Abstract
Human Herpesvirus 6B (HHV-6B) is the causative agent of roseola infantum. HHV-6A and 6B can reactivate in immunosuppressed individuals and are linked with severe inflammatory response, organ rejection and central nervous system diseases. About 0.85% of the US and UK population carries an integrated HHV-6 genome in all nucleated cells through germline transmission. We have previously reported that the HHV-6A genome integrated in telomeres of patients suffering from neurological dysfunction and also in telomeres of tissue culture cells. We now report that HHV-6B also integrates in telomeres during latency. Detailed mapping of the integrated viral genomes show that a single HHV-6 genome integrates and telomere repeats join the left end of the integrated viral genome. When HEK-293 cells carrying integrated HHV-6A were exposed to the histone deacetylase inhibitor Trichostatin A, circularization and/or formation of concatamers were detected and this assay could be used to distinguish between lytic replication and latency.
INTRODUCTION
Herpesviruses are ubiquitous and have adapted a balance of lytic and latent infection within the infected host. Human Herpesvirus 6 (HHV-6) was isolated from B-lymphocytes of patients with lymphoproliferative disorders while coinfected with HIV and HTLV (Salahuddin et al., 1986). Further molecular studies as well as sequencing of clinical isolates determined that humans carry two different viral species: HHV-6A and HHV-6B (Ahlqvist et al., 2005; Dominguez et al., 1999; Gompels et al., 1995; Yamanishi et al., 1988).
Human Herpesvirus 6B (HHV-6B) primary infection in young children is the etiological agent of exanthema subitum (roseola infantum), which is characterized by high fever, diarrhea, and a mild skin rash along the trunk, neck, and face (Asano et al., 1994; Yamanishi et al., 1988). Serologic studies have found that by the age of two, greater than 90% of children have acquired a primary HHV-6A/B infection (Okuno et al., 1989; Zerr et al., 2005). HHV-6A has been associated with several adult diseases and various neurological disorders including encephalitis, ataxia, seizures, multiple sclerosis, Hashimoto's thyroiditis, and has been implicated as a cofactor in AIDS progression (Ablashi et al., 2010; Ahlqvist et al., 2005; Cameron et al., 2010; Caselli et al., 2012; De Bolle et al., 2005; Donati et al., 2003; Jones et al., 1994; Lusso et al., 2007; Lusso et al., 1989; Yamashita and Morishima, 2005; Yao et al., 2010). Nevertheless, the causal link between human disease and HHV-6A infection remains to be fully elucidated.
The viral genome of HHV-6B is 162 kbp long, encompassing 119 unique open reading frames (ORF), while HHV-6A genome contains 119 unique ORF and is 159 kbp long (Dominguez et al., 1999; Gompels et al., 1995). The genome of HHV-6A and HHV-6B is organized into two major regions (Fig. 1a). The unique (U) region of the genome is 143 kbp and comprises seven major core gene blocks that are conserved amongst Herpesviridae. The second major genomic regions of HHV-6A and HHV-6B are the ~8 kbp of left and right direct repeats (DR) located at the termini of the U region (Dominguez et al., 1999; Gompels et al., 1995). Within the right end direct repeat (DRR) and the left end direct repeat (DRL) the HHV-6A/HHV-6B genome encodes perfect and imperfect TTAGGG telomere repeat arrays.
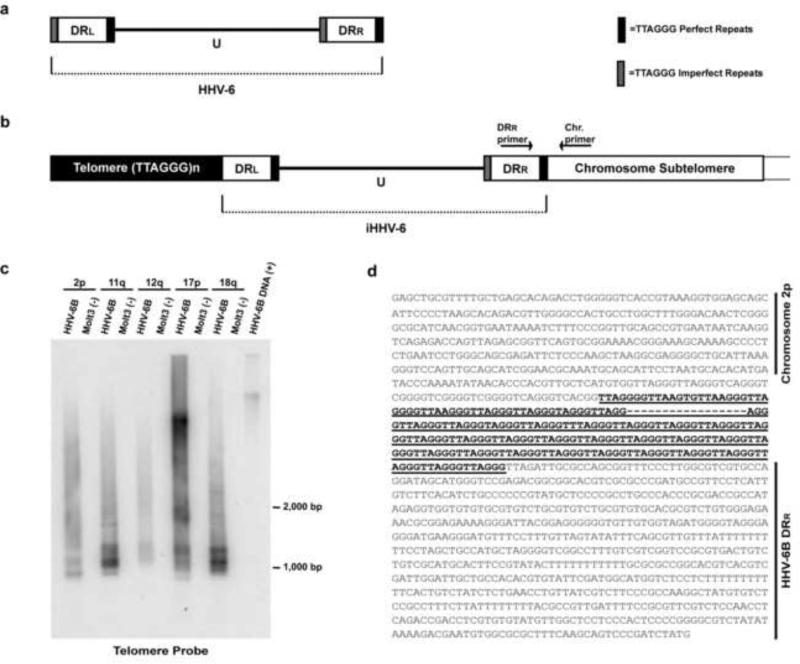
Fig. 1. Amplification of subtelomere-HHV-6B junction fragments from in vitro-infected Molt3 cells.
(a) Genome structure of HHV-6B and HHV-6A in which unique (U) region is surrounded by left and right direct repeats (DRL and DRR) containing perfect (dark bars) and imperfect (shaded bars) telomere repeats. (b) The putative model of the integrated HHV-6-chromosome structure. Arrows indicate position of primers derived from the DRR of the HHV-6 genome and from a specific chromosome subtelomere. (c) DNA from HHV-6B (strain Z29) infected and uninfected Molt3 cells were subjected to amplification by PCR primers derived from HHV-6B DRR and subtelomere primers 2p, 11q, 12q, 17p, and 18q (primer positions depicted in Fig. 1b). PCR products from virus-chromosome telomere junction were separated by electrophoresis and analyzed by Southern blot hybridization utilizing a 32P-labeled telomere oligonucleotide probe. (d) DNA sequencing results of cloned fragments (n=3) of chromosome 2p subtelomere (upper) joined with TTAGGG telomere repeats (bold and underlined), and HHV-6B DRR sequence (lower) during viral integration into chromosome telomere. Additional sequencing results are shown in Fig. S1.
