Abstract
Autophagy is an evolutionarily conserved process of cellular self-digestion that serves as a mechanism to clear damaged organelles and recycle nutrients. Since autophagy can promote cell survival as well as cell death, it has been linked to different human pathologies, including cancer. Although mono-allelic deletion of autophagy-related gene BECN1 in breast tumors originally indicated a tumor suppressive role for autophagy in breast cancer, the intense research during the last decade suggests a role for autophagy in tumor progression. It is now recognized that tumor cells often utilize autophagy to survive various stresses, such as oncogene-induced transformation, hypoxia, endoplasmic reticulum (ER) stress and extracellular matrix detachment. Induction of autophagy by tumor cells may also contribute to tumor dormancy and resistance to anticancer therapies, thus making autophagy inhibitors promising drug candidates for breast cancer treatment. The scientific endeavors continue to define a precise role for autophagy in breast cancer. In this article, we review the current literature on the role of autophagy during the development and progression of breast cancer, and discuss the potential of autophagy modulators for breast cancer treatment.
Keywords: Autophagy, breast cancer, transformation, hypoxia, ER stress, tumor microenvironment, metabolism, metastasis, apoptosis, cancer therapy
Introduction
Breast cancer is the second leading cause of cancer-related deaths among women in the United States, with over 229,000 new cases and over 39,000 deaths estimated for the year 2013 [1]. Concerted efforts have been made in recent years to develop therapies that are able to treat various forms of breast cancer more effectively. Recent studies suggest that the process of autophagy or self-cannibalism plays an important role in the development, progression and response of breast cancer cells to chemotherapy [2]. Thus, it is believed that understanding the context specific roles of autophagy could revolutionize the field of breast cancer research.
Autophagy (or macroautophagy) is a cellular process by which cells recycle their components, such as long-lived proteins and damaged organelles, using the lysosomal degradation mechanisms of the cell [3,4]. The proteins and organelles that have been tagged for destruction are sequestered within a vesicle known as an autophagosome. The autophagosome then fuses with the lysosome, forming a complex known as an autolysosome, where the cargo is degraded into its constituent molecules and then released [3]. The important component proteins involved in the execution of autophagy have been grouped as Atg(s) (autophagy-related) in yeast [5].
Successful completion of the complex process of autophagy requires coordinated function of a number of proteins/complexes at different steps, namely: (i) Atg1 (Unc-51-like kinase 1 and -2 or ULKs in mammals) complex mediates the induction step [6-8], (ii) class III PI3K (phosphatidylinositol 3-kinase) complex is required for nucleation of the autophagosomal membrane [9], (iii) p62/sequestosome 1 targets ubiquitinated proteins to autophagosomes [10], (iv) Atg5-Atg12 and LC3-phosphatidylethanolamine are two ubiquitin-like conjugating systems that carry out the expansion of the growing autophagosome membrane [11-13]. Tight regulation of each of these steps by upstream modulators aids in fine-tuning the autophagic process [14]. The mammalian target of rapamycin (mTOR) complex, which is activated by growth factors and amino acids, inhibits autophagy during fed state by inhibiting ULK1 complex [7,15-17]. AMP-activated protein kinase (AMPK), in contrast, is activated by low energy state in the cell and positively regulates autophagy directly by phosphorylating ULK1 [18-20] or indirectly by inhibiting mTOR [21]. The Bcl-2 family proteins, especially Bcl-2, Bcl-xL and Mcl-1, inhibit autophagy by disrupting the interaction of Beclin 1/Atg6 with class III PI3K complex thus inhibiting the early stages of autophagosome formation [22-24]. Deregulation at any of these steps results in too much or too little autophagy which is associated with a number of disorders, including cancer [25].
Autophagy is believed to promote cancer by allowing cells to survive under conditions of metabolic and genotoxic stress [26,27]. Unfavorable conditions of hypoxia and acidity in the tumor microenvironment can cause cells to be metabolically stressed [28]. Furthermore, many chemotherapeutic drugs and radiotherapies function by blocking access to essential nutrients or by damaging the genome of a tumor cell [29]. In this situation, autophagy may be triggered within the cells of the neoplasm, allowing the cells to survive the conditions induced by the chemotherapy and radiotherapy, in essence giving the tumor resistance to the treatment. Under these circumstances, it is believed that inhibiting autophagy within the neoplastic cells may chemosensitize and radiosensitize them, thus increasing the efficacy of the treatment [30,31].
