed between dissimilar growth states (i.e., anaerobic vs. aerobic; wild-type vs. metal-stressed). For a bacterium with a genome of about 4 million base pairs, it is possible to measure up to ~ 2500-3500 proteins of the possible ~4000 proteins, thus characterizing the proteome in remarkable depth. These datasets provide information that can be used to interrogate metabolic activities by synthesizing the protein information into broader regulatory pathways. For example, it was possible to identify the key proteins involved in chromate reduction by Shewanella oneidensis 13,14, which provided the information necessary to design more detailed gene knockout experiments.15
Prior successful work on microbial isolates has spawned an acute interest in extension of the methodology to more complex samples, such as consortia found in natural environments. In this case, the level of organismal diversity is substantially greater than that of a laboratory cultivable isolate. As a starting point, relevant metagenomic information for the community to be characterized by proteomics is required. Such information provides the catalog of all possible proteins that could be present in a sample at any given time. Interrogation of the metaproteome seeks to elucidate the metabolic activities employed by the community at the moment of sampling. While challenges remain in comprehensive metaproteomic characterization for natural environmental samples, as might be expected, initial work has been focused on lower complexity microbial communities, such as biofilms found in acid mine drainage 16-18, sludge water bioreactor systems 19, and synthetic communities in gnotobiotic mice.20 These systems provided an excellent starting point to evaluate, develop, and optimize advanced proteomic methods (and associated informatics) for more complex systems. Even though there are notable challenges in these systems, remarkable progress has been made in demonstrating proteomic approaches for even more advanced querying, such as species strain-level resolution 21 and peptide-inferred genome typing.22
Inspection of metaproteome datasets reveals information about microbial community structure, dynamics, and functional activities that are important for a better understanding of various community aspects, such as microbial recruiting, how participating organisms cooperate and compete for nutrient resources, and how these organisms distribute metabolic activities across the community (including defense systems). At a slightly higher level, such information will be crucial for the characterization of host/microbe interactions, such as bacterial/plant or bacterial/human interfaces (i.e. the human gut ecosystem) – two research areas poised to make substantial scientific contributions in the not so distant future.
In total, this environmental metaproteomic approach has established a “proof-of-concept” that is beginning to facilitate applications to a variety of important research areas, including:
Bioremediation – characterizing how microbial species might help to arrest/remediate toxic metal contamination in soils, sediments, and ground waters.
Carbon cycling – characterizing the role that microbial species have in the flow of carbon in a given ecosystem.
Bioenergy – characterizing how microbial species might help convert cellulosic material to biofuels (bioethanol/biodiesel/biohydrogen).
Human health – characterizing how microbial species impact/control disease vs. health in various body sites (i.e. oral, gastrointestinal, genital).
This article will summarize the current state of metaproteomic research, highlighting experimental and bioinformatic details, giving several working examples, and providing an outlook for where this field is headed. It is not intended to serve as a comprehensive review, but rather to provide lay readers with a broader perspective of the experimental approach, the driving scientific questions, and what can be learned from the resulting massive datasets.
Experimental approach for complex metaproteome samples
Proteomic measurements can be accomplished by a variety of mass spectrometry-based approaches, all of which center around the unambiguous identification of the range of proteins (or peptides) that exist in a given sample. The success of a metaproteome measurement relies on three factors: efficient protein extraction from a complex environmental sample, peptide/protein separation/fractionation prior to detection, and subsequent high-throughput unambiguous peptide/protein identification, as illustrated in the flowchart of Figure 1. We acknowledge that there are other versions of the experimental protocol for metaproteome measurements, but we chose here to highlight the technique that we feel provides the deepest level of information. Clearly, protein extraction protocols are designed to be unbiased, but in reality they suffer similar biases to DNA, where the fidelity of extraction is not 100%. This is not to say that proteomics measurements are invalid, but rather that the results of both environmental proteomics and genomics must be evaluated honestly in the reality of the sample preparation processes. Although the bulk of the measurements made during the infancy of proteomics were largely protein cataloguing, the field has matured to a more hypothesis-driven experimental approach. These proteomic measurements are now able to provide information regarding the differential expression of proteins based on environmental conditions or time points, their partitioning into various subcellular structures, their post-translational modifications (PTM), their involvement in protein-protein interactions, and even their specific molar quantities.
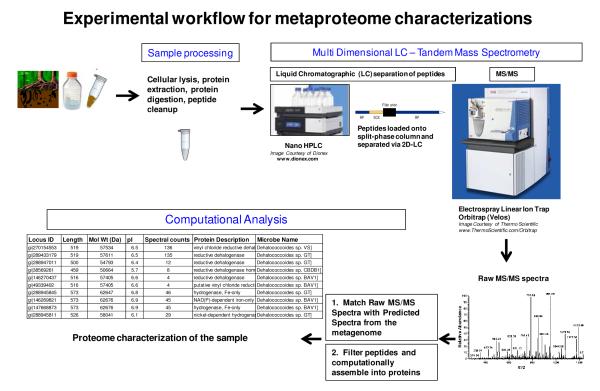
Figure 1.
Experimental flowchart for sample preparation and measurement in a metaproteomic experiment. Sample collection and processing steps must be optimized to match the nature of the material to be analyzed, in terms of biomass amount and complexity, matrix composition, sample heterogeneity, etc. The resulting proteome sample is digested with trypsin and loaded onto a bi-phasic HPLC column for concomitant 2D-separation and MS analysis via nanoelectrospray-based ionization of eluting peptides. Acquisition of parent peptide ion (MS1) mass and fragmentation (MS/MS or MS2) information provides an experimental dataset containing hundreds of thousands of spectra that can be computationally matched to the predicted proteome obtained from the metagenome information.
There are essentially two basic types of LC-MS-based proteomic measurement protocols: top-down and bottom-up. The top-down strategy is conceptually straightforward; whole proteins are separated via liquid chromatography by exploiting hydrophobicity and/or charge, and analyzed directly by MS and tandem mass spectrometry (MS/MS).23-26 The resulting MS information obtained over the course of the separation is evaluated against a protein database to identify the proteins. These types of measurements are useful in that the exact mass of the protein, along with fragmentation information, can provide details of not only the identity of the protein, but also its intact molecular form, the presence of isoforms, and potential modifications.27 However, the same characteristic that makes this method straightforward also complicates the analysis when complex changes to the protein’s predicted mass occur, i.e. post-translational modifications (PTMs), truncations, and single nucleotide polymorphisms (SNPs). In practice, measurement of the exact mass is suggestive of a potential protein identification, but is complicated by the range of possible protein modifications that can alter predicted molecular masses from the genome database. Clearly, mass measurement by itself has some limitation on what it can provide, especially for an uncharacterized mixture for which many of the potential proteins are not even predictable. Additionally, technical challenges involving the separation and detection of larger proteins (> 50 kDa) still exist. These challenges, coupled with the increased complexity, homology, and species variability intrinsic to metaproteomes, limit the extensive deployment of top-down measurements to characterize complex environmental samples at this point in time.