Primary infection of HHV-6A/B leads to an increase in viral copies present in peripheral blood and subsequent decrease in viral copies during latency (Asano et al., 1994; De Bolle et al., 2005; Okuno et al., 1989; Yamanishi et al., 1988; Zerr et al., 2005). Following primary infection, the genome of most herpesviruses establishes latency as multi-copy circular episomes within the nucleus of the infected cell (reviewed by Pellett and Roizman, 2007). The expression of latent viral genes allows the stable replication and persistence of the latent viral episomes. However, no multi-copy episomes were detected in latently infected HHV-6A and 6B cells (Arbuckle et al., 2010). Through the use of fluorescent in situ hybridization (FISH), Gardella gel, Southern hybridization, and direct sequencing of viral integration sites from patient tissues and establishment of in vitro integrated cell lines our laboratory has demonstrated that HHV-6A specifically integrates into telomeres of chromosomes (Arbuckle et al., 2010; Arbuckle and Medveczky, 2011). These experiments suggest that homologous recombination occurs between the perfect TTAGGG telomere repeat arrays encoded in DRR of HHV-6A genome with the subtelomere of the human chromosome.
In approximately 0.8% of the US and United Kingdom general population high copies of HHV-6 genomes can be detected in peripheral blood measuring greater than 0.5 million copies per ml of blood and referred to as chromosomally integrated HHV-6 or CIHHV-6 (Arbuckle et al., 2010; Hall et al., 2008, Leong et al., 2007; Ward et al., 2006; Ward et al., 2005). In 1993 Luppi et al. identified a unique high molecular weight viral fragment present in PBMCs of three patients with elevated copies of HHV-6 (Luppi et al., 1993). This high molecular weight fragment resolved by pulse field gel electrophoresis demonstrated one of the earliest observations of HHV-6 integration into the human genome. Subsequently, several other labs have suggested integration of HHV-6 into human chromosomes through FISH and HHV-6 sequences were detected in hair follicles, lymph node, spleen, kidney, brain, liver, and cardiac tissues by PCR (Daibata et al., 1998; Jarrett et al., 1988; Nacheva et al., 2008; Strenger et al., 2010; Ward et al., 2006; Arbuckle et al. 2010).
In this study we set out to further map the genome structure of chromosome integrated HHV-6A and 6B. Here we show that similarly to HHV-6A, the HHV-6B genome integrates into telomeres of human chromosomes near the subtelomere via the perfect TTAGGG telomere repeat array encoded in DRR of the viral genome. Single telomere length analyses (STELA) suggest that the physical end of chromosome harboring the integrated virus comprises telomeric repeats of variable length similar to those found in normal chromosomes. Finally, latently infected HEK-293 cells containing integrated viral genome were exposed to the histone deacetylase inhibitor Trichostatin A while circularization and/or formation of concatamers were detected by a novel PCR assay indicating reactivation of HHV-6 from latency.
RESULTS
HHV-6B integrates in telomeres in vitro
The genomic architecture of both HHV-6A and HHV-6B are organized into two major regions (Fig. 1a). The unique (U) region of the genome (~143 kb) contains seven major core gene blocks that are conserved amongst herpesviruses (Dominguez et al., 1999; Gompels et al., 1995). The second major genomic region of HHV-6A/B is the ~8 kb left and right direct repeats (DR) located at the termini of the unique region (Dominguez et al., 1999; Gompels et al., 1995). Within the right end direct repeat (DRR) and the left end direct repeat (DRL) the HHV-6A and HHV-6B genomes encode a perfect TTAGGG telomere repeat array and an imperfect TTAGGG repeat array.
Since we have previously demonstrated the genome of HHV-6A integrates into telomeres of human chromosomes (Arbuckle et al., 2010), we hypothesized that the genome of HHV-6B would as well integrate into the telomeres of chromosomes during the course of infection. Furthermore, the T-cell line Molt3 is routinely used to propagate HHV-6B, yet we observed that cell lysis was often incomplete and many cells survived after the peak of productive infection possibly due to latent infection. DNA was isolated from HHV-6B infected Molt3 cells and PCR amplified for the putative viral genome-chromosome telomere junctions using subtelomere based primers (2p, 11q, 12q, 17p, and 18q) (Lusso et al., 1989; Martin et al., 1991) and a primer annealing to the right end of HHV-6B DRR (Fig. 1b).
The primer to DRR was utilized since our previously published results determined that HHV-6A in vivo and in vitro integration occurs through the linkage of perfect telomere array encoded in DRR with the telomere sequence directly adjacent to the end of the chromosome (Fig. 1b) (Arbuckle et al., 2010). Amplified fragments from the subtelomeres of 2p, 11q, 12q, 17p, and 18q were fractionated on an agarose gel and Southern hybridization was performed with [γ32P]-ATP-radiolabeled TTAGGG probe that hybridizes to both chromosome and HHV-6B telomere sequences located at their termini. As illustrated in Fig. 1c, PCR amplification coupled with Southern hybridization identified the linkage of the DRR of HHV-6B genome with the telomere of specific chromosomes during in vitro infection of Molt3 cells.
To confirm the specific integration of HHV-6B into the telomere of chromosomes 2p, 11q, and 18q, we first cloned the PCR amplified fragments through pCR®4-TOPO® cloning system. Following transformation of competent cells, positive clones were identified by colony hybridization with [γ32P]-ATP-radiolabeled HHV-6B and telomere oligonucleotide probes. Sequencing of clones confirmed in vitro infection of Molt3 cells resulted in the HHV-6B genome covalently linked with the telomere of chromosomes 2p, 11q, and 18q (Figs. 1d and S1). Therefore, the results from these experiments illustrate that similar to HHV-6A (Arbuckle et al., 2010), HHV-6B also specifically integrates into telomeres of chromosomes during the course of infection through the perfect telomere repeat array encoded in the DRR of the viral genome.
Mapping the DRL from the integrated HHV-6 genome
Results in the previous section and publication by our group illustrated that integration of HHV-6B and HHV-6A occurs through recombination with the perfect telomere repeats encoded in the DRR (Fig. 1) (Arbuckle et al., 2010; Arbuckle and Medveczky, 2011). Therefore, we sought to further characterize the genomic structure of integrated HHV-6A and HHV-6B through mapping of the DRL region and determine whether the tandem array of telomere repeats [(TTAGGG)n] was extended beyond the DRL (Figs. 1b and 2a). During the process of integration into chromosome telomeres, it is unknown whether recombination occurs with the perfect or imperfect telomere repeats of the DRL (Figs. 1a and 1b). If recombination occurs through the perfect telomere repeats, the DRL could potentially be lost during the process of telomere integration. To map various portions of the integrated viral genome in patients with inherited HHV-6, we developed two PCR based assays to determine whether the entire DRL is present or absent from their PBMCs (Fig. 2a).