In addition to its ability to promote cell survival, excessive autophagy or self-eating may paradoxically lead to a type of programmed cell death (PCD) known as autophagic cell death, or type II PCD, a form of death that is considered to be distinct from apoptosis and necrosis [32]. Unlike apoptosis, there is early degradation of organelles and a lack of caspase activation or DNA fragmentation and preservation of cytoskeleton elements in autophagy [33,34]. Furthermore, autophagy is characterized by lack of the inflammatory response seen in necrosis. It is believed that inducing autophagic cell death within transformed cells that are resistant to apoptosis may be a viable therapeutic strategy for these types of cancers and that the induction of autophagy could potentially be utilized as a prophylactic mechanism in high-risk patients [35].
Targeted therapy in breast cancer is dictated by the expression of specific molecules such as hormone or growth factor receptors. For example, triple-negative breast cancer (TNBC) cells lack estrogen, progesterone, and human epidermal growth factor 2 (ErbB2 or HER2) membrane receptors and, as such, cannot be treated with hormonal therapies (e.g. tamoxifen) or herceptin (trastuzumab), a humanized antibody against HER2 [36]. Furthermore, many types of breast cancer possess distinct abnormalities in the apoptotic pathway, such as caspase deficiencies and Bcl-2 overexpression, which confer resistance to many forms of chemotherapy [37]. Because of the fundamental importance of autophagy in the development and progression of cancer and its ability to influence treatment response, there has been an explosion of research on the molecular regulation and signal transduction mechanisms that control autophagy. There are several excellent review articles published in the area of autophagy and cancer. In this review, we primarily focused on how autophagy contributes to the development and progression of breast cancer, and its impact on various therapeutic options. Although it is intended for the breast cancer aficionados, it should also benefit investigators engaged in other areas of research related to autophagy.
Autophagy and malignant transformation in breast cancer
Beclin 1, autophagy and breast cancer
Beclin 1 was originally identified as a Bcl-2-interacting protein that is structurally similar to the yeast Atg6 protein [38], which plays an essential role in nitrogen deprivation-induced autophagy [39]. Binding of Bcl-2 inhibits the association of Beclin 1 with class III PI3K and hence prevents autophagy vesicle membrane nucleation [22]. Subsequent studies mapped BECN1 close to the BRCA1 region on chromosome 17, a locus that is frequently deleted in breast, ovarian and prostate cancers [38]. Support for a tumor suppressive role of autophagy in breast cancer came from the identification of Beclin 1 as a haploinsufficient (Beclin 1+/-) tumor suppressor [38]. Furthermore, mice harboring mono-allelic deletion of Beclin 1 were highly susceptible to mammary hyperplasia and had an increased incidence of spontaneous tumors at various sites [40,41]. Consistent with these studies, breast cancer tissues show a decrease in Beclin 1 expression compared to normal breast tissue, and the ectopic expression of Beclin 1 in MCF-7 breast cancer cells, which are haploinsufficient for Beclin 1, decreased their proliferation and in vivo tumor formation [42]. Furthermore, Ras-induced cell death in MCF-7 cells was associated with Beclin 1 upregulation [43]. It has been suggested that autophagy-independent functions of Beclin 1 may also play a role in tumor suppression. However, the expression of a wild-type, but not an autophagy-deficient mutant of Beclin 1, was able to decrease proliferation and tumorigenecity of MCF-7 cells [44]. Nevertheless, the autophagy-independent roles of Beclin 1 need to be considered when assessing its tumor suppressive function [45].
While the mono-allelic deletion of Beclin 1 increased spontaneous tumors in mice, its complete genetic deletion was embryonically lethal [40,41]. What then is the requirement for retaining a single copy of Beclin 1 or the strict haploinsufficiency in tumors? It has been suggested that transformed cells may not be able to tolerate complete loss of Beclin 1 [46] and hence maintain a single copy and low levels of the Beclin 1 protein to ensure that the pro-survival autophagy machinery is intact to overcome the stressful conditions often encountered by cancer cells [47]. Furthermore, incomplete whole body disruption of a second essential autophagy gene Atg5 in mice gave rise only to hepatomas, suggesting that tumor suppression by autophagy may be limited to the liver [48]. These observations suggest that the role of autophagy in cancer may be context-dependent and requires an in-depth analysis of its tissue-specific roles in cancer development and progression.