Bottom-up or shotgun proteomics 28-30, on the other hand, employs additional sample processing and analysis that greatly expand the ability to attain deep proteomic measurements. In this strategy, proteins are first digested to peptides via proteases such as trypsin. These peptides are then chromatographically separated and analyzed by MS (parent molecular mass) and MS/MS techniques (fragmentation/sequence information). The resulting fragmentation spectrum serves as a type of barcode that provides a means to uniquely identify a given peptide.31 Combined with its initial mass, fragmentation data, and in some cases its chromatographic retention time 32, the peptide sequence is computationally determined, and in most cases assigned back to a specific protein.
Although bottom-up proteomics is the standard protocol for LC-MS/MS-based protein identification, its very design creates a unique problem; the resulting homogenized mixture of proteolytic peptides must be computationally linked back to a specific protein. Thus redundant, homologous, or isobaric peptide species can complicate the analysis, as they may be potentially assigned to multiple proteins in a given genome database. This issue is exacerbated with metaproteomic analyses, as the ratio of ambiguous peptide IDs increase at the expense of unique ones, both of which affect the fidelity of the final protein call.
Despite these minor issues, the power of bottom-up proteomics has been successfully demonstrated in microbial isolates, both cultured and uncultured, and more recently in the study of environmental communities with established metagenomes.33-35 As might be expected, there was early recognition in the value of integrating top-down with bottom-up proteomics to exploit the power of combining the two approaches.36
The experimental heart of any extensive proteome measurement rest on two factors: effective peptide/protein separation and unambiguous detection. For complex environmental samples, these factors become even more crucial. For example, not all proteins are expressed and maintained at similar or consistent levels – this is true even for a microbial isolate. In fact, the dynamic range of proteins, defined as the range between the protein of highest abundance to those of the lowest abundance, can be on the order of 104-106 for microbial isolates37 and perhaps significantly larger for environmental community samples. This exceedingly wide range of protein expression challenges the very notion of complete proteomic characterization. However, coupling extensive separation (i.e. gel electrophoresis and/or liquid chromatography) with sensitive detectors (i.e. mass spectrometers) provides the best opportunity to accomplish this goal.
One powerful separation technology that has become widely employed in the proteomic arena is high performance liquid chromatography (HPLC or LC). Like 2D-PAGE, LC separations can also be multidimensional, employing properties such as hydrophobicity and charge to separate biomolecules.29 HPLC is an attractive option for MS-based proteomic detection, as separation of complex mixtures can be performed directly online with the mass spectrometer.38-39 This simple, yet powerful capability reduces sample handling/processing, is automatable, generally more high-throughput, and, most importantly, enables very robust and reproducible separations at time scales compatible with MS measurements. This latter point is related to the duty cycle of the instrument, in that most current mass spectrometers can scan sufficiently fast with respect to liquid chromatographic elution profiles to handle incomplete separations, but still greatly benefit from the reduced complexity per unit time afforded by the LC separations, thus enhancing more comprehensive peptide identifications from complex samples.
Several emerging fractionation methodologies/technologies are beginning to play a critical role in metaproteomics. Advances in online separation such as ultra-high pressure liquid chromatography (UPLC) and/or stationary phase modifications (monolithic, sub-2μm, or microparticle shell technologies) provide increased chromatographic resolution, sensitivity, and speed of online LC-MS measurements.6,40-43 In addition, application and development of offline separations serve to simplify the complex mixture upfront. Two powerful offline methodologies, IEF and GELFrEE 44,45 separate complex protein mixtures by isoelectric point and molecular size, respectively, and provide a starting point for three-dimensional LC separations. Both methodologies are compatible with bulk separations at sample amounts in line with current procedures. Coupled together, these strategies outline a multidimensional approach that better separates complex environmental mixtures, thus augmenting protein detection.
Likewise, improvements to current mass spectrometers, such as increasing their duty-cycle, selectivity, and sensitivity46, provide additional enhancements to dynamic range. In addition, the increased commercial availability of high mass accuracy, high resolution instruments6,47-48 provides the means to discriminate between homologous proteins of cohabitating microorganisms, as well as detect subtle differences and/or modifications to expressed proteins, i.e. sequence polymorphisms and/or PTMs.
Although most LC-MS/MS-based proteomics studies thus far have focused on microbial isolates, transitioning the methodology to the study of metaproteomes mandates careful consideration of the unique challenges posed by more complex samples. For instance, as opposed to laboratory-cultivable isolates that provide an essentially inexhaustible amount of biological material, environmentally-derived samples are often biomass-limited, partly due to complications in sample acquisition and/or restrictions due to time and expense of sampling. In addition to biomass limitations, proper experimental design revolves around several factors, such as the type of MS-based experiments to be performed (protein cataloguing, differential expression in various growth states, quantitation, etc.), the separation/fractionation required (tailored to the complexity of the sample), the level of mass accuracy and resolution needed for unambiguous protein identification,49-50and measurement replication for proper statistics (both technical and biological replications.
Bioinformatic considerations
The use of high throughput multidimensional LC-MS/MS measurements for proteomics clearly provides an impetus to develop robust bioinformatic approaches to convert the raw spectral data to peptide sequence information, thus identifying the proteins from which each peptide spectrum was derived. For example, an online multidimensional LC-MS/MS experiment acquires molecular masses and fragmentation information for tens of thousands of tryptic peptides over the course of an extended chromatographic run (which can be greater than 24 hours) for each sample. This process results in the generation of hundreds of thousands of fragmentation spectra, most of which can be coupled with the measured mass of the parent peptide and assigned to a specific peptide sequence. Clearly, manual determination of the peptide sequences from these MS/MS spectra is impractical. However, as powerful a technique as tandem mass spectrometry is, the resulting data is essentially a series of peak lists that, with appropriate bioinformatic processing, can be quickly analyzed and consolidated into a meaningful output that is both concise and informative.
