Abstract
Background
Bromelain, ficin and papain are cysteine proteases from plants that produce itch upon injection into skin. Their mechanism of action has not been considered previously.
Objectives
To determine the mechanism by which these proteases function.
Methods
The ability of these proteases to activate protease-activated receptors was determined by ratiometric calcium imaging.
Results
We show here that bromelain, ficin and papain activate protease-activated receptors 2 and 4.
Conclusions
Bromelain, ficin and papain function as signalling molecules and activate protease-activated receptors. Activation of these receptors is the likely mechanism by which these proteases evoke itch.
Keywords: cysteine protease, itch, protease-activated receptor, pruritus
It was demonstrated in the 1950s that several endopeptidases, or proteases that cleave internal residues of proteins, caused itching, often associated with a burning sensation when injected in the skin.1 The itch-inducing proteases included bromelain from pineapples, ficin from figs, papain from papaya, mucunain, also known as cowhage, from pods of a bean plant, and trypsin from animals. The molecular mechanism by which these proteases evoked pruritus was not understood. The onset of itch was rapid, occurring within seconds, and was thought to result from direct or almost direct activation of sensory fibres. The absence of a weal or flare suggested that the itch was independent of histamine. It is now known that the sensation of peripheral itch is mediated by two distinct nonoverlapping populations of cutaneous nerve fibres that evoke comparable degrees of itch. One set of fibres, the mechanoinsensitive population, is more responsive to histamine than to cowhage. The other set is mechanosensitive and is more responsive to cowhage than to histamine.2,3
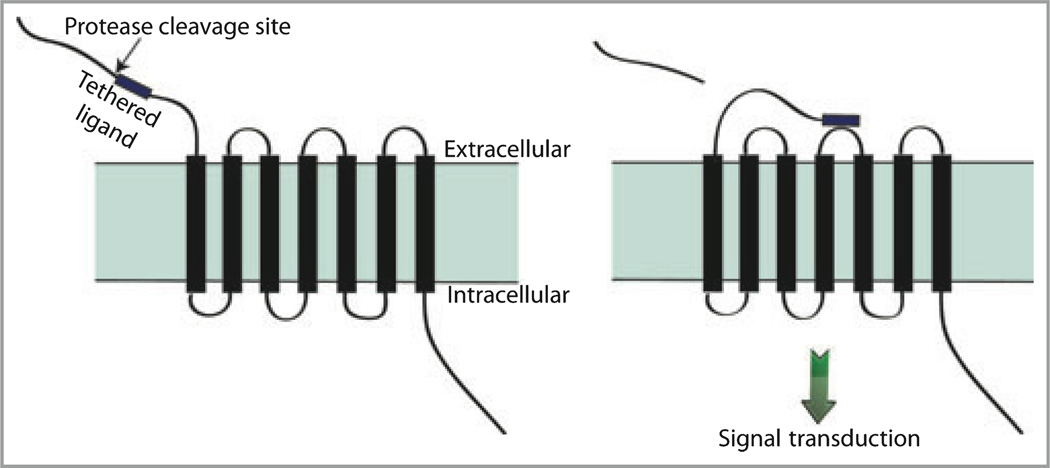
An important development with respect to a new mechanism of protease action was the identification of a family of G-protein coupled receptors (GPCRs) called protease-activated receptors or PARs.4 The family consists of four members, PAR1–PAR4, which are activated by proteolysis. GPCRs are conventionally activated by a ligand binding to an extracellular portion of the receptor leading to a conformational change with subsequent coupling to a cytoplasmic G-protein and downstream signalling. In contrast, with PARs, a residue near the N-terminus of the receptor itself is cleaved by a protease to expose a ‘tethered ligand’. This newly exposed peptide ligand folds back on to the PAR leading to activation (Fig. 1). Certain proteases were thus identified as participating in signal transduction. The only endogenous proteases known to activate PARs were serine proteases, i.e. proteases with serine residues at their active sites, including thrombin, trypsin, tryptase and several kallikreins.5,6 Exogenous cysteine proteases shown to activate PAR2 included a cockroach allergen, the dust mite allergen der p 1 and gingipain from a bacterium associated with gingivitis.7,8 The recent observations that mucunain, which is the active component of cowhage and an exogenous cysteine protease, and cathepsin S, an endogenous cysteine protease, evoke itch and activate PAR2 extended observations that PAR2 activation was associated with itch.9,10 These findings led us to consider the possibility that bromelain, ficin and papain activate PARs, providing a mechanism by which they may cause pruritus.