Role of autophagy in Ras-driven transformation
Given its role in cell survival, there had been earlier speculations about a causal role for autophagy in oncogenic transformation [47,49]. The role of autophagy in Ras transformation has been studied extensively in pancreatic, lung and colon cancer models, especially because Ras mutation is frequently observed in these tumors [50,51]. Although Ras mutation is not observed in large proportions of breast tumors, there is considerable evidence that the Ras pathway is activated in breast cancer cell lines [52]. Ras hyperactivation may also occur downstream of ErbB2 signaling, which is often amplified in breast cancers [53,54]. Lock et al. showed that autophagy is required for Ras-mediated adhesion-independent transformation in H-Ras-transformed non-malignant breast epithelial MCF-10A cells, as well as in human breast cancer cell lines that carry K-Ras mutations [55]. Similarly, Kim et al. showed that K-Ras transformation in MCF-10A cells resulted in increased basal autophagy [56]. In addition, autophagy was required for the growth of Ras-transformed cells in soft agar and tumor formation in nude mice. Involvement of autophagy in tumorigenesis was also confirmed in the polyoma middle T (PyMT) mouse model of mammary cancer, where abrogation of autophagy by genetic deletion of FIP200 (focal adhesion kinase family-interacting protein of 200 kD) resulted in decreased tumor growth [57].
What leads to autophagy upon oncogenic transformation is not precisely known. Kim et al. provided evidence for the involvement of the ROS (reactive oxygen species)-induced JNK (c-Jun N-terminal kinase) pathway [56]. According to this model, Ras transformation leads to increase in ROS, which results in the induction of Atg5 and Atg7 via activation of JNK [56]. Alternate mechanisms that mediate oncogene-induced autophagy activation remain to be elucidated.
Remarkably, autophagy promoted tumorigenesis by enhancing proliferation of transformed cells rather than increasing cell survival [55,57]. The exact mechanism by which autophagy promotes cell proliferation remains to be established. Present studies have led to the “autophagy addiction” model of transformed cells [46]. According to this model, oncogenic transformation alters cellular metabolism in a way that makes autophagy indispensable for tumor progression. Transformed cells, in which autophagy is inhibited by genetic manipulation, are characterized by accumulation of defective mitochondria and defects in glucose metabolism. Intriguingly, different pathways of glucose metabolism are affected in different model systems. In breast cancers, autophagy helps in glucose uptake and facilitates glycolysis in transformed cells, and a defect in autophagy leads to reduced glycolysis [55-57]. However, in pancreatic and kidney cancer models, the requirement of autophagy comes from its ability to sustain the tricarboxylic acid (TCA) cycle [50,51]. It has been speculated that autophagy helps maintain a pool of healthy mitochondria by causing the degradation of defective mitochondria (i.e. mitophagy) and thus supports the TCA cycle. In addition, autophagy-supplied substrates might also be essential for the TCA cycle [46]. White et al. have suggested several insightful mechanisms on how autophagy might regulate the metabolism of transformed cells [46].
Autophagy and tumor cell survival during starvation, hypoxia and endoplasmic reticulum (ER) stress
Tumor progression is characterized by a highly proliferative dense tumor core. Neoangiogenesis tries to cater to the increasing needs of the growing tumor, but fails after a point. This leads to nutrient and growth factor deprivation and reduced oxygen supply in the tumor core. Autophagy is perhaps the only process that is promptly induced in response to starvation. Nutrient recycling during starvation is a function of autophagy that is conserved from yeast to mammals [58,59]. Thus, autophagy is expected to be an important survival mechanism for tumor cells during starvation. Autophagy is induced by growth factor deprivation and nutrient starvation in breast cancer cell lines and promotes cell survival [60,61]. Whether autophagy promotes cell survival under starvation in animal models of breast cancer remains to be studied. Nevertheless, autophagy was shown to be increased in patient-derived mammary tumors and was associated with decreased long-term patient survival [62].
In order to combat reduced oxygen availability, tumor cells induce hypoxia-inducible factors (HIFs). HIF-1α is overexpressed in tumors and helps in tumor cell adaptation to hypoxia by altering metabolism, pH, angiogenesis, erythropoiesis, cell migration, invasion, and inflammation [63,64]. Recent studies have shown that hypoxia can also lead to the induction of autophagy [65-67]. Hypoxia-induced autophagy was mediated via Bnip3 [65], a Bcl-2-interacting BH3-only protein that was initially shown to function as a cell death effector [66]. Subsequent studies, however, showed that Bnip3 induces autophagy, as a survival mechanism, by disengaging Bcl-2 from Beclin 1 [65,67]. Thus, HIF-1α/Bnip3 represents the primary axis that mediates hypoxia-induced autophagy.