In the context of environmental metaproteomics and downstream bioinformatic processing, a predicted protein database constructed from metagenomic information is required to properly assign peptide sequence information, as inferred from MS/MS-derived fragmentation patterns, to the proteins to which the peptides were derived. Therefore, the quality of metaproteomic data is inextricably linked to the quality of the metagenomic analysis. Certainly, the transition in DNA sequencing from Sanger approaches to 454 and now Illumina has had dramatic impacts on metaproteome measurements. In particular, the shorter reads accessible with the newer sequencing approaches initially confounded metaproteome measurements, but have become addressable with improved informatics and assembly methods.51 Obviously the best metaproteome identifications will be dependent on searching a comprehensive and relevant predicted database from the metagenome for the exact same sample. Interestingly, the reduced cost and wide-spread availability of high throughput DNA sequencing is revolutionizing (meta)genome availability. Some researchers argue that we now should require matched metagenomes for every sample. However, this is still not completely feasible, and in cases where the metagenome for the exact same sample is not available or possible, related metagenomic data as well as synthetic metagenome databases are also valid approaches.52, 53 The inability to get exact metagenome information, along with biological complexities intrinsic to environmentally-derived samples, i.e. homologous proteins/domains, horizontal gene transfer, and/or strain variation, require additional proteomic measurement constraints in order to maximize the number of protein identifications while controlling the false discovery rate (FDR). One solution employs the use of high mass accuracy, high resolution mass spectrometers, such as FTICR or Orbitrap instrumentation, that have the discriminatory power (< 5 parts-per-million mass accuracy) to resolve both nominally isobaric and co-eluting peptide species of similar mass to charge ratios, as shown in Figure 2. Note that the ability to simultaneously measure and resolve both mass and charge for the peptide ions permits high fidelity assignments even in the complex regions of the chromatogram. In fact, the application of high mass accuracy allows one to achieve extraordinarily low FDR levels (< 0.1%).54
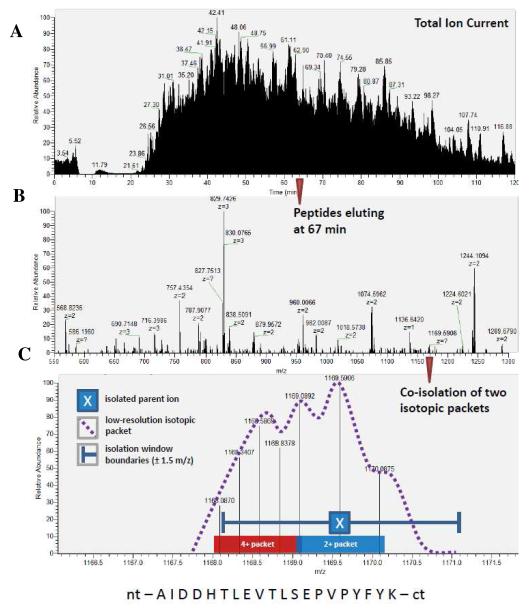
Figure 2.
Experimental data from 2D-LC-MS/MS of a simulated microbial consortium consisting of Rhodopseudomonas palustris, Escherichia coli, Ignicoccus hospitalis, and Nanoarchaeum equitans. A) total ion chromatogram of the 2-hour reverse phase measurement of salt pulse #2; B) parent ion mass spectrum (MS1) of the peptides eluting at 67.0 min from the chromatogram in ‘A’ above. All the ions are recorded with mass resolutions of 30,000 and mass accuracies of < 5 ppm. The charge state of each peptide is denoted by ‘z’. Undeciphered charge states, represented by ‘?’, occur when the instrument cannot fully distinguish overlapping isotopic packets; C) zoom expansion of isobaric ion region at nominal m/z 1169, revealing two isotopic packets (manually verified as overlapping 2+ and 4+ ions). Note that a low resolution measurement would not have distinguished the presence of two distinct ions here. The resulting MS/MS measurement revealed the sequence identity (listed below) of the 2+ ion, without confusing it with the co-eluting 4+ ion, even though both isotopic packets we co-isolated and fragmented creating a hybrid MS/MS spectrum.
The field of proteome bioinformatics encompasses a range of computational operations, including protein database searching and filtering of raw mass spectra, peptide-spectrum matching followed by peptide-to-protein assembly, data mining, graphical representation, and data dissemination. Within the past 3-5 years, there has been a tremendous increase in the diversity and availability of software architectures to accomplish these functions.55 While the field is too vast to adequately detail in this feature report, it is worth noting that some of the earlier versions of database searching 56 and peptide scoring 57 methodologies have been replaced with much more advanced bioinformatic approaches, most of which have become quite standardized and fairly routine, leaving more attention to be paid to other aspects of data mining and analyses.
While database searching and protein identifications are fairly straight-forward for microbial isolates, moving to a metaproteome measurement also raises some other challenging aspects. For example, most proteome database search algorithms parse peptide identifications into two categories; unique and non-unique. Aptly named, unique peptides can unambiguously be assigned to one specific protein within the database, whereas non-unique are shared between two or more proteins. For microbial community samples, this designation can be too restrictive; there are “semi-unique” peptides that correspond to a class of microbes whose genomes are quite similar, but in fact are designated as separate species. For example, several species of Bacteroides exist in the human gut microbiome. While it is easy to resolve peptides between Clostridium and Bacteroides genera, it is more difficult to differentiate peptides between closely-related Bacteroides species (or strains). If one relies only on the traditional unique vs. non-unique classification system, it is easy to misrepresent the datasets. To address this issue, some informatics approaches cluster or group similar proteins together, and then report unique “protein groups” rather than unique proteins. While this challenges the biological interpretation, it permits a better and more accurate treatment of the measurements and identifications.
Although database searching may be the most widely used strategy for analyzing proteomic data, its most notable drawback is its reliance on a user provided database. With regard to a sequenced isolate, this is normally acceptable. However, what type of database would one search against to identify proteins from an environmental sample if the metagenome was not available or was incomplete? Often, the microbial species that populate an environmental sample are uncultivable and thus lack complete genome sequence. This begs the question, how does one deal with naturally underrepresented species? Is it possible that less prevalent, undetected community members could be contributing proteins to the metaproteome? These proteins, if abundant enough to exist within the sample’s defined dynamic range, could potentially go undetected as there would be no representative sequence in the provided metagenomic database. These interesting dilemmas are perhaps addressable using another bioinformatic approach termed de novo sequencing.
De novo sequencing is an alternative approach for analyzing peptide fragmentation data. Instead of matching an observed spectrum to a database-derived theoretical spectrum, de novo algorithms determine the sequence of a peptide directly from the data provided in its tandem mass spectrum.58 The major advantage of this approach is, of course, the identification of peptides that are non-existent in a given database as well as those with polymorphisms or post-translational modifications. With regard to environmental samples, this is extremely advantageous, as complete metagenome sequences are difficult to acquire for numerous samples. While there is considerable interest in these approaches, the accuracy and speed of de novo analyses for metaproteome measurements are still limiting for wide-spread implementation. The emergence of rapid-scanning high resolution mass spectrometers will likely favorably impact this area, as the availability of high resolution, high mass accuracy measurements will greatly aid both the throughput and accuracy of the de-novo approaches.
Recent exponential growth in metaproteomics research
Success in proteomics for microbial isolates has prompted the biological research community to push this experimental approach to natural environmental samples in which microbial components compete and cooperate with each other. The focus of the following section is not intended as a comprehensive review, as this has been featured recently elsewhere 34-35, 59-63, but rather is meant to provide a synopsis of current work and recent developments in this field.
The development of multidimensional LC-MS/MS technology for characterizing microbial isolates has greatly expanded the accessible proteome range of 2D-GE work, thereby opening a new regime of proteome characterization 38,64 that enables the identification of several thousand proteins from individual microorganisms.65-66 This provides an approximate order of magnitude increase in the range of the measured proteomes, which permits a much deeper and thus more comprehensive glimpse into the molecular activities of microorganisms.