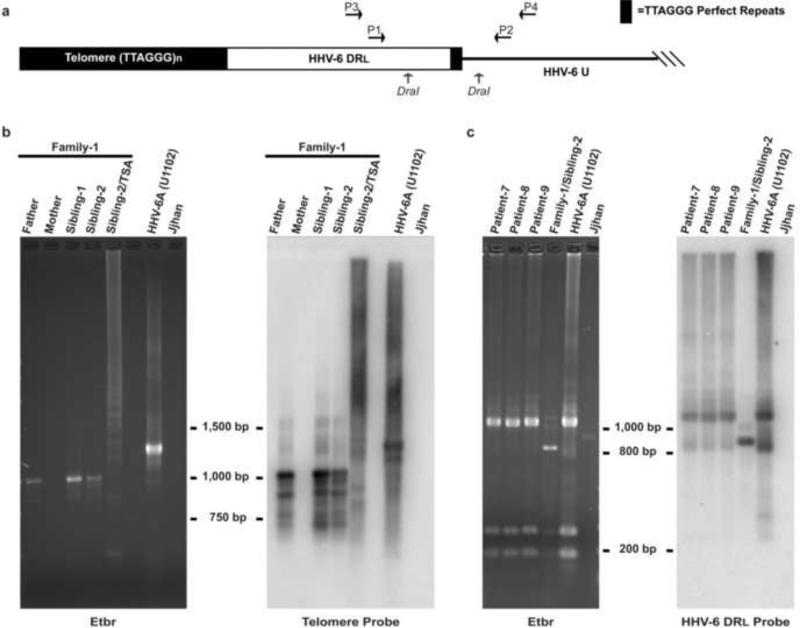
Fig. 2. Amplification and restriction enzyme digestion of DRL fragments from DNA isolated from PBMC of members of a family with inherited HHV-6.
DNA from PBMC of members of Family-1 (Arbuckle et al., 2010) carrying HHV-6A integrated in telomere of chromosome 18q, Patients 7 - 9 carrying HHV-6B integrated, and from Jjhan cell line infected with HHV-6A U1102 was subjected to PCR amplification using primer pairs as depicted. (a) Diagram for the amplification of DRL in which primer 1 (P1) and 3 (P3) bind to regions inside DRL and primer 2 (P2) and 4 (P4) binds within the U of HHV-6. DraI restriction sites are indicated by arrows. (b) Amplification of DRL with primers P1 and P2 from members of Family-1 results in the predicted size of 1,198 bp (HHV-6A U1102) as shown in the ethidium bromide (Etbr) staining (left panel). Southern hybridization with telomere oligonucleotide probe (right panel). Controls: HHV-6A (U1102) positive control while Family-1/Mother and Jjhan were negative controls. (c) Amplification and restriction digestion with DraI results in the predicted 1027, 217, 186 bp (HHV-6A U1102) as shown in the ethidium bromide (Etbr) staining (left panel). Southern hybridization with HHV-6 DRL oligonucleotide probe hybridizes with DRL 1027 bp band (right panel). Controls: HHV-6A (U1102) infected Jjhan DNA positive control and Jjhan negative control.
In the first experiment, amplification of the DRL was completed by using primers that annealed to the DRL and unique region of the integrated genome (Fig. 2a, Primer 1= P1 and Primer 2= P2), which effectively amplified across the perfect telomere repeat (Fig. 2a, short dark square). Genomic DNA isolated from Family-1 members with inherited HHV-6A (Arbuckle et al., 2010) was subjected to this PCR based assay. The DRL was successfully amplified as shown by Southern hybridization with telomere specific oligonucleotide probe (Fig. 2b).
To confirm the results in the previous experiment, we evaluated the DRL in a second group of patients with inherited HHV-6 and utilized a second primer set (P3 and P4) in combination with DraI restriction enzyme digestion (Fig. 2a). The PCR amplified DRL fragment has two DraI restriction sites that lead to the expected fragment sizes of 1027, 217, 186 bp according to the HHV-6A genome (strain U1102) (Gompels et al., 1995). Similarly to the results in Fig. 2b, the 1027 bp DRL fragment was present in the inherited HHV-6 genome of all patients as this fragment hybridized with the oligonucleotide probe corresponding to the DraI- DraI fragment (Fig. 2c). In summary, there results demonstrate that the DRL is present in the integrated HHV-6A and HHV-6B genome.
Telomere repeats are covalently linked to the DRL of integrated HHV-6 genome
The next series of experiments were designed to determine whether a tandem array of telomere repeats [(TTAGGG)n] at the end of the chromosome is linked to the DRL of the integrated HHV-6 genome. We adapted the methodology of Baird et al. single telomere length analysis (STELA) assay (Baird et al., 2003; Britt-Compton et al., 2006) to amplify and estimate the length of a telomere from a specific chromosome end as depicted in Fig. 3a. Genomic DNA was isolated from PBMCs of patients with inherited HHV-6A and HHV-6B. Next, a oligonucleotide linker (Tellorette-2) containing a single TTAGGG repeat was ligated to the end of the telomere by incubating 60 ng and 150 ng of genomic DNA with T4 ligase for 13 hours at 35°C (Fig. 3a). Linker-ligated DNA was amplified with a high fidelity Taq polymerase, linker annealing primer (P1), and a HHV-6 primer annealing to the DRL (P2). The specific amplification of the chromosome telomere was identified through Southern hybridization with [γ32P]-ATP-radiolabeled telomere oligonucleotide probe (Fig. 3b, 3c, and 3d). Subsequently, the nitrocellulose blot was then stripped and re-hybridized with PCR amplified fragment from the viral DRL. Co-hybridization of bands with telomere and DRL probes confirmed the covalent linkage of chromosome telomere with the DRL of inherited HHV-6 genome (Fig. 3a). Moreover, similar results were also obtained in STELA analysis of HEK-293 cell line harboring integrated HHV-6A (Fig. 3d). Heterogeneous banding pattern was observed when DNA from uninfected HEK-293 and Jjhan cells were subjected to STELA (Fig. S2). The heterogeneity of PCR amplified telomere fragments with the STELA assay has also been previously observed by Baird et al. in control human cells (Baird et al., 2003; Britt-Compton et al., 2006); this heterogeneity is likely due to the amplification of telomeres from a mixed populations of cells that contain variable lengths of telomeric sequence. The wide range of telomere length heterogeneity has been described by several other methods (reviewed by Lauzon et al., 2000).