HIF-1α is overexpressed in a subset of breast cancers. Its expression was associated with ER/PR-negative and ErbB2-positive status [68,69]. HIF-1α expression is also increased in familial breast cancers in comparison to sporadic tumors [70]. Moreover, transcriptional activation of HIF-1α target genes was shown to be required for breast cancer progression [71]. HIF-1α mediates pro-survival autophagy in breast cancer cells in response to PPARγ (peroxisome proliferator-activated receptor γ) activation [72]. Hypoxia-induced autophagy promotes cell survival in breast cancer cells [73]. However, HIF-1α is not the only mechanism by which hypoxic cells induce autophagy. Hypoxia can also induce autophagy via activation of AMPK or inhibition of mTOR, independent of HIF-1α [74,75].
Rapidly growing tumor cells also display increased ER stress due to overload on protein synthesis machinery. When the load on protein folding machinery is overwhelming, there can be accumulation of unfolded proteins. As a result, the cell triggers UPR (unfolded protein response). Key enzymes activated during UPR are PERK (PKR-like ER kinase), IRE1 (inositol requiring-1α) and ATF6 (activating transcription factor-6) [76]. These enzymes attempt to rescue cells by reducing the rate of new protein synthesis and increasing the expression of chaperon proteins. When the ER stress is excessive and unmanageable, UPR triggers ER-associated protein degradation (ERAD), which is responsible for degradation of unfolded proteins. The proteosomal degradation system was the only recycle system known to be triggered in response to ER stress. Recently, autophagy has been shown to work as an alternate pathway to aid protein degradation during ER stress.
ER stress can induce autophagy through several different mechanisms. PERK-mediated phosphorylation of eIF2α and Atg12 was shown to be required for autophagy induction in response to polyglutamine aggregates [77]. Similarly, PERK-eIF2α mediated radiation-induced autophagy [78]. On the other hand, IRE1 induced autophagy in response to a variety of ER stressors by activating JNK [79]. Inhibition of Akt and mTOR has also been implicated in ER stress-induced autophagy [80]. ER stress and the UPR pathway can also regulate autophagy during starvation and hypoxia [65,73]. Such a cooperative regulation of autophagy is expected, as different stresses prevail concomitantly in the tumor core.
A number of studies suggest that inhibition of ER stress-induced autophagy could be exploited for the treatment of breast cancer [81,82]. In MCF-7 and MDA-MB-231 breast cancer cell lines, capsaicin treatment induced autophagy, causing resistance to ER stress-mediated apoptosis [81]. Such protective autophagy was not induced in non-malignant MCF-10A cells [81]. Moreover, ER stress and autophagy were shown to form the basis of Bortezomib resistance in MCF-7 cells, and inhibition of autophagy increased Bortezomib sensitivity [82].
While most studies suggest a protective role for autophagy, some reports show that autophagy may act as a cell death mechanism in response to stress. Autophagy was shown to promote cell death in apoptosis-deficient cells [83]. Using Bax/Bak double knockout MEFs, Ullman et al. showed that ER stress induced autophagy in both apoptosis-competent as well as apoptosis-deficient cells [83]. However, while autophagy was cytoprotective in apoptosis-competent cells, it led to necrotic cell death in apoptosis-deficient cells [83]. Inhibition of autophagy using 3-methyladenine (3-MA), as well as by genetic knockdown of Atg5, resulted in decreased cell survival in response to ER stress [83]. Similarly, ER stress-mediated autophagy in response to radiation was cytotoxic in caspase-3/7-deficient cells [78]. Hypoxia on the other hand was shown to induce autophagic cell death in various cancer cell lines, including apoptosis-competent MDA-MB-231 and ZR-75 breast cancer cell lines [84]. Thus, the cross-talk between apoptosis and autophagy appears to be stimulus-dependent and warrants further investigation in order to exploit this interaction to achieve maximum killing of cancer cells.
Autophagy in tumor microenvironment
It is being increasingly appreciated that the tumor microenvironment plays a critical role in tumor growth [85]. The tumor microenvironment consists, apart from cancer cells, of non-malignant epithelial cells, fibroblasts, blood vessels, immune cells and extracellular matrix. There is an active interaction between these different factors that govern overall tumor outcome [85]. How other cell types, like fibroblasts, support tumor cell survival is an area of intense investigation [86]. Since autophagy can serve as a crucial survival mechanism, it is important to understand how the process of autophagy is regulated in cancer cells versus stromal cells, and how this interaction governs the fate of tumor cells.