One of the first large-scale whole community proteome measurements involved an native microbial consortium from acid mine drainage.18 Extension of this approach was used for the strain-resolved characterization of the dominant microbial species22,67, and revealed genome recombination as a crucial component for adaptation to specific ecological niches.21 Further studies of this system uncovered the ecological distribution of member organisms and provided information about initial microbial colonization, subsequent recruitment of microbial membership, and aging/maturing of the biofilms.68-69 More recent work has been directed at various quantification approaches (both label-free and stable isotope labeling) for metaproteomics, including the demonstration of extensive isotopic labeling of an environmental biofilm community in the lab.70-71 Additionally, the use of stable isotope probing (SIP) has been demonstrated for metaproteomics, allowing for the characterization of microbial and protein turn-over in a microbial community.72-75
Metaproteomic measurements have tended to focus on three major types of ecosystems: (1) aqueous (lakes and oceans), (2) terrestrial (soils, sediments), and (3) eukaryotic host microbiomes (termites, mice, plants, and humans). For aqueous ecosystems, metaproteomics has been used to decipher biological information about microbial populations in highly productive or nutrient-limited ocean ecosystems. For example, investigation of a highly productive coastal upwelling system revealed abundant microbial proteins involved in the prevention of oxidative damage and protein refolding.76 Related work revealed how nutrient transport functions dominate the SAR11 metaproteome at nutrient-limited locations in the Sargasso Sea77; this general approach was extended to investigate ocean-scale shifts in microbial nutrient utilization and energy transduction.78 These studies illustrate the power of metaproteomics for characterizing the functional protein signatures that reveal metabolic information for microbes directly in their natural environmental ecosystems, thereby providing insight into how nutrients are utilized, how microbial life cycles over time, and how microbes cooperate and compete with other members of their ecosystems. Metaproteomics has also begun to play a role in bioengineering systems by providing key microbial metabolic information that can be used to custom design and fine-tune industrial operations. For example, metaproteomics has been used as a critical research component for investigating and optimizing sewage sludge treatment by biological agents.5, 79-80
For terrestrial ecosystem research, most work is focused on characterization of microbes in soil in an effort to better understand carbon/nitrogen flow and contaminant remediation.81 For example, metaproteomics has helped characterize microbial metabolic activities relevant for bioremediation at nutrient-stimulated82-84, xenobiotic85, hydrocarbon86-87, and heavy metal-contaminated sites.88-89 These studies provide not only important information about how microbes can be used in industrial clean-up operations, but also provide a level of information that can be used in a predictive fashion to custom-design microbes for specific applications.
Recent advances in in-situ proteome extraction techniques from soils have opened the door to a much deeper level of metaproteome measurement.90-93 Soil metaproteomics has become a high interest research target area, although significant challenges with regard to microbial diversity, environmental matrices, and limited metagenomic information complicate this issue at present. Exciting new applications are beginning to emerge in the characterization of permafrost soils,94 as well as a broader characterization of the earth’s soil microbiome (the Terragenome project; http://www.terragenome.org/about/).
In perhaps the most complex level of microbial metaproteomics, there is substantial interest in understanding the symbiotic/pathogenic relationships between microbes and their eukaryotic hosts. For example, to better understand the basis of cellulose degradation by termites, metaproteomics was used to characterize the functional activities of uncultivable symbiotic microbes in the termite hindgut.95 Metaproteomics has also been used to investigate factors mediating plant-microbial interactions, in particular focusing on the proteome differences between lab cultured microbes vs. their plant symbiont counterparts 96, or factors influencing crop rhizosphere communities.97-98
The last 3-5 years has seen an explosion of research interest in the human microbiome, fueled primarily by health-related issues. Microbes vastly outnumber human cells in even healthy individuals. This necessitates a thorough understanding of both normal (symbiotic) and diseased (dysbiotic) states, specifically with regard to microbiome functional dynamics. In response to the early interest in characterizing a possible microbial basis for periodontal disease, a novel 3D peptide fractionation method was used with tandem MS to characterize human salivary microbiota99, and proteomics was used to study Porphyromonas gingivalis as part of a model oral microbiome community.100
One of the crucial microbe-host ecosystems is the human gut, which provides the impetus for focused research in integrated microbial metagenomics and metaproteomics. For example, metaproteomics has been employed to examine the microbiota in the developing human infant GI tract, although the measurement depth was very limited in this case.101 This approach has also revealed temporal stability of a core proteome for an established intestinal microbiome of a healthy, adult human.53 A more recent extensive metaproteome study focused on the adult gut microbiomes for healthy, matched, human twins, and provided a glimpse into the highly integrated relationship between microbial and human proteins.102 Numerous proteins were identified from the most dominant bacterial members, a portion of which are shown in Table 1, revealing a remarkable insight into which bacteria are metabolically active and, more specifically, which metabolic activities are most prevalent. As might be expected, the functional distribution of COGs (clusters of orthologous groups) predicted from the metagenome were somewhat distinct from what was actually observed in the metaproteome, lending credence to the notion that metagenome analysis alone is not sufficient to capture the actual metabolic activities in progress at the time of sampling. Interestingly, several human proteins were detected in these enriched bacterial samples, and provided information about evidence of host innate immunity response to the microbiome. The reader is referred to reference #102 for further details on specific human proteins and their relation to human host respond to the microbiome. The integrated metagenomic/metaproteomic approach has now been extended to examine healthy vs. diseased conditions in the human gut microbiome, specifically focusing on matched twins that were either healthy or had Crohn’s disease.103 Notable differences in bacterial species as well as functional signatures were observed, and provided insight into the relationship between intestinal inflammation and microbiome structure / function.
Table 1. Abundant microbial proteins in two human fecal samples (healthy twins).
Partial list of abundant proteins (and corresponding microbes) identified in fecal samples from two matched, healthy, human twins. (extracted from Supplemental Table S2 of reference #88). Note the range of microbial species identified and their respective proteins, which provide some insight into at least the most dominant metabolic activities underway in this sample. The second column of normalized spectra abundance factors (NSAF) provides relative peptide abundances in each case. Clearly, the microbiome signatures from the two individuals differ, indicating that even among normal “healthy” individuals, there are some specific differences in the microbiome composition and activities.