Fig 1.
Protease-activated receptors (PARs) are activated by proteolytic cleavage. Proteases cleave PARs near the extracellular N-terminus resulting in exposure of a previously cryptic tethered ligand that folds back on to the receptor to become active.
Materials and methods
Materials
Plant proteases and the irreversible cysteine protease inhibitor E64 were obtained from Sigma (St Louis, MO, U.S.A.) and had the following catalogue numbers: bromelain, B4882; ficin, F6008; papain, P4762; E64, E3132. Thrombin was obtained from Enzyme Research Laboratories (South Bend, IN, U.S.A.). Recombinant cathepsin S was prepared in our laboratory. Sephadex G-50 spin columns were used to purify the crude preparations of bromelain and ficin. Protein concentrations were estimated by a noninterfering protein assay kit (G-Biosciences, St Louis, MO, U.S.A.). For the experiments using calcium imaging, proteases were dissolved in 100 mmol L−1 NaCl, 50 mmol L−1 sodium acetate, 10 mmol L−1 β mercaptoethanol, pH 6·0. E64 was dissolved in water.
Human PAR cDNAs were obtained from commercial sources and cloned into the pcDNA3·1(−) vector from Invitrogen (Carlsbad, CA, U.S.A.) and used as described previously.9
Protease-activated receptor activation as determined by calcium mobilization
Additional details are given in Reddy et al.9 HeLa cells were obtained from the American Type Culture Collection (Manassas, VA, U.S.A.) and grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. These cells were transiently transfected with the individual PAR cDNAs using Lipofectamine 2000 (Invitrogen), and incubated for 1 h with 2 µmol L−1 fura-2 in DMEM containing 10% fetal bovine serum. The medium was then aspirated and replaced with 20 mmol L−1 HEPES, 115 mmol L−1 NaCl, 5·4 mmol L−1 KCl, 2 mmol L−1 CaCl2, 0·8 mmol L−1 MgCl2 and 13·8 mmol L−1 glucose, pH 7·4. Cells were stimulated with bromelain, ficin and papain and examined on a Zeiss Axiovert 200M microscope platform (Carl Zeiss, Thornwood, NY, U.S.A.). In experiments that included E64, this protease inhibitor was mixed with the proteases immediately before the latter were added to the cells, to achieve a final concentration of 10 µmol L−1 of the inhibitor. Calcium mobilization was determined using ratiometric imaging of the cells at 340/380 nm coupled with Axiovision software, version 4.6. Ten microlitres of the proteases was used to provide a final concentration of 0·05 mg mL−1. This corresponds to approximately 2 µmol L−1 for each protease as their molecular weights are similar. This concentration was used because each plant protease gave a response that was optimal at 0·05 mg mL−1. No response was detected in nontransfected cells. Positive controls included thrombin and cathepsin S.10 Proteases were added at 20 s after the start of the excitation procedure and images were taken every 5 s, including at zero time, over a 3-min period. The software later analysed 37 images taken in each 3-min excitation. The 340/380 nm fluorescence ratio of a single cell in each image was calculated and the data points were plotted against time using Delta-Graph 4·5 software (Red Rock Software, Salt Lake City, UT, U.S.A.). The data in the figures represent typical single cells. Between 10 and 15 cells were imaged from each well with each protease and the experiments were repeated at least three times.