Recent studies have provided insights into the role of autophagy in tumor-associated stromal cells and its effect on heterotypic signaling between stroma and tumor cells [87,88]. Using immortalized fibroblast and breast cancer cell co-culture system, it was shown that breast cancer cells increase oxidative stress in the cancer-associated fibroblasts (CAFs) [89], and that increased oxidative stress leads to induction of autophagy in CAFs [90,91]. The autophagy then provides recycled nutrients, which support the growth of cancer cells. Moreover the CAFs lose their mitochondria through autophagy and hence switch to anaerobic glycolysis, resulting in the excess production of lactate, pyruvate and ketone bodies [92,93]. These molecules are then utilized by the cancer cells via the TCA cycle to provide energy for anabolic processes in cancer cells [94-96]. Thus, anaerobic glycolysis in CAFs supports aerobic oxidation in tumor cells, a phenomenon that the authors term as the “reverse Warburg effect” [97].
The autophagy in CAFs is associated with the loss of caveolin-1 (Cav-1) [92]. Cav-1 is a scaffolding protein which is found in the caveolae membranes of most cell types and has roles in receptor-independent endocytosis [98,99]. Cav-1 loss is associated with early tumor recurrence, increased lymph node metastasis and tamoxifen resistance in breast cancer patients [100-103]. Lisanti et al. showed that Cav-1 loss leads to further increase in autophagy in a feed-forward fashion [87]. Cav-1 loss and ROS stimulate autophagy in CAFs and induce hypoxia-like responses, such as upregulation of HIF-1α, Beclin 1, BNIP3 and BNIP3L in the tumor microenvironment [90].
Breast cancer stem cells have been implicated in tumor promotion [104,105]. Recent studies have shown that cancer stem cells utilize autophagy for their survival [106,107]. Cufi et al showed that autophagy inhibition by knockdown of autophagy genes was associated with a decrease in the number of cells with CD44+CD24-/low expression, while it enhanced the expression of the CD24 gene [107]. Similarly, serum-deprived mesenchymal stem cells (SD-MSC) were shown to survive starvation stress via upregulation of autophagy [108]. Moreover, SD-MSC also inhibited apoptosis and promoted survival of breast cancer cells during serum/nutrient deprivation in vitro as well as in xenograft models [108]. Thus, autophagy in the stromal cells of a tumor’s microenvironment presents a new druggable target for cancer treatment.
Autophagy and metastasis
Metastasis involves detachment of tumor cells from the primary tumor, intravasation into the blood stream, transportation to the secondary site and colonization and re-growth of the secondary tumor [109]. Given the versatile nature of autophagy as well as that of metastasizing tumor cells, studying their relation is a fascinating area of investigation. As discussed before, autophagy was shown to be induced during extracellular matrix (ECM) detachment [110,111]. It is generally believed that when adherent cells are deprived of ECM attachment, they undergo detachment-induced cell death, or anoikis [112]. However, autophagy is also induced in matrix attachment-deprived cells in suspension culture, as wells as in the luminal cells in three-dimensional (3D) culture [113]. Moreover inhibition of integrin β1, the key molecule mediating matrix attachment, also induced autophagy [111]. Although it was initially believed that autophagy in 3D culture helps in the luminal clearance [114], a subsequent study showed autophagy as a survival mechanism in the attachment-deprived cells [110]. While it is conceivable that autophagy helps detached cancer cells survive while they traverse through the circulation, it remains to be tested in an in vivo setting.
Another important process that usually correlates with metastasis is epithelial to mesenchymal transition (EMT), which is characterized by the loss of epithelial phenotype (i.e. loss of tight junctions) and the gain of mesenchymal phenotype, which includes spindle shaped cells that have increased migratory and invasive potential [115]. A recent study showed that induction of autophagy by death-effector domain-containing DNA-binding protein (DEDD) correlates with EMT inhibition in breast cancer cells [116]. Moreover, the knockdown of BECN1 gene increased the EMT markers in MCF-7 cells [116]. This study implies a negative regulation of EMT by autophagy. A recent study showed that EMT helps metastasizing tumor cells to migrate to a secondary site, where they revert back to epithelial phenotype (mesenchymal to epithelial transition, MET), which is essential for the growth of the secondary tumor [117,118]. It is possible that at both the primary and secondary site, autophagy helps tumor cells grow by maintaining their epithelial phenotype.