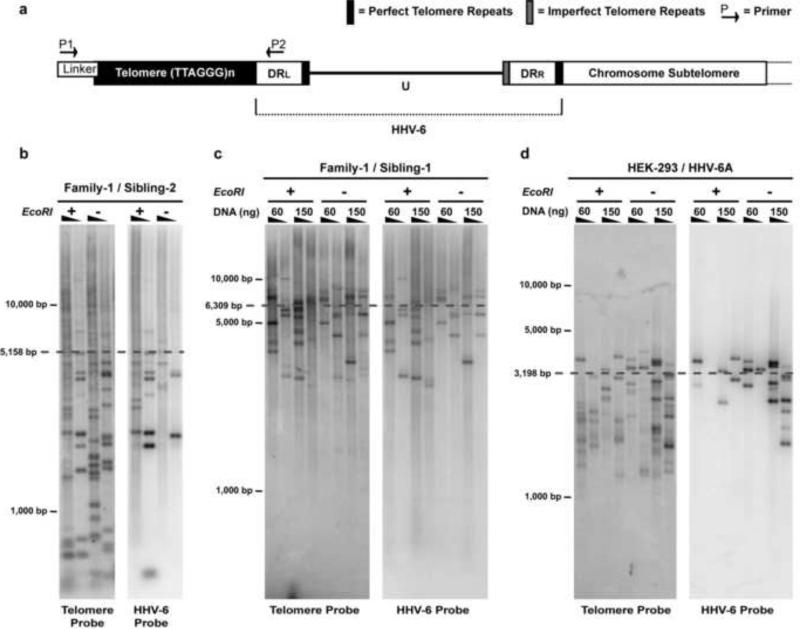
Fig. 3. STELA of DRL adjoining telomere in PBMCs of in vivo integrated HHV-6 and in vitro integrated HEK-293 cells.
(a) Diagram illustrating the strategy of single telomere length analysis (STELA) (Baird et al., 2003; Britt-Compton et al., 2006). To amplify the specific chromosome telomere extended beyond the DRL of the integrated HHV-6 genome a linker (Telorette-2) oligonucleotide is ligated to the end of the telomere. The Telorette-2 ligation reaction contained 60 or 150 ng genomic DNA (EcoRI digested +/- for DNA solubilization). From this initial ligation, both ligation reactions are then serially diluted two-fold indicated by the triangles in panels b, c and d and subsequently amplified by PCR with primers P1 and P2. Genomic DNA isolated from PBMCs from CIHHV-6A individuals mapped to chromosome 17p13.3 was subjected to STELA; (b) sibling-2, and (c) sibling-1 of Family-1 (Arbuckle et al. 2010). (d) STELA was also performed on HEK-293 cells harboring telomere-integrated HHV-6A strain U1102 (Arbuckle et al. 2010). In each figure, mean telomere length is calculated from co-hybridization of telomere (left panel) and HHV-6 probes (right panel).
Next using the STELA assay we determined the specific telomere length of DNA isolated from PBMCs of patients with inherited HHV-6A integrated into chromosome 17p. This was calculated by determining the mean molecular weight of all bands co-hybridizing with telomere and DRL (HHV-6A) radiolabeled probes from eight different PCR reactions according to the protocol of Baird et al., 2003 and Britt-Compton et al., 2006. The mean telomere length of Sibling-2 and Sibling-1 was 5,158 bp and 6,309 bp, respectively (Figs. 3b and 3c). Therefore, the structure of the in vivo and in vitro integrated HHV-6A/B genome is as follows: chromosome-subtelomere-(TTAGGG)n-DRR-U-DRL-(TTAGGG)n (Fig. 3a).
Trichostatin A promotes formation of circular/catenated viral genomes in latently infected cells
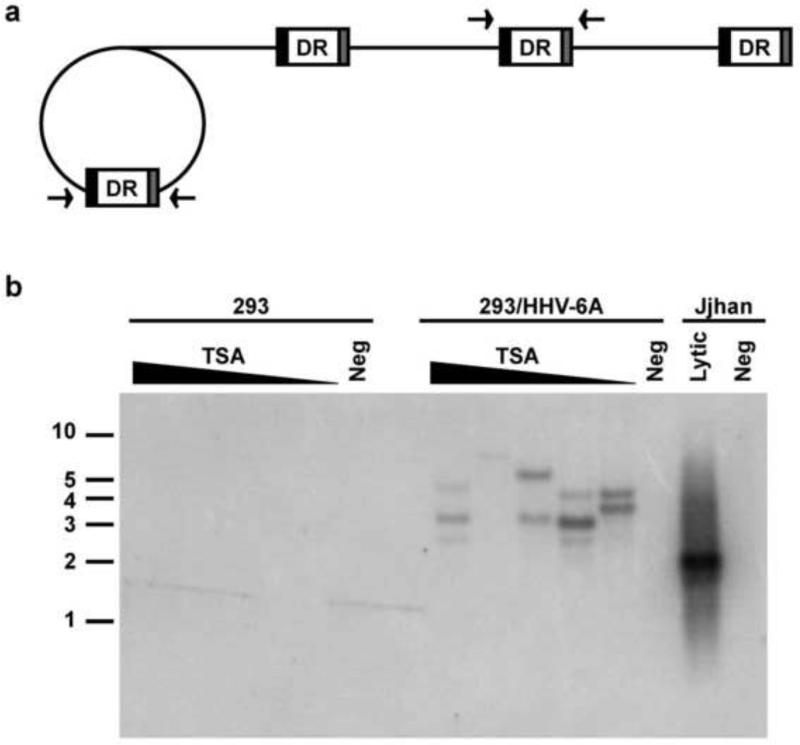
In our previous study we have demonstrated the histone deacetylase inhibitor Trichostatin A (TSA) can induce reactivation of the integrated HHV-6A viral genome from individuals’ PBMCs as well as latently infected cell lines as evidenced by increase in viral DNA copies upon exposure to the TSA (Arbuckle et al., 2010). We hypothesized that reactivation of integrated HHV-6 genome resulted in formation of circular replication intermediates and/or tandem catenated forms of the genome as described for HHV-6 (Dominguez et. al, 1999) and with other herpesviruses (Jacob et al., 1979; Poffenberger and Roizman, 1985). To test this hypothesis, HEK-293 cells latently infected and harboring integrated HHV-6A (strain U1102) (Arbuckle et al., 2010) were cultured in the presence of TSA at concentrations ranging from 10 ng/ml to 160 ng/ml for 3 days. Detection of viral episomes and/or concatemers was performed by PCR amplification of the DR by two primers that annealed to either end of the unique region of HHV-6A (Fig. 4a). Thus, amplification of DR would only occur during circularization or catenation of the viral genome in a head-to-tail conformation (Borenstein and Frenkel, 2009; Borenstein et al., 2010; Jacob et al., 1979; Martin et al., 1991; Poffenberger and Roizman, 1985; Severini et al., 2003). PCR products were subjected to sequential Southern hybridization with oligonucleotide probes to telomere, DR, and U regions of HHV-6A. The autoradiograms revealed that all three probes hybridized with PCR products of variable length from DNA isolated from cells that were treated with TSA (Fig. 4b, hybridization with telomere probe is shown) and not from control unstimulated or uninfected cells. Furthermore, similar variable fragments were also detected in control HHV-6A infected Jjhan cells. The amplification of variable size bands is likely due to insertion of variable number of telomere repeats in the DR during reactivation mediated by homologous recombination with telomere sequences. The absence of catenated genomes in latently infected cells also indicates that only a single genome integrates in telomeres. We studied two additional HEK-293 cell lines latently infected with HHV-6 and harboring telomere integrated U1102 virus described by Arbuckle et al., 2010, but no evidence for circularization/catenation was detected after TSA induction presumably due to integration of defective viral genomes (data not shown).