Results
Partial purification of bromelain and ficin
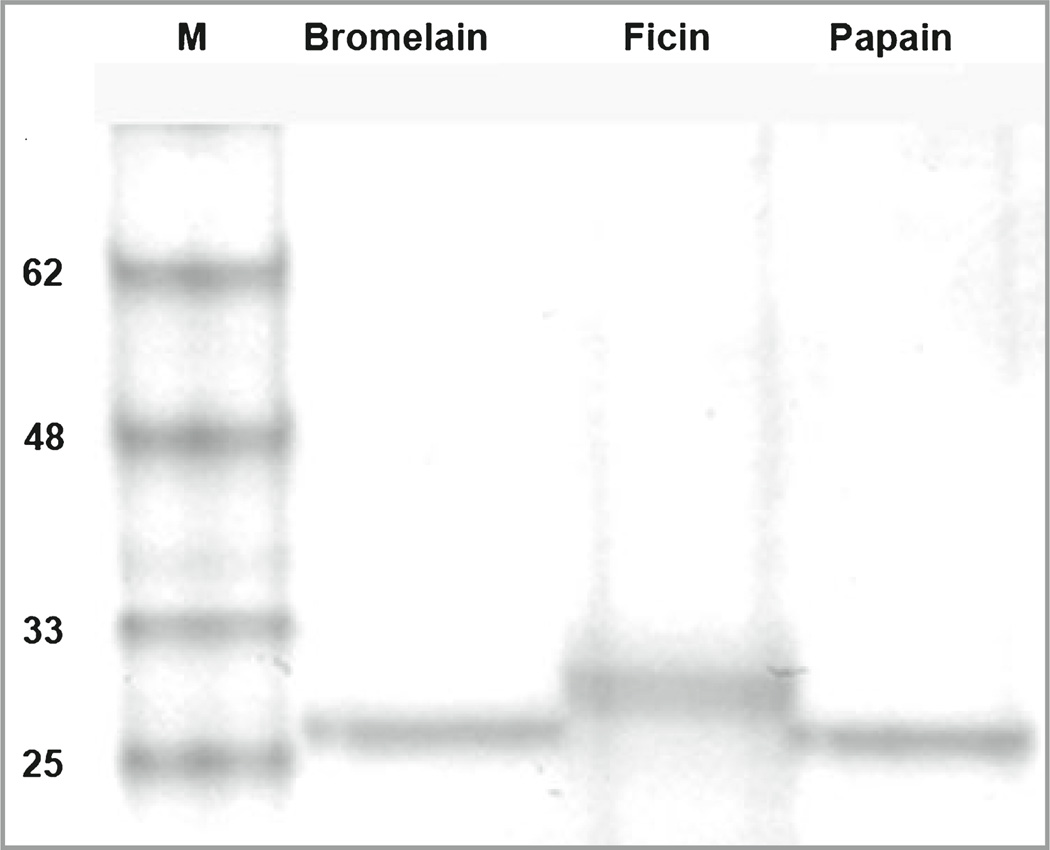
Bromelain and ficin were available as lyophilized powders from pineapple stem and fig tree latex, respectively. As enzymes, their associated descriptions noted enzymatic activity rather than purity as protein concentration would not be of primary concern for conventional use. Because gel electrophoresis revealed a smear at the expected molecular weight, the preparations of bromelain and ficin were at least partially purified over Sephadex, resulting in sharp bands as shown in Figure 2. Papain was supplied in a pure crystallized form, providing a distinct band as expected.
Fig 2.
Polyacrylamide gel electrophoresis of plant proteases. Extracts containing bromelain and ficin were passed over G-50 Sephadex columns to remove most contaminants. Papain was a cleaner preparation, having been obtained in crystalline form. Molecular weight and proteases are indicated. These preparations were used in the receptor activation studies.