A critical aspect of recurrent metastatic tumors is dormancy. Isolated tumor cells exit the cell cycle and reside in quiescence, thus evading cell killing by chemotherapeutic drugs that target proliferating cells [119]. After long disease-free periods, regrowth of these dormant cells can result in cancer relapse. Recently, autophagy was shown to be required for the survival of the dormant ovarian cancer cells [120]. Since inhibition of integrin β1 is known to induce autophagy [111], and it was shown to promote dormancy in the PyMT model of mouse mammary tumor [121], it has been speculated that the induction of autophagy, upon integrin β1 inhibition, promotes survival of dormant breast tumor cells [122]. Another mechanism by which autophagy can contribute to tumor dormancy is via its ability to cause TNF-related apoptosis inducing ligand (TRAIL) resistance [123]. Autophagy was shown to be cytoprotective against TRAIL-induced apoptosis [123]. Moreover detached breast tumor cells can remain in bone marrow for extended periods before making their way to bone. Since bone marrow has elevated levels of TRAIL [124], there have been speculations that autophagy in detached tumor cells fights TRAIL cytotoxicity in bone marrow and thus helps in bone metastasis of breast cancer [122].
Autophagy modulators in breast cancer treatment
Due to its dual ability to be cytoprotective and cytotoxic, autophagy has been described as a double-edged sword in cancer therapy [125]. Autophagy was found to be induced by most of the cancer therapeutic agents, since they result in cellular stress [126,127]. Because some inducers of autophagy augmented cell death in response to anti-cancer drugs, it was believed that chemotherapy leads to autophagic cell death [128-132]. Consistent with this notion, inhibition of autophagy by the knockdown of the specific autophagy genes was cytoprotective in several settings [133,134]. Studies examining the role of autophagy as a cell death mechanism had suffered from the lack of a specific autophagy inducer. A recent report has identified a cell-permeable autophagy-inducing peptide, which is derived from an evolutionarily conserved domain of Beclin 1 [135]. This peptide induced autophagy and showed anti-viral activity in mice. Development of this peptide is an important advancement in the field of autophagy and could be an attractive therapeutic approach in scenarios where autophagy induction is desired.
A number of studies also implicate autophagy in therapy resistance. Indeed, autophagy is induced in response to targeted therapy, endocrine therapy, chemotherapy, as well as radiation therapy [126,127,136]. Endocrine or hormonal therapy includes antiestrogens (e.g., tamoxifen) or aromatase inhibitors (e.g., exemestane). It is the first-line of therapy in the treatment of hormone receptor-positive breast cancers. Tumor cells often develop resistance to hormonal therapy. Autophagy inhibition was shown to sensitize breast cancer cells to tamoxifen treatment [137]. ErbB2 is another important target for breast cancer therapy. However trastuzumab, the most widely used monoclonal antibody against ErbB2, is not sufficient in reducing the tumor burden. The combination of trastuzumab with the autophagy inhibitor 3-MA increased the anticancer effect of trastuzumab, in ErbB2-positive breast cancer cells [138]. Autophagy inhibition also showed promise in treating triple-negative breast cancers that are difficult to treat due to the lack of any molecular targets [139-141]. AMPK activators, as well as histone deacetylase inhibitors, induced autophagy in TNBCs [139,140]. Moreover, inhibition of autophagy by chloroquine increased the cytotoxic activity of both agents [139,140]. Cytoprotective autophagy is also induced in response to radiation therapy [142,143]. Inhibition of autophagy using chemical inhibitors or by genetic manipulation of ATG genes sensitized breast cancer cells to ionizing radiation [142,143]. The radiosensitizing effect of autophagy inhibition is seen in hormone receptor-positive as well as TNBC cells [142,143].
Although autophagy inhibitors present attractive therapeutic agents, caution must be exercised while interpreting the effects of these inhibitors. A usual concern with chemical inhibitors is their lack of selectivity. For example, 3-methyladenine, which is used to inhibit autophagy by inhibition of class III PI3K, can also inhibit class I PI3K [144]. Similarly, lysosomal inhibition by chloroquine or bafilomycin A1 can block other cellular processes like receptor endocytosis and phagocytosis [145]. In fact, chloroquine has been shown to reduce cell viability independent of autophagy [146]. Hence, the specific contribution of autophagy to the effect of these pharmacological agents is yet to be determined. Moreover, none of the inhibitors inhibit autophagy at every single step. While 3-MA inhibits autophagy at an early step, the lysosomal inhibitors chloroquine and bafilomycin A1 inhibit autophagy at later steps. Inhibition of autophagy at different stages might have different consequences. Nevertheless, chloroquine and hydrochloroquine have shown promise and are in clinical trials for the treatment of a variety of cancers, including breast cancer [126,145].