| Subject 7 | ||
|---|---|---|
| Protein | NSAF | Description |
| Blon_NCC2705:637328939 | 0.0057 | NP_696945 BL1798 DNA-binding protein Hu [Bifidobacterium longum NCC2705] |
| Lmon_EGD-e:637220958 | 0.0046 | NP_464684 lmo1159 hypothetical protein [Listeria monocytogenes EGD-e] |
| Sent_ParA_ATCC_9150:637601100 | 0.0046 | YP_150128 SPA0826 putative propanediol utilization protein PduJ [Salmonella enterica enterica sv Paratyphi A ATCC 9150] |
| Cbei_NCIMB_8052:638818377 | 0.00424 | ZP_00907197 CbeiDRAFT_4763 Propanediol utilization: polyhedral bodies [Clostridium beijrincki NCIMB 8052] |
| Bado-ATCC_15703:633763489 | 0.0042 | YP_909415 BAD_0552 DNA-binding protein HB1 [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703.633763205 | 0.00368 | YP_909148 BAD_0285 5OS ribosomal protein L7/L12 [Bifidobacterium adolescents ATCC 15703] |
| Cbei_NCIMS_8052:638013307 | 0.00364 | ZP_009C7207 CbeiDRAFT_4773 Propanediol utilization: polyhedral bodies [Closlndium beijerincki NCIMB 8052] |
| Bthe_VPI-5482:637412747 | 0.0034 | NP_813174 BT4263 glyceraldehyde 3-phosphate dehydrogenase [Bacteroides thetaiotaomicron VPI-5482] |
| Bthe_VPI-5482:637409127 | 0.00333 | NP_809628 BTD715 ATP synthase c subunit [Baderoides thetaiotaomicron VPI-5482] |
| Bthe_VPI-5482:637409804 | 0.00263 | NP_810286 BTI373 ferritin A [Bacteroides thetaiotaomicron VPI-5482] |
| Bado_ATCC_15703:639764448 | 0.00241 | YP_910346 BAD_1483 30S ribosomal protein S6 [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703:639763236 | 0.00226 | YP_909195 BAD_0332 50S ribosomal protein L24 [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703:639763244 | 0 00191 | YP_909103 BAD_0320 30S ribosomal protein S10 [Bifidobacterium adolescentis ATCC 15703] |
| Bfra_NCTC_9343:637229119 | 0.00182 | YP_213042 BF3437 glutamate dehydrogenase [Baderoides fragilis NCTC 9343] |
| Bado_ATCC_15703:639763254 | 0.0018 | YP_909193 BAD 0330 30S ribosomal protein S17 [Bifidobacterium adolescentis ATCC 15703] |
| Bfra_NCTC_9343:637228728 | 0.0018 | YP_212659 BF3045 hypolhetical protein [Baderoides fraqilis NCTC 9343] |
| Bthe_VPI-5482:637411194 | 0.00162 | NP_811648 BT2736 ribosomal protein L10 [Bacteroides thetaiotaomicron VPI-5482] |
| Bthe-VPI-5482:637411311 | 0.00156 | NP_811756 BT2844 hypotbelical protein [Bacteroides thelaiotaomicron VPI-5482] |
| Bthe_VPI-5482:637410267 | 0.00155 | NP_810743 BT1830 co-chaperonin GroES [Bacteroides thetaiotaomicron VPI-5482] |
| Bthe_VPI-5482:637411522 | 0.00152 | NP_811966 BT3054 succinate dehydrogenase [Baderoides thetaiotaomicron VPI-5482] |
| Bado_ATCC_15703:639763468 | 0.00149 | YP_909394 BAD_0531 30S ribosomal protein S7 [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703:635763780 | 0.00144 | YP_909693 BAD_0830 transaldolase [Bifidobacterium adolescentis ATCC 15703] |
| Subjects 8 | ||
| Protein | NSAF | Description |
| Bado_ATCC_15703:639763205 | 0.00393 | YP_909148 BAD_0285 58S ribosomal protein L7/L12 [Bifidobacterium adolescentis ATCC 15703] |
| Bfra_NCTC_9343:637228728 | 0.00301 | YP_212659 BF3045 hypothetical protein [Baderoides fragilis NCTC 9343] |
| Cper_ATCC_13124:638082321 | 0:00278 | YP_695886 CPF_1441 hypothetical protein [Clostidium perfringens ATCC 13124] |
| Cbei_NCIMB_8052:638818471 | 0.00269 | ZP_00907288 CbeiDRAFT_4854 conserved hypothetical protein [Clostridium beijerincki NCIMB 8052] |
| Msmi_ATCC_35061:640592178 | 0.00238 | YP_001272963 Msm_0390 hypothetical protein Msm_0390 [Metbanobrevibacter smithii ATCC 35061] |
| Bthe_VPI-5482:637409127 | 0:00218 | NP_809626 BT0715 ATP synthase C subunit [Bacteroides thetaiotaomicron VPI-5482] |
| Bfra_NCTC_9343:637227901 | 0.00218 | YP_211854 BF2231 ATP synthase C chain [Bacteroides fraqilis NCTC 9343J |
| Lmon_EGD-e:637220958 | 0.00197 | NP_464684 lmo1159 hypothetical protein [Lisleria monocytogenes EGO-e] |
| Sent_ParA_ATCC_9150:637601100 | 0.00197 | YP_150128 SPA0826 putative propanediol utilization protein PduJ [Salmonella enterica enterica sv Paratyphi A ATCC 9150] |
| Blon_NCC2705:637328939 | 0.00192 | NP_696945 BL1798 DNA-binding protein Hu [Bifidobacterium longum NCC2705] |
| Bthe_VPI-5482:637409804 | 0.00175 | NP_810286 BTI373 ferritin A [Bacteroides thetaiotaomicron VPI-5482] |
| Bado_ATCC_15703:633763489 | 0.00171 | YP_909415 BAD_0552 DNA-binding protein HB1 [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703:639763780 | 0.00163 | YP_909693 BAD_0830 transaldolase [Bifidobacterium adolescentis ATCC 15703] |
| Bado_ATCC_15703:639763256 | 0.00161 | YP_909195 BAD_0332 50S ribosomal protein L24 [Bifidobacterium adolescentis ATCC 15703] |
| Spne_D39:639679311 | 0.0016 | YP_817231 SPD_1823 glyceraldehyde-3-phosphate dehydroqenase, type I [Streptococcus pneumoniae D39] |
| Bado_ATCC_15703:639763468 | 0.0014 | YP_909394 BAD_0531 30S ribosomal protein S7 [Bifidobacterium adolescentis ATCC 15703] |
| Bfra_NCTC_9343.637229696 | 0.00135 | YP_213598 BF4D19 50S ribosomal protein L11 [Baderoides fraqilis NCTC 9343] |
| Cbei_NCIMB_8052:638818377 | 0.0013 | ZP_009C7197 CbeiDRAFT_4763 Propanediol utilization polyhedral bodies [Clostndrum beijerincki NCIMB 8052] |
| Bado_ATCC_15703:639764448 | 0.00129 | YP_910346 BAD_1483 30S ribosomal protein S6 [Bifidobacterium adolescentis ATCC 15703] |
| Cdif_630:640157746 | 0.00126 | YP_001088425 CD1918 putative ethanolamine/propanediol utilization protein [Clostridium difficile 630] |
| Bado_ATCC_15703:633763125 | 0.0012 | YP_909072 BAD_0209 30S ribosomal protein S16 [Bifidobacterium adolescentis ATCC 15703] |
| Bthe_VPI-5482:637411194 | 0.0012 | NP_811648 BT2736 ribosomal protein L10 [Bacteroides thetaiotaomicron VPI-5482] |
Outlook
With regard to whole community proteome measurements, there are obvious concerns about the complexity and species/protein dynamic range of these systems. However, these challenges provide the spark to development of the next generation of proteomic approaches with a specific focus on superior separation and measurement technologies. Based on the explosion of analytical technology in the past five years, there is no reason to expect a slow down. Better chromatographic methods are continuing to emerge and likewise, faster scanning, higher performance mass spectrometers are becoming common-place, thereby providing important new capabilities that can be integrated into proteomic workflows as they become available. In fact, the incorporation of such technologies into existing workflows often provides dramatic improvements, even in the absence of other changes specific to sample acquisition or preparation. Similarly, advancements in multidimensional chromatography and depletion approaches (for abundant proteins) are dramatically increasing the accessible dynamic range of proteome measurements, which will no doubt lead to more robust and biologically informative measurements.