Plant cysteine proteases activate protease-activated receptors
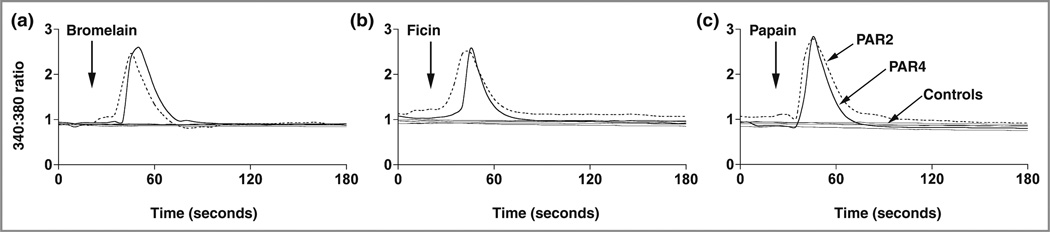
Bromelain, ficin and papain each activated PAR2 and PAR4 (Fig. 3). The capacity of the plant proteases to activate PARs as measured by calcium mobilization is shown. The data are similar to those for mucunain and cathepsin S.9,10 Each protease activated PAR2 and PAR4 to a similar degree. No effect on nontransfected, PAR3- (not shown) or PAR1-transfected cells was observed. E64 prevented each of the proteases from activating PARs.
Fig 3.
Plant proteases activate human protease-activated receptors (PARs). Single-cell imaging of (a) bromelain (2 µmol L−1), (b) ficin (1.5 µmol L−1) and (c) papain (2.17 µmol L−1) induced responses in HeLa cells transfected with either PAR2 or PAR4 as measured by ratiometric calcium imaging in cells loaded with fura-2.9 Labels of curves for bromelain and ficin correspond to those for papain. The responses to PAR2 and PAR4 were blocked by the protease inhibitor E64 (10 µmol L−1). Other negative controls included cells transfected with either salmon sperm DNA or PAR1. Positive controls for PAR2 and PAR4 included cathepsin while thrombin served as a positive control for PAR1 and PAR4 (not shown). Proteases were added as indicated.
Discussion
Plant proteases have long been known to cause itching following introduction to skin but the associated mechanism underlying the sensation of pruritus was not known. The possibility that these proteases could function as signalling molecules was not considered previously. The data presented here reveal that bromelain, ficin and papain are signalling molecules and function via activation of human PAR2 and PAR4. PAR2 has been implicated in signalling associated with the sensation of itch by other proteases and it follows that these plant proteases probably participate using the same mechanism. It has been suggested that from an evolutionary standpoint, at least one function of these proteases is to provide defence,11 perhaps by leading to irritation in animals that might otherwise eat the plant. PAR4 has not been shown to have a significant presence in skin and it is therefore unlikely that the effect of these proteases on this receptor is functionally relevant with respect to itch.
Cowhage, which also activates PARs, is used experimentally as a tool to study nonhistamine itch.2 It is a useful tool because it consistently elicits nociceptive responses and does not require injection as the spicules act as an effective delivery vehicle. Bromelain and ficin are not used routinely in itch studies, perhaps because they have not been readily available in purified forms, in contrast to papain. Dermatologists should be aware that topical preparations containing papain, although no longer available in the U.S.A., and separately bromelain, have been used clinically for the debridement of wounds or burns.12 The sensation of itch is often present in burns. While we are not aware of itch from these preparations, itch might be expected. PAR activation is associated with pain and is likely to explain papain-induced pain. The potential effect of repeated topical application and systemic absorption, or systemic administration of these proteases, on nociception is not readily predictable.
It is possible that cowhage is more effective at activating PARs and inducing itch compared with these other plant-derived proteases. Future studies could address the relative potencies of these proteases on both PAR activation and itch induction as it has been suggested that papain is less potent than bromelain and ficin at evoking itch.1 Studies of relative PAR activation by these proteases could be accomplished with formal concentration–effect curves as opposed to the single concentrations presented here. These studies could also take into account the observations that while proteases can turn on or ‘arm’ PARs by cleavage at certain sites, cleavage at other sites can result in the receptor being ‘disarmed’ such that it cannot be activated.5 While trypsin reliably cleaves C-terminal to arginine and lysine, such specificity is unusual among proteases. The cleavage site(s) for cysteine protease activation of PARs remain(s) to be determined. Comparing and contrasting the likely receptor cleavage sites of plant cysteine proteases and human cysteine and serine proteases will lead to an improved understanding of protease specificity and receptor activation. Such studies may aid in the development of antipruritics that target PARs.