Conclusions
While autophagy modulation is an attractive approach for breast cancer therapy, the area is still premature. Autophagy plays important roles in multiple stages of breast cancer as described in Figure 1. The prevailing idea is that autophagy prevents tumor initiation but favors the progression of established tumors. However, existing studies show that generalization of the role of autophagy in cancers from different tissues of origin would be nearsighted. An area that could potentially shed more light on the complex relationship between autophagy and breast cancer is its role in breast cancer prevention by caloric restriction, the physiologically relevant inducer of autophagy [147]. In established tumors that are metabolically dependent on their microenvironment, how can autophagy in the microenvironment of the mammary tumor be targeted? Given the important role of basal autophagy in development and tissue homeostasis, what would be the effects of autophagy inhibition on normal tissue? For example, autophagy inhibition can impair the flux through the ubiquitin-proteasome system leading to the accumulation of toxic, aggregate-prone proteins [148]. Also, in this age of personalized medicine, will autophagy modulation depend on the status of the apoptotic machinery in the tumor of a particular patient? These questions suggest that modulating autophagy for cancer therapy is highly context-dependent, and warrant further research into the pathways that activate it and the molecules that execute it.
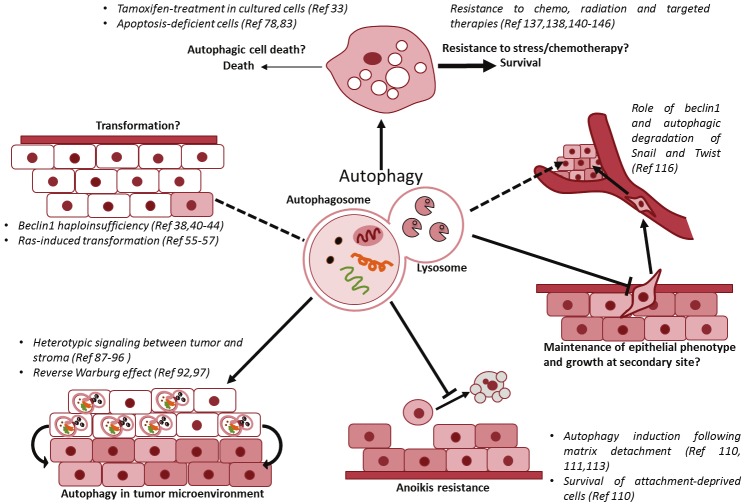
Figure 1.
Role of autophagy in breast cancer initiation and progression. Autophagy plays an important role in several aspects of breast cancer, such as tumor initiation or transformation of mammary epithelial cells, resistance of breast cancer cells to detachment-induced cell death facilitating metastatic spread and maintenance of epithelial phenotype to possibly promote tumor growth at secondary sites. While excessive self-eating can promote death, low levels of autophagy activated in response to cellular stress is believed to promote resistance of breast cancer cell to chemotherapy, radiation and targeted therapy in most settings. Another area where autophagy is gaining importance is in the heterotypic signaling between the mammary tumor and stroma to sustain the growth of cancer cells.
Acknowledgements
This project was supported by the pre-doctoral bridge funding (KJ and SS) from the UNT Health Science Center. We sincerely apologize if we inadvertently left out any major contribution in this field.
Disclosure of conflict of interest
The authors declare no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Debnath J. The multifaceted roles of autophagy in tumors-implications for breast cancer. J Mammary Gland Biol Neoplasia. 2011;16:173–87. doi: 10.1007/s10911-011-9223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 13.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sridharan S, Jain K, Basu A. Regulation of autophagy by kinases. Cancers. 2011;3:2630–2654. doi: 10.3390/cancers3022630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1. ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T, Inoki K. Spatial regulation of the mTORC1 system in amino acids sensing pathway. Acta Biochim Biophys Sin (Shanghai) 2011;43:671–679. doi: 10.1093/abbs/gmr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440:283–291. doi: 10.1042/BJ20101894. [DOI] [PubMed] [Google Scholar]
- 20.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. doi: 10.1038/cddis.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 26.Mathew R, White E. Autophagy, stress, and cancer metabolism: what doesn’t kill you makes you stronger. Cold Spring Harb Symp Quant Biol. 2011;76:389–396. doi: 10.1101/sqb.2012.76.011015. [DOI] [PubMed] [Google Scholar]
- 27.Qiang L, Wu C, Ming M, Viollet B, He YY. Autophagy controls p38 activation to promote cell survival under genotoxic stress. J Biol Chem. 2013;288:1603–1611. doi: 10.1074/jbc.M112.415224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey KM, Wojtkowiak JW, Hashim AI, Gillies RJ. Targeting the metabolic microenvironment of tumors. Adv Pharmacol. 2012;65:63–107. doi: 10.1016/B978-0-12-397927-8.00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chabner BA, Roberts TG Jr. Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 30.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 31.Palumbo S, Comincini S. Autophagy and ionizing radiation in tumors: the “survive or not survive” dilemma. J Cell Physiol. 2013;228:1–8. doi: 10.1002/jcp.24118. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bursch W, Hochegger K, Torok L, Marian B, Ellinger A, Hermann RS. Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J Cell Sci. 2000;113:1189–1198. doi: 10.1242/jcs.113.7.1189. [DOI] [PubMed] [Google Scholar]