The ability to conduct environmental metaproteomic measurements, in particular for unculturable organisms, is already generating a new standard of molecular level interrogation. Because it is possible to get metagenomic information on unculturable organisms, this opens the door to other omics techniques of characterization as well. The main point here is that systems biology science can be taken to the field, and is not limited to lab-based cultures for study. The simultaneous development, advancement, and integration of closely related metagenomic and metaproteomic approaches are paving the way towards the characterization of lower complexity microbial consortia, as discussed in detail in the previous section. Such information provides a detailed look into how communities assemble (even at the strain-resolved level), how they distribute metabolic activities, how they progress and mature with time, and how they respond to environmental perturbations. Though moderate challenges lie ahead in applying this proteomic approach to very complex communities, such as those found in the human gut or in soils, the incredible progress over the past five or so years in metaproteomics research suggests that the ground-work has been laid for enhancing the success of these studies as well.
While it is difficult to speculate too far into the future, it would be remiss to end this article without a discussion of where metaproteomics is headed and what is likely to be delivered by this approach. Based on present activity in the field, it is reasonable to expect that very detailed, perhaps even comprehensive, proteome maps of many major microbial communities will be available within the next 5-10 years. This undoubtedly will revolutionize our understanding of microbial ecology, in particular for metabolic activities, regulatory processes (and interactions), and species interaction dynamics. When integrated with other systems biology-based tools (i.e other omics technologies), it may be possible to obtain extensive enough molecular-level information to permit the construction of detailed, high-resolution regulatory maps of biological function. This would have an enormous impact on a variety of research fields. For example, it would be possible to quickly ascertain the functional potential of microbes for various biotechnological applications such as bioenergy production, environmental cleanup, carbon sequestration, chemical and pharmaceutical synthesis, and even help unravel the nature of microbes - human host relationships. With regards to the latter, there is, at present, a very poor understanding of not only how microbes interact with each other, but also how they interact with their various hosts, i.e. plant-microbe (endophytic or pathogenic) or human-microbe (beneficial vs. pathogenic).
Although often ignored due to their microscopic sizes, microbes comprise the overwhelming number of living organisms on earth. The effect they have on influencing their environments, including the healthy human “ecosystem,” seems obvious, but remains largely unexplored. Metaproteomics holds tremendous potential to be one of the key approaches to help unravel this information, even in unculturable environmental systems.
Acknowledgements
Financial support was provided by the U.S. Department of Energy, Biological and Environmental Research Division, Genome Sciences Program (for content related to the environmental metaproteomics) and by the National Institutes of Health, Human Microbiome Project, grant UH2DK83991 (for content related to the human microbiome metaproteomics). Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy.
Author biographies
Dr. Robert Hettich is a Distinguished Research Staff Scientist at the Oak Ridge National Laboratory. His research interests focus on demonstration of high performance mass spectrometry methodologies for proteome characterizations, in particular for natural microbial communities. Dr. Chongle Pan is a research staff scientist at the Oak Ridge National Laboratory. His research focuses on integration of experimental and computational methodologies for proteome quantification in microbial systems. Dr. Karuna Chourey is a research staff scientist at the Oak Ridge National Laboratory. Her research involves design of experimental MS-based approaches for characterizing the microbial metaproteomes in natural soil ecosystems. Dr. Richard Giannone is a research staff scientist at the Oak Ridge National Laboratory. His research is centered on MS-based proteome approaches for characterizing metabolic activities and symbiotic interactions in microbial systems.
Footnotes
Notice: This manuscript has been authored by UT-Battelle, LLC, under contract with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
References
- 1.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Singh J, Behal A, Singla N, Joshi A, Birbian N, Singh S, Bali V, Batra N. Biotechnol J. 2009;4:480–494. doi: 10.1002/biot.200800201. [DOI] [PubMed] [Google Scholar]
- 4.Banfield JF, VerBerkmoes NC, Hettich RL, Thelen MP. Omics. 2005;9:301–333. doi: 10.1089/omi.2005.9.301. [DOI] [PubMed] [Google Scholar]
- 5.Wilmes P, Bond PL. Environ. Microbiol. 2004;6:911–920. doi: 10.1111/j.1462-2920.2004.00687.x. [DOI] [PubMed] [Google Scholar]
- 6.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 7.Beverley SM, et al. Philos Trans R Soc Lond B Biol Sci. 2002;357:47–53. doi: 10.1098/rstb.2001.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klose J. Humangenetik. 1975;26:231–243. doi: 10.1007/BF00281458. [DOI] [PubMed] [Google Scholar]
- 9.O’Farrell PH. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 10.Unlu M, Morgan ME, Minden JS. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 11.Delahunty CM, Yates JR., 3rd Biotechniques. 2007;43:563–567. [PubMed] [Google Scholar]
- 12.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Prot. Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 13.Brown SD, Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Zhou J, Hettich RL, Thompson DK. Mol Cell Proteomics. 2006;5:1054–1071. doi: 10.1074/mcp.M500394-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Thompson DK, Hettich RL. J Prot. Res. 2007;6:1745–1757. doi: 10.1021/pr060502x. [DOI] [PubMed] [Google Scholar]
- 15.Chourey K, Thompson MR, Shah M, Zhang B, VerBerkmoes NC, Thompson DK, Hettich RL. J Prot. Res. 2009;8:59–71. doi: 10.1021/pr800776d. [DOI] [PubMed] [Google Scholar]