What’s already known about this topic?
Certain human proteases implicated in itch have been shown to activate protease-activated receptors.
Mucunain, a cysteine protease derived from cowhage, evokes itch and has recently been shown to activate these receptors.
The plant proteases discussed here have been reported more than 50 years ago to evoke itch but their mechanism of action has not been examined.
What does this study add?
This study establishes the likely mechanism by which bromelain, ficin and papain evoke itch.
This study establishes that these plant proteases can act as signalling molecules.
Acknowledgments
This study was supported by an agreement between Massachusetts General Hospital and the Shiseido Co., Ltd.
Footnotes
Conflicts of interest
None declared.
References
- 1.Arthur RP, Shelley WB. The role of proteolytic enzymes in the production of pruritus in man. J Invest Dermatol. 1955;25:341–346. doi: 10.1038/jid.1955.138. [DOI] [PubMed] [Google Scholar]
- 2.Johanek LM, Meyer RA, Friedman RM, et al. A role for polymodal C-fiber afferents in nonhistaminergic itch. J Neurosci. 2008;28:7659–7669. doi: 10.1523/JNEUROSCI.1760-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namer B, Carr R, Johanek LM, et al. Separate peripheral pathways for pruritus in man. J Neurophysiol. 2008;100:2062–2069. doi: 10.1152/jn.90482.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vu TK, Hung DT, Wheaton VI, et al. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 5.Oikonomopoulou K, Hansen KK, Saifeddine M, et al. Proteinase-activated receptors, targets for kallikrein signaling. J Biol Chem. 2006;281:32095–32112. doi: 10.1074/jbc.M513138200. [DOI] [PubMed] [Google Scholar]
- 6.Steinhoff M, Corvera CU, Thoma MS, et al. Proteinase-activated receptor-2 in human skin: tissue distribution and activation of keratinocytes by mast cell tryptase. Exp Dermatol. 1999;8:282–294. doi: 10.1111/j.1600-0625.1999.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 7.Ebeling C, Lam T, Gordon JR, et al. Proteinase-activated receptor-2 promotes allergic sensitization to an inhaled antigen through a TNF-mediated pathway. J Immunol. 2007;179:2910–2917. doi: 10.4049/jimmunol.179.5.2910. [DOI] [PubMed] [Google Scholar]
- 8.Lourbakos A, Yuan YP, Jenkins AL, et al. Activation of proteaseactivated receptors by gingipains from Porphyromonas gingivalis leads to platelet aggregation: a new trait in microbial pathogenicity. Blood. 2001;97:3790–3797. doi: 10.1182/blood.v97.12.3790. [DOI] [PubMed] [Google Scholar]
- 9.Reddy VB, Iuga AO, Shimada SG, et al. Cowhage-evoked itch is mediated by a novel cysteine protease: a ligand of protease-activated receptors. J Neurosci. 2008;28:4331–4335. doi: 10.1523/JNEUROSCI.0716-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy VB, Shimada SG, Sikand P, et al. Cathepsin S elicits itch and signals via protease-activated receptors. J Invest Dermatol. 2010;130:1468–1470. doi: 10.1038/jid.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaller A. A cut above the rest: the regulatory function of plant proteases. Planta. 2004;220:183–197. doi: 10.1007/s00425-004-1407-2. [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg L, Lapid O, Bogdanov-Berezovsky A, et al. Safety and efficacy of a proteolytic enzyme for enzymatic burn debridement: a preliminary report. Burns. 2004;30:843–850. doi: 10.1016/j.burns.2004.04.010. [DOI] [PubMed] [Google Scholar]