Fig. 4. Treatment of HHV-6A latently integrated HEK-293 cells with the histone deacetylase inhibitor TSA induces novel PCR products consistent with episome and/or concatemer formation and induction of lytic viral DNA replication.
(a) Schematic illustration of rolling-circle replication of HHV-6 results in the formation of concatemers of viral genomes. Arrows indicate the location of primers utilized in the amplification of DR. (b) Latently infected, single-cell cloned and uninfected control HEK-293 cells were cultured with TSA at concentrations of 160 ng/ml, 80 ng/ml, 40 ng/ml, 20 ng/ml, and 10 ng/ml and without drug treatment for 3 days. DR amplification and Southern hybridization with DR oligonucleotide probe demonstrate the induction of DNA fragments amplifiable with the primers in DNA from cells treated with TSA. No amplification was detected in untreated 293/HHV-6A or 293 cells (Negative controls). Bands of variable size were also detected in DNA from Jjhan cells cultures infected with HHV-6A.
DISCUSSION
HHV6-B and HHV-6A can integrate into the telomeres of human peripheral mononuclear cells in vivo and following in vitro infection of Jjhan T-cells, Molt3 T-cells, and HEK-293 cells (Fig. 1) (Arbuckle et al., 2010; Arbuckle and Medveczky, 2011). It appears that in cell lines capable of supporting productive, lytic infection, some of the cells quickly become latently infected through chromosomal integration, and remain viable. Thus, HHV-6B and HHV-6A integration is not an unusual dead-end phenomenon as described for other herpesviruses, but an alternative mechanism of achieving latency.
These results suggest that HHV-6B and HHV-6A integrate into the host genome via homologous recombination with human telomeres. In particular, integration of HHV-6B and HHV-6A occurs through recombination of the perfect telomere repeats encoded in the DRR juxtaposed to the subtelomeric end of the chromosome (Figs. 1, S1, and 2) (Arbuckle et al., 2010). Moreover, the tandem array of telomere repeats [(TTAGGG)n] at the end of chromosomes was shown to extend beyond the DRL of the HHV-6 genome (Fig. 3). Therefore, the structure of the telomere integrated HHV-6 is as follows: chromosome-subtelomere-(TTAGGG)n-DRR-U-DRL-(TTAGGG)n (Fig. 3a) (Arbuckle et al., 2010).
Delecluse et al. previously demonstrated the latent telomere-integrated Marek's Disease Virus (MDV) of chickens can also reactivate (Delecluse et al., 1993). MDV integrants were detected as long concatamers suggesting that MDV integration requires initiation of DNA replication and catenation. In contrast, our data show that a single HHV-6 genome integrates immediately following infection. Taken together, these observations suggest the mechanism of integration of the HHV-6/B and MDV are mediated by different mechanisms.
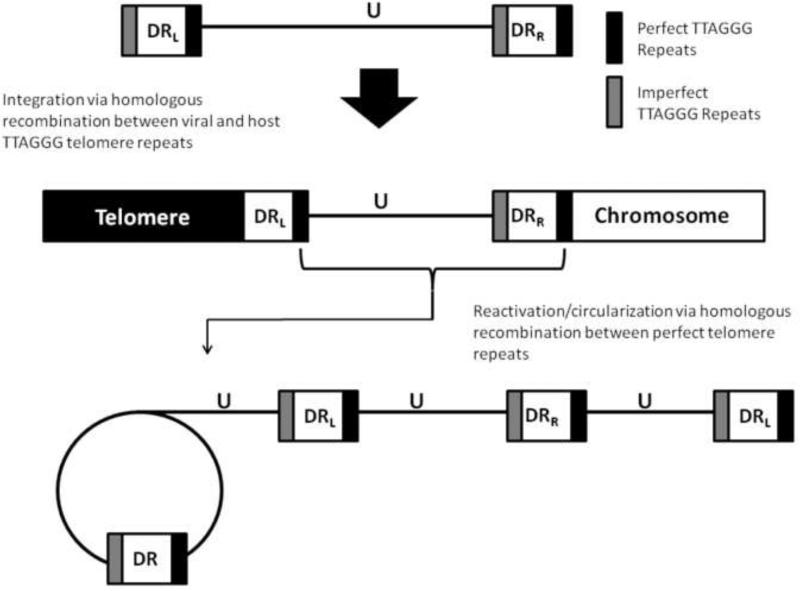
Current diagnostic assays for the detection of reactivated are based on quantitative PCR to determine viral copy number, however quantitative PCR cannot distinguish between actively replicating HHV-6 or latency. Demonstration of catenation/circularization of replicating virus as described in this work can be used in clinical studies to verify that the virus is indeed replicating (Fig. 5).
Fig. 5. Proposed model for latent HHV-6 in vitro reactivation.
During latency, HHV-6A and HHV-6B can integrate into the telomeres during in vivo and in vitro infection. However, reactivation of integrated HHV-6 can be achieved through culturing of latently integrated cells with TSA, or alternatively, TPA plus hydrocortisone. (b) Reactivation of the integrated HHV-6 results in genome replication through rolling-circle replication.
Telomere position effects (TPE) is characterized by transcriptional repression of telomere adjacent genes through the enrichment of H3K9 methylation and decrease in activating H3K4 methylation emanating from the constitutive heterochromatin state of the telomere (Ottaviani et al., 2008). The epigenetic regulation by TPE is well characterized in S. cerevisiae (Gottschling et al., 1990; Ottaviani et al., 2008; Tham and Zakian, 2002; Wyrick et al., 1999). The use of reporter systems (GFP, luciferase, URA3) has identified TPE in excess of 20 kbp downstream of the telomere sequence in S. cerevisiae. However less established in human cells, Baur et al. observed a 10 fold decrease in the expression of telomere adjacent luciferase reporter when compared to reporter integrated near the centromere (Baur et al., 2001). Moreover, the heterochromatin state of TPE was reversible after treatment of human cells with TSA. This resulted in the suppression of HDACs and transcriptional activation of the luciferase reporter, which parallels the TSA mediated reactivation of HHV-6A from its latent integrated state (Arbuckle et al., 2010). The impact of TPE on HHV-6 and cellular gene expression is unknown. In particular, it is intriguing whether TPE plays a role in transcriptional repression of lytic HHV-6 genes thus maintaining viral latency during integration. Furthermore, it is unknown whether integration of the ~160kb HHV-6 genome would alter expression of nearby cellular genes due to interruption/alteration of TPE.