- 16.Baker BJ, Banfield JF. FEMS Microbiol Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 17.Singer SW, Chan CS, Zemla A, VerBerkmoes NC, Hwang M, Hettich RL, Banfield JF, Thelen MP. App. Env. Microbiology. 2008;74:4454–4462. doi: 10.1128/AEM.02799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, 2nd, Shah M, Hettich RL, Banfield JF. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- 19.Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B, Hettich RL, Bond PL, VerBerkmoes NC, Banfield JF. ISME J. 2008;2:853–864. doi: 10.1038/ismej.2008.38. [DOI] [PubMed] [Google Scholar]
- 20.Mahowald MA, et al. Proc Natl Acad Sci U S A. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo I, Denef VJ, VerBerkmoes NC, Shah MB, Goltsman D, DiBartolo G, Tyson GW, Allen EE, Ram RJ, Detter JC, Richardson P, Thelen MP, Hettich RL, Banfield JF. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- 22.Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, Hettich RL, Banfield JF. Environ. Microbiol. 2009;11:313–325. doi: 10.1111/j.1462-2920.2008.01769.x. [DOI] [PubMed] [Google Scholar]
- 23.Kelleher NL. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 24.Parks BA, Jiang L, Thomas PM, Wenger CD, Roth MJ, Boyne MT, 2nd, Burke PV, Kwast KE, Kelleher NL. Anal Chem. 2007;79:7984–7991. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid GE, McLuckey SA. J Mass Spectrom. 2002;37:663–675. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 26.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KJ, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton P, Thomas PM, Kelleher NL. Nature. 2012;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siuti N, Kelleher NL. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 29.Wolters DA, Washburn MP, Yates JR., 3rd. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 30.Yates JR., 3rd. Annu Rev Biophys Biomol Struct. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- 31.Eng JK, Mccormack AL, Yates JR., 3rd. J. Amer. Soc. Mass Spect. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Yang L, Freitas MA. BMC Bioinformatics. 2008;9:347. doi: 10.1186/1471-2105-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elias DA, Monroe ME, Marshall MJ, Romine MF, Belieav AS, Fredrickson JK, Anderson GA, Smith RD, Lipton MS. Proteomics. 2005;5:3120–3130. doi: 10.1002/pmic.200401140. [DOI] [PubMed] [Google Scholar]
- 34.Keller M, Hettich R. Microbiol Mol Biol Rev. 2009;73:62–70. doi: 10.1128/MMBR.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF. Nat Rev Microbiol. 2009;7:196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- 36.Strader M, VerBerkmoes N, Tabb D, Connelly H, Barton J, Bruce B, Pelletier D, Davison B, Hettich RL, Larimer F, Hurst G. J Prot. Res. 2004;3:965–978. doi: 10.1021/pr049940z. [DOI] [PubMed] [Google Scholar]
- 37.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Proc Natl Acad Sci U S A. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 39.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Chem Rev. 2007;107:3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 40.Anspach JA, Maloney TD, Colon LA. J. Sep Sci. 2007;30:1207–1213. doi: 10.1002/jssc.200600535. [DOI] [PubMed] [Google Scholar]
- 41.Everley RA, Croley TR. J. Chromatogr A. 2008;1192:239–247. doi: 10.1016/j.chroma.2008.03.058. [DOI] [PubMed] [Google Scholar]
- 42.Kay RG, Gregory B, Grace PB, Pleasance S. Rapid Comm. Mass Spect. 2007;21:2585–2593. doi: 10.1002/rcm.3130. [DOI] [PubMed] [Google Scholar]
- 43.Motoyama A, Venable JD, Ruse CI, Yates JR., 3rd. Anal Chem. 2006;78:5109–5118. doi: 10.1021/ac060354u. [DOI] [PubMed] [Google Scholar]
- 44.Lee JE, et al. J. Amer Soc Mass Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran JC, Doucette AA. Anal Chem. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 46.Olsen JV, Nielsen ML, Damoc NE, Griep-Raming J, Moehring T, Makarov A, Schwartz J, Horning S, Mann M. Mol. Cell. Proteomics. 2009:S40–S40. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. J. Mass Spect. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 48.Scigelova M, Makarov A. Proteomics. 2006:16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 49.Bradshaw RA, Burlingame AL, Carr S, Aebersold R. Mol Cell Proteomics. 2006;5:787–788. doi: 10.1074/mcp.E600005-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Taylor CF, et al. Nat Biotechnol. 2008;26:860–861. doi: 10.1038/nbt0808-860. [DOI] [PubMed] [Google Scholar]
- 51.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle C, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton M, Williams KH, Long PE, Banfield JF. Science. 2012;337:1661–1662. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 52.Rooijers K, Kolmeder C, Juste C, Dore J, de Been M, Boeren S, Galan P, Beauvallet C. BMC Genomics. 2011;12:6. doi: 10.1186/1471-2164-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolmeder CA, de Been M, Nikkila J, Ritamo I, Matto J, Valmu L, Salojarvi J, Palva A, Salonen A, de Vos WM. PLoS ONE. 2012;7:e29913. doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michalski A, Neuhauser N, Cox J, Mann M. J. Proteome Res. 2012;11:5479–5491. doi: 10.1021/pr3007045. [DOI] [PubMed] [Google Scholar]
- 55.Wright JC, Hubbard SJ. Comb. Chem. High Throughput Screening. 2009;12:194–202. doi: 10.2174/138620709787315508. [DOI] [PubMed] [Google Scholar]
- 56.Tabb D, Narasimhan C, Strader M, Hettich RL. Anal. Chem. 2005;77:2464–2474. doi: 10.1021/ac0487000. [DOI] [PubMed] [Google Scholar]
- 57.Razumovskaya J, Olman V, Xu D, Uberbacher E, VerBerkmoes N, Hettich RL, Xu Y. Proteomics. 2004;4:961–969. doi: 10.1002/pmic.200300656. [DOI] [PubMed] [Google Scholar]
- 58.Allmer J. Expert Rev Proteomics. 2011;8:645–57. doi: 10.1586/epr.11.54. [DOI] [PubMed] [Google Scholar]
- 59.Hettich RL, Sharma R, Chourey K, Giannone RJ. Curr Opin Microbiol. 2012;3:373–80. doi: 10.1016/j.mib.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 60.Siggins A, Gunnigle E, Abram F. FEMS Microbiol Ecol. 2012;80:265–80. doi: 10.1111/j.1574-6941.2011.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seifert J, Taubert M, Jehmlich N, Schmidt F, Volker U, Vogt C, Richnow H, von Bergen M. Mass Spectrom Rev. 2012;31:683–97. doi: 10.1002/mas.21346. [DOI] [PubMed] [Google Scholar]
- 62.Armengaud J. Environ. Microbiol. 2013;15:12–23. doi: 10.1111/j.1462-2920.2012.02811.x. [DOI] [PubMed] [Google Scholar]
- 63.Schneider T, Riedel K. Proteomics. 2010;10:785–98. doi: 10.1002/pmic.200900450. [DOI] [PubMed] [Google Scholar]
- 64.Cravatt BF, Simon GM, Yates JR., 3rd. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 65.Gupta N, et al. Genome Research. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VerBerkmoes NC, Shah MB, Lankford PK, Pelletier DA, Strader MB, Tabb DL, McDonald WH, Barton JW, Hurst GB, Hauser L, Davison BH, Beatty JT, Harwood CS, Tabita FR, Hettich RL, Larimer FW. J. Prot.Res. 2006;5:287–298. doi: 10.1021/pr0503230. [DOI] [PubMed] [Google Scholar]