Integration of the viral genome may also alter stability of telomeres of the affected chromosome. We thus far have shown that the viral DRR of the genome is integrated within 5 - 41 telomere repeats from the end of the chromosome (Figs. 1 and S1) (Arbuckle et al., 2010). The proximity of the ~160 kbp HHV-6 genome integrated between the end of the chromosome and tandem array of telomere repeats suggests that the virus has a minimal impact on the telomere/shelterin complex. However, if the HHV-6 genome were to integrate near the end of the telomere, one would expect progressive shortening of the telomere and the viral genome. Subsequently, this loss may lead to chromosome instability as the result of chromosome fusion, replicative senescence, or the inability of the shelterin complex to bind the non-telomere coding sequence of the virus to form the protective cap at the end of telomere. These intriguing questions are currently being addressed.
The studies presented here and earlier (Arbuckle et al., 2010; Arbuckle and Medveczky, 2011) lead to some important questions across fields of virology, telomere biology, DNA recombination/repair, and inheritance. Future experiments are needed to determine the overall clinical impact of germ-line integrated HHV-6.
To conclude, we have provided further results supporting that HHV-6A and HHV-6B are unique among human herpesviruses: they have the potential to specifically integrate into telomeres of chromosomes as described for CIHHV-6 individuals and also in vitro under experimental conditions. Moreover, the integrated viral genome is capable of reactivation to produce infectious virions (Arbuckle et al., 2010) and here we designed a PCR-based assay to test clinical specimens for actively replicating HHV-6. More work is required to evaluate whether integration of HHV-6A and 6B is an unusual dead-end phenomenon, or a novel mechanism of achieving latency.
MATERIALS AND METHODS
Primary T-cells, cell lines, and viruses
Heparinized peripheral blood from patients with germ-line inherited HHV-6 was obtained from the HHV-6 Foundation, after the subjects gave informed consent (Arbuckle et al., 2010). Peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoprep™, according to the manufacturer's protocol. PBMCs were incubated in RPMI-1640 medium containing 10% FBS, 50 μg/ml gentamycin and 5 μg/ml phytohemagglutinin (PHA, Sigma-Aldrich) for 72-hours, followed by culturing in 100 U/μl IL-2 medium. T-cell lines Jjhan (HHV-6 Foundation), and Molt3 (ATCC) were maintained in RPMI-1640 medium containing 10% FBS and 50 μg/ml gentamycin. Human embryonic kidney-293 cells (HEK-293) and HHV-6A in vitro integrated cells previously established in our lab (Arbuckle et al., 2010) were maintained in DMEM supplemented with 10% FBS. HHV-6A (U1102 strain) and HHV-6B (Z29 strain) viruses were obtained from Dr. Philip Pellett (Wayne State University).
Infection of cell lines with HHV-6 in vitro to study integration
After removal of dead cells using Lymphoprep™ isotonic density gradient, 107 T-cells were infected with HHV-6A (U1102) and HHV-6B (Z29) at 1.0 MOI in 1 ml of RPMI-1640 complete for 2 hrs at 37°C. Specifically, the T-cell lines Jjhan and Molt3, were infected with HHV-6A and HHV-6B, respectively. Unabsorbed virus was removed by centrifugation for 5 min at 470×g, 25°C (Fisher accuSpin 1R). Cell pellet was resuspended to 106 cells/ml and incubated at 37°C, 5% CO2.
Reactivation of chromosome integrated HHV-6
PBMCs were cultured in RPMI-1640 medium containing 10% FBS and various concentrations of Trichostatin (TSA), or alternatively, 12-O-Tetradecanoyl-13 acetate (TPA) final concentration of 10ng/ml and 10 M hydrocortisone as previously described by Arbuckle et al. (Arbuckle et al., 2010). DNA was prepared from cells following the time of peak cytopathic effect (5 – 14 days).
Chromosome-specific PCR for HHV-6B integration
The PCR amplification of HHV-6B integration sites included: 400 ng genomic DNA, 50 μM HHV-6 DRR and chromosome specific primers, 500 μM dNTPs, Expand 20 kbPLUS PCR system (Roche Diagnostics, Indianapolis, IN), 10 X Taq buffer, and final volume was increased to 25 μl with H2O. Samples were amplified with the Peltier Thermal Cycler PCR (PTC-220, MJ Research Inc, Waltham, Ma) and subjected to the following PCR conditions: 94°C for 2 min, which is then followed by 35 cycles of 94°C for 10 sec, 64°C for 30 sec, and 68°C for 10 min, and finished with a final extension of 68°C for 7 min. Agarose gel electrophoresis and Southern blot hybridization with telomere, HHV-6, and chromosome specific [γ32P]-ATP-radiolabeled oligonucleotide probes was utilized for identifying DNA bands representing the putative viral-chromosomal DNA junction. Bands were gel extracted with Perfectprep® Gel Cleanup Kit (Eppendorf, Westbury, NY) and cloned with pCR®4-TOPO® (TOPO TA Cloning® Kit for Sequencing, Invitrogen, Carlsbad, CA). A minimum of three clones from each sample was sequenced.
Single Telomere Length Analysis (STELA)
Genomic DNA was isolated from PBMCs of patients with inherited HHV-6A and HHV-6B and from HEK-293 cells carrying integrated HHV-6A (Arbuckle et al., 2010). Oligonucleotide linker (Tellorette-2) containing a single TTAGGG repeat was ligated to the end of the telomere by incubating 60 – 150 ng of genomic DNA, 60 - 100 μM of oligonucleotide Telorette-2, and 4.5 U of T4 DNA ligase for 13 hrs at 35°C. The ligation reaction was then purified through phenol chloroform extraction, ethanol precipitation, and resuspended in 15 μl TE. Serial dilution of Telorette-2-ligated-DNA was amplified with a high fidelity taq polymerase (Expand 20 kbPLUS PCR System, Roche), 25 μM linker annealing primer (Teltail-P1), 25 μM HHV-6 STELA-P2. Cycling conditions were 92°C for 2 min, 27 cycles of 92°C for 15 sec, 59°C for 30 sec, 68°C 13.5 min or 18 min, and a final extension of 68°C for 7 min. Amplification products were separated through electrophoresis on a 0.8% agarose gel and Southern hybridized with [γ32P]-ATP-radiolabeled telomere oligonucleotide probe and PCR amplified (primers P3 + P4) HHV-6 DRL random primer [α32P]-dATP-radiolabeled probe. Telomere length linked to integrated HHV-6 was then calculated based upon the mean molecular weight of bands co-hybridizing with telomere and HHV-6 DRL probes.
Reactivation of integrated HHV-6
PBMCs were cultured in RPMI-1640 medium containing 10% FBS and various concentrations of Trichostatin (TSA), or alternatively, 12-O-Tetradecanoyl-13 acetate (TPA) final concentration of 10ng/ml and 10 μM hydrocortisone as described by Arbuckle et al. (Arbuckle et al., 2010).
Primer sequences
Oligonucleotides for chromosome-specific PCR and probe
HHV-6 DRR: 5’-CATAGATCGGGACTGCTTGAAAGCGC-3’ (Arbuckle et al., 2010)
HHV-6 DRR-probe: 5′-GCGGAGACACATAGCCTTGGCGGGAAGAC-3′
Chromosome 2p: 5’-GAGCTGCGTTTTGCTGAGCAC (Baird et al., 2003)
Chromosome 11q: 5’-CAGACCTTGGAGGCACGGCCTTCG-3’ (Baird et al., 2003)
Chromosome 17p: 5’-AACATCGAATCCACGGATTGCTTTGTGTAC-3’ (Baird et al., 2003)
Chromosome 18q: 5’-CTCATGTCCTCGGTCTCTTGCCTC-3’ (Arbuckle et al., 2010)
Telomere Probe: 5′-TTAGGGTTAGGGTTAGGGTTAGGG-3′ (Arbuckle et al., 2010)
DRL PCR
P1: 5’-ATGTGGCGATCATCCAATCAACGG-3’
P2: 5’-AAATGTCTGCGGAAAGGTCAACCG-3’
P3: 5’-CATAGATCGGGACTGCTTGAAAGCGG-3’
P4: 5’- AAATGTCTGCGGAAAGGTCAACCG-3’
HHV-6 DRL Probe: 5’-CCCCAACGCGCGCGCGCACGC-3’
Oligonucleotides for STELA PCR and probe
Tellorette-2 (Linker): 5′-TGCTCCGTGCATCTGGCATCTAACCCT-3′ (Baird et al., 2003)
P1-Teltail: 5′-TGCTCCGTGCATCTGGCATC-3′ (Baird et al., 2003)
P2-HHV6 STELA: 5’-AAGGATGGTAGGGTTTAGGGTCGAACC-3’
P3-HHV6 STELA: 5’-CCCTAACACTAATCCTCGCATCCGC-3’
XpYpE2: 5’-TTGTCTCAGGGTCCTAGTG-3’ (Baird et al., 2003)
XpYpB2: 5’-TCTGAAAGTGGACCTATCAG-3’ (Baird et al., 2003)
Supplementary Material
Fig. Supplementary 2. Chromosome Xp single telomere length analysis of uninfected Jjhan and HEK-293 cells. (a) Diagram illustrating the strategy of single telomere length analysis (STELA). To amplify the telomere of chromosome Xp, a linker (Telorette-2) oligonucleotide is ligated to the end of the telomere. Amplification of the telomere is then preceded with primers P1 and P3. Serial dilution of 150 ng of linker-ligated DNA isolated from the (b) T-cell line Jjhan and (c) HEK-293 cells. In each figure, mean telomere length is based upon co-hybridization of telomere (left panel) and chromosome Xp probes (right panel).
HHV 6B integrates in telomeres during latency > The left end of both HHV 6A and HHV 6B is joined by telomere repeats > A single viral genome integrates in telomeres > Histone deacetylase Trichostatin A induces circularization/catenation of the integrated viral genome
ACKNOWLEDGMENTS
We thank Dr. Philip Pellett providing HHV-6A and 6B stocks. This work was supported by a grant from the HHV-6 Foundation and the National Institute of Health 5R01CA111196 for PGM.
Abbreviations
- HHV-6
human herpesvirus 6
- CIHHV-6
chromosome integrated human herpesvirus 6
- PBMCs
peripheral blood mononuclear cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ablashi DV, Devin CL, Yoshikawa T, Lautenschlager I, Luppi M, Kuhl U, Komaroff AL. Review Part 3: Human herpesvirus-6 in multiple non-neurological diseases. J Med Virol. 2010;82:1903–1910. doi: 10.1002/jmv.21860. [DOI] [PubMed] [Google Scholar]
- Ahlqvist J, Fotheringham J, Akhyani N, Yao K, Fogdell-Hahn A, Jacobson S. Differential tropism of human herpesvirus 6 (HHV-6) variants and induction of latency by HHV-6A in oligodendrocytes. J Neurovirol. 2005;11:384–394. doi: 10.1080/13550280591002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle JH, Medveczky PG. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect. 2011;13:731–741. doi: 10.1016/j.micinf.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Yoshikawa T, Suga S, Kobayashi I, Nakashima T, Yazaki T, Kajita Y, Ozaki T. Clinical features of infants with primary human herpesvirus 6 infection (exanthem subitum, roseola infantum). Pediatrics. 1994;93:104–108. [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science (New York, N.Y. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- Biesecker LG. The end of the beginning of chromosome ends. Am J Med Genet. 2002;107:263–266. doi: 10.1002/ajmg.10160. [DOI] [PubMed] [Google Scholar]
- Borenstein R, Frenkel N. Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19138–19143. doi: 10.1073/pnas.0908504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein R, Zeigerman H, Frenkel N. The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines. Journal of virology. 2010;84:2648–2656. doi: 10.1128/JVI.01951-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum Mol Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- Cameron B, Flamand L, Juwana H, Middeldorp J, Naing Z, Rawlinson W, Ablashi D, Lloyd A. Serological and virological investigation of the role of the herpesviruses EBV, CMV and HHV-6 in post-infective fatigue syndrome. J Med Virol. 2010;82:1684–1688. doi: 10.1002/jmv.21873. [DOI] [PubMed] [Google Scholar]
- Caselli E, Zatelli MC, Rizzo R, Benedetti S, Martorelli D, Trasforini G, Cassai E, degli Uberti EC, Di Luca D, Dolcetti R. Virologic and immunologic evidence supporting an association between HHV-6 and Hashimoto's thyroiditis. PLoS pathogens. 2012;8:e1002951. doi: 10.1371/journal.ppat.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daibata M, Taguchi T, Taguchi H, Miyoshi I. Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br J Haematol. 1998;102:1307–1313. doi: 10.1046/j.1365-2141.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse HJ, Schuller S, Hammerschmidt W. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. Journal of virology. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati D, Akhyani N, Fogdell-Hahn A, Cermelli C, Cassiani-Ingoni R, Vortmeyer A, Heiss JD, Cogen P, Gaillard WD, Sato S, Theodore WH, Jacobson S. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology. 2003;61:1405–1411. doi: 10.1212/01.wnl.0000094357.10782.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM. Fields virology. 5th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Hall CB, Caserta MT, Schnabel K, Shelley LM, Marino AS, Carnahan JA, Yoo C, Lofthus GK, McDermott MP. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics. 2008;122:513–20. doi: 10.1542/peds.2007-2838. [DOI] [PubMed] [Google Scholar]
- Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. Journal of virology. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett RF, Gledhill S, Qureshi F, Crae SH, Madhok R, Brown I, Evans I, Krajewski A, O'Brien CJ, Cartwright RA, et al. Identification of human herpesvirus 6-specific DNA sequences in two patients with non-Hodgkin's lymphoma. Leukemia. 1988;2:496–502. [PubMed] [Google Scholar]
- Jones CM, Dunn HG, Thomas EE, Cone RW, Weber JM. Acute encephalopathy and status epilepticus associated with human herpes virus 6 infection. Dev Med Child Neurol. 1994;36:646–650. doi: 10.1111/j.1469-8749.1994.tb11903.x. [DOI] [PubMed] [Google Scholar]
- Lauzon W, Sanchez Dardon J, Cameron DW, Badley AD. J Flow cytometric measurement of telomere length. Cytometry. 2000;15:159–64. doi: 10.1002/1097-0320(20000615)42:3<159::aid-cyto1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Leong HN, Tuke PW, Tedder RS, Khanom AB, Eglin RP, Atkinson CE, Ward KN, Griffiths PD, Clark DA. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- Lusso P, Crowley RW, Malnati MS, Di Serio C, Ponzoni M, Biancotto A, Markham PD, Gallo RC. Human herpesvirus 6A accelerates AIDS progression in macaques. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5067–5072. doi: 10.1073/pnas.0700929104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusso P, Ensoli B, Markham PD, Ablashi DV, Salahuddin SZ, Tschachler E, Wong-Staal F, Gallo RC. Productive dual infection of human CD4+ T lymphocytes by HIV-1 and HHV-6. Nature. 1989;337:370–373. doi: 10.1038/337370a0. [DOI] [PubMed] [Google Scholar]
- Martin ME, Thomson BJ, Honess RW, Craxton MA, Gompels UA, Liu MY, Littler E, Arrand JR, Teo I, Jones MD. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. The Journal of general virology. 1991;72(Pt 1):157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- Nacheva EP, Ward KN, Brazma D, Virgili A, Howard J, Leong HN, Clark DA. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J Med Virol. 2008;80:1952–1958. doi: 10.1002/jmv.21299. [DOI] [PubMed] [Google Scholar]
- Okuno T, Takahashi K, Balachandra K, Shiraki K, Yamanishi K, Takahashi M, Baba K. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J Clin Microbiol. 1989;27:651–653. doi: 10.1128/jcm.27.4.651-653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90:93–107. doi: 10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Pellett PE, Roizman B. The Family Herpesviridae: A Brief Introduction. In: Knipe David M., Howley Peter M., Griffin Diane E., Lamb Robert A., Straus Stephen E., Martin Malcolm A., Roizman Bernard., editors. Field's Virology. Lippincott Williams; Philadelphia, PA: 2007. p. 2481. [Google Scholar]
- Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. Journal of virology. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science (New York, N.Y. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- Severini A, Sevenhuysen C, Garbutt M, Tipples GA. Structure of replicating intermediates of human herpesvirus type 6. Virology. 2003;314:443–450. doi: 10.1016/s0042-6822(03)00451-3. [DOI] [PubMed] [Google Scholar]
- Strenger V, Aberle SW, Wendelin G, Pfurtscheller K, Nacheva EP, Zobel G, Nagel B. Chromosomal integration of the HHV-6 genome as a possible cause of HHV-6 detection in cardiac tissues. J Clin Pathol. 2010;63:1129–1130. doi: 10.1136/jcp.2010.079277. [DOI] [PubMed] [Google Scholar]
- Tham WH, Zakian VA. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21:512–521. doi: 10.1038/sj.onc.1205078. [DOI] [PubMed] [Google Scholar]
- Ward KN, Leong HN, Nacheva EP, Howard J, Atkinson CE, Davies NW, Griffiths PD, Clark DA. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J Clin Microbiol. 2006;44:1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KN, Thiruchelvam AD, Couto-Parada X. Unexpected occasional persistence of high levels of HHV-6 DNA in sera: detection of variants A and B. J Med Virol. 2005;76:563–570. doi: 10.1002/jmv.20399. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Morishima T. HHV-6 and seizures. Herpes. 2005;12:46–49. [PubMed] [Google Scholar]
- Yao K, Crawford JR, Komaroff AL, Ablashi DV, Jacobson S. Review part 2: Human herpesvirus-6 in central nervous system diseases. J Med Virol. 2010;82:1669–1678. doi: 10.1002/jmv.21861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Meier AS, Selke SS, Frenkel LM, Huang ML, Wald A, Rhoads MP, Nguy L, Bornemann R, Morrow RA, Corey L. A population-based study of primary human herpesvirus 6 infection. N Engl J Med. 2005;352:768–776. doi: 10.1056/NEJMoa042207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. Supplementary 2. Chromosome Xp single telomere length analysis of uninfected Jjhan and HEK-293 cells. (a) Diagram illustrating the strategy of single telomere length analysis (STELA). To amplify the telomere of chromosome Xp, a linker (Telorette-2) oligonucleotide is ligated to the end of the telomere. Amplification of the telomere is then preceded with primers P1 and P3. Serial dilution of 150 ng of linker-ligated DNA isolated from the (b) T-cell line Jjhan and (c) HEK-293 cells. In each figure, mean telomere length is based upon co-hybridization of telomere (left panel) and chromosome Xp probes (right panel).
HHV 6B integrates in telomeres during latency > The left end of both HHV 6A and HHV 6B is joined by telomere repeats > A single viral genome integrates in telomeres > Histone deacetylase Trichostatin A induces circularization/catenation of the integrated viral genome