- 67.Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, VerBerkmoes NC, Hettich RL, Banfield JF. Proc. Natl. Acad. Sci. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mueller RS, Denef VJ, Kalnejais LH, Suttle KB, Thomas BC, Wilmes P, Smith RL, Nordstrom DK, McCleskey RB, Shah MB, VerBerkmoes NC, Hettich RL, Banfield JF. Mole. Sys. Biol. 2010;6:374. doi: 10.1038/msb.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mueller RS, Dill BD, Pan CL, Belnap CP, Thomas BC, VerBerkmoes NC, Hettich RL, Banfield JF. Environ. Microbiol. 2011;13:2279–2292. doi: 10.1111/j.1462-2920.2011.02486.x. [DOI] [PubMed] [Google Scholar]
- 70.Belnap CP, Pan C, Denef VJ, Samatova NF, Hettich RL, Banfield JF. ISME J. 2011;5:1152–1161. doi: 10.1038/ismej.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belnap CP, Pan C, VerBerkmoes NC, Power ME, Samatova NF, Carver RL, Hettich RL, Banfield JF. ISME J. 2010;4:520–530. doi: 10.1038/ismej.2009.139. [DOI] [PubMed] [Google Scholar]
- 72.Pan CL, Fischer CR, Hyatt D, Bowen BP, Hettich RL, Banfield JF. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006049. Issue: 4 Article Number: 006049 DOI: 10.1074/mcp.M110.006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taubert M, Jehmlich N, Vogt C, Richnow HH, Schmidt F, von Bergen M, Seifert J. Proteomics. 2011;11:2265–2274. doi: 10.1002/pmic.201000788. [DOI] [PubMed] [Google Scholar]
- 74.Jehmlich N, Schmidt F, Taubert M, Seifert J, Bastida F, von Bergen M, Richnow HH, Vogt C. Protocols. 2010;5:1957–1966. doi: 10.1038/nprot.2010.166. [DOI] [PubMed] [Google Scholar]
- 75.Taubert M, Vogt C, Wubet T, Kleinsteuber S, Tarkka MT, Harms H, Buscot F, Richnow H-H, von Bergen M, Seifert J. ISME J. 2012;6:2291–2301. doi: 10.1038/ismej.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sowell SM, Abraham PE, Shah M, VerBerkmoes NC, Smith DP, Barofsky DF, Giovannoni SJ. ISME J. 2011;5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 78.Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS. ISME J. 2010;4:673–685. doi: 10.1038/ismej.2010.4. [DOI] [PubMed] [Google Scholar]
- 79.Kuhn R, Benndorf D, Rapp E, Reichl U, Palese LL. Proteomics. 2011;11:2738–2744. doi: 10.1002/pmic.201000590. [DOI] [PubMed] [Google Scholar]
- 80.Wilmes P, Wexler M, Bond PL. PLoS ONE. 2008;3:e1778. doi: 10.1371/journal.pone.0001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bastida F, Moreno JL, Nicolas C, Hernandez T, Garcia C. European J. Soil Sci. 2009;60:845–859. [Google Scholar]
- 82.Williams MA, Taylor EB, Mula HP. Soil Biol. Biochem. 2010;42:1148–1156. [Google Scholar]
- 83.Wilkins MJ, VerBerkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H, N’Guessan AL, Thomas BC, Nicora CD, Shah MB, Abraham P, Lipton MS, Lovley DR, Hettich RL, Long PE, Banfield JF. App. Env. Microbiology. 2009;75:6591–6599. doi: 10.1128/AEM.01064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Callister SJ, Wilkins MJ, Nicora CD, Williams KH, Banfield JF, VerBerkmoes NC, Hettich RL, N’Guessan L, Mouser PJ, Elifantz H, Smith RD, Loyley DR, Lipton MS, Long PE. Environ. Sci. Tech . 2010;44:8897–8903. doi: 10.1021/es101029f. [DOI] [PubMed] [Google Scholar]
- 85.Desai C, Pathak H, Madamwar D. Bioresource Tech. 2010;101:1558–1569. doi: 10.1016/j.biortech.2009.10.080. [DOI] [PubMed] [Google Scholar]
- 86.Bastida F, Nicolas C, Moreno JL, Hernandez T, Garcia C. Pedosphere. 2010;20:479–485. [Google Scholar]
- 87.Guazzaroni ME, Herbst F-A, Lores I, Tamames J, Pelaez AI, Lopez-Cortes N, Alcaide M, Del Pozo MV, Vietes JM, von Bergen M, Gallego JLR, Bargiela R, Lopez-Lopez A, Pieper DH, Rossello-Mora R, Sanchez J, Seifert J, Ferrer M. ISME J. 2013;7:122–136. doi: 10.1038/ismej.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Halter D, Cordi A, Gribaldo S, Gallien S, Goulhen-Chollet F, Heinrich-Salmeron A, Carapito C, Pagnout C, Montaut D, Seby F, Van Dorsselaer A, Schaeffer C, Bertin PN, Bauda P, Arsene-Ploetze F. Res. in Microbiol. 2011;162:877–887. doi: 10.1016/j.resmic.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 89.Lacerda CMR, Choe LH, Reardon KF. J. Prot. Res. 2007;6:1145–1152. doi: 10.1021/pr060477v. [DOI] [PubMed] [Google Scholar]
- 90.Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria K, Tom L, Brodie E, Hettich RL. J. Prot. Res. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- 91.Taylor EB, Williams MA. Microbial Ecol. 2010;59:390–399. doi: 10.1007/s00248-009-9593-x. [DOI] [PubMed] [Google Scholar]
- 92.Leary DH, Hervey WJ, Li RW, Deschamps JR, Kusterbeck AW, Vora GJ. Anal Chem. 2012;84:4006–13. doi: 10.1021/ac203315n. [DOI] [PubMed] [Google Scholar]
- 93.Keiblinger KM, Wilhartitz IC, Schneider T, Roschitzki B, Schmid E, Eberl L, Riedel K, Zechmeister-Boltenstern S. Soil Biol. Biochem. 2012;54:14–24. doi: 10.1016/j.soilbio.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. Nature. 2011;480:368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 95.Hongoh Y. Cell. Mol. Life Sciences. 2011;68:1311–1325. doi: 10.1007/s00018-011-0648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knief C, Delmotte N, Vorholt JA. Proteomics. 2011;11:3086–3105. doi: 10.1002/pmic.201000818. [DOI] [PubMed] [Google Scholar]
- 97.Wang HB, Zhang ZX, Li H, He HB, Fang CX, Zhang AJ, Li QS, Chen RS, Guo XK, Lin HF, Wu LK, Lin S, Chen T, Lin RY, Peng XX. J. Proteome Res. 2011;10:932–940. doi: 10.1021/pr100981r. [DOI] [PubMed] [Google Scholar]
- 98.Wu LK, Wang HB, Zhang ZX, Lin R, Zhang ZY, Lin WX. PLoS ONE. 2011;6:e20611. doi: 10.1371/journal.pone.0020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rudney JD, Xie H, Rhodus NL, Ondrey FG, Griffin TJ. Mole. Oral Microbiol. 2010;25:38–49. doi: 10.1111/j.2041-1014.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuboniwa M, Hendrickson EL, Xia QW, Wang TS, Xie H, Hackett M, Lamont RJ. BMC Microbiol. 2009;9 doi: 10.1186/1471-2180-9-98. DOI: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klaassens ES, de Vos WM, Vaughan EE. App.Env. Microbiology. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.VerBerkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson J. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 103.Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, Shah M, Halfvarson J, Tysk C, Henrissat B, Raes J, VerBerkmoes NC, Fraser CM, Hettich RL, Jansson JK. PloS ONE. 2012 doi: 10.1371/journal.pone.0049138. 10.1371/journal.pone.0049138df. [DOI] [PMC free article] [PubMed] [Google Scholar]