- 34.Blommaart EF, Luiken JJ, Meijer AJ. Autophagic proteolysis: control and specificity. Histochem J. 1997;29:365–385. doi: 10.1023/a:1026486801018. [DOI] [PubMed] [Google Scholar]
- 35.Espina V, Wysolmerski J, Edmiston K, Liotta LA. Attacking breast cancer at the preinvasion stage by targeting autophagy. Womens Health (Lond Engl) 2013;9:157–170. doi: 10.2217/whe.13.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 38.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 39.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 40.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 43.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 45.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang C, Avery L. To be or not to be, the level of autophagy is the question: dual roles of autophagy in the survival response to starvation. Autophagy. 2008;4:82–84. doi: 10.4161/auto.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, Der CJ. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 2004;64:4585–4592. doi: 10.1158/0008-5472.CAN-04-0396. [DOI] [PubMed] [Google Scholar]
- 53.Birnbaum D, Sircoulomb F, Imbert J. A reason why the ERBB2 gene is amplified and not mutated in breast cancer. Cancer Cell Int. 2009;9:5. doi: 10.1186/1475-2867-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias-Romero LE, Chernoff J. p21-activated kinases in Erbb2-positive breast cancer: A new therapeutic target? Small GTPases. 2010;1:124–128. doi: 10.4161/sgtp.1.2.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, Hwang SG, Yoon G, Lee SJ. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 59.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 60.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 61.Santi SA, Lee H. Ablation of Akt2 induces autophagy through cell cycle arrest, the downregulation of p70S6K, and the deregulation of mitochondria in MDA-MB231 cells. PloS One. 2011;6:e14614. doi: 10.1371/journal.pone.0014614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, Gatter KC, Giatromanolaki A. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am J Pathol. 2010;176:2477–2489. doi: 10.2353/ajpath.2010.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 65.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Kaya AO, Gunel N, Benekli M, Akyurek N, Buyukberber S, Tatli H, Coskun U, Yildiz R, Yaman E, Ozturk B. Hypoxia inducible factor-1 alpha and carbonic anhydrase IX overexpression are associated with poor survival in breast cancer patients. J BUON. 2012;17:663–668. [PubMed] [Google Scholar]
- 69.Chen CL, Chu JS, Su WC, Huang SC, Lee WY. Hypoxia and metabolic phenotypes during breast carcinogenesis: expression of HIF-1alpha, GLUT1, and CAIX. Virchows Arch. 2010;457:53–61. doi: 10.1007/s00428-010-0938-0. [DOI] [PubMed] [Google Scholar]
- 70.Saponaro C, Malfettone A, Ranieri G, Danza K, Simone G, Paradiso A, Mangia A. VEGF, HIF-1alpha expression and MVD as an angiogenic network in familial breast cancer. PloS One. 2013;8:e53070. doi: 10.1371/journal.pone.0053070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci U S A. 2012;109:E3367–3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, Lisanti MP, Lanza-Jacoby S, Pestell RG, Wang C. PPARgamma activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol. 2009;41:2334–2342. doi: 10.1016/j.biocel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29:4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 74.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 75.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol Sci. 2012;33:53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 77.Kouroku Y, Fujita E, Jimbo A, Kikuchi T, Yamagata T, Momoi MY, Kominami E, Kuida K, Sakamaki K, Yonehara S, Momoi T. Polyglutamine aggregates stimulate ER stress signals and caspase-12 activation. Hum Mol Genet. 2002;11:1505–1515. doi: 10.1093/hmg/11.13.1505. [DOI] [PubMed] [Google Scholar]
- 78.Kim KW, Moretti L, Mitchell LR, Jung DK, Lu B. Endoplasmic reticulum stress mediates radiation- induced autophagy by perk-eIF2alpha in caspase-3/7-deficient cells. Oncogene. 2010;29:3241–3251. doi: 10.1038/onc.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- 81.Choi CH, Jung YK, Oh SH. Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol. 2010;78:114–125. doi: 10.1124/mol.110.063495. [DOI] [PubMed] [Google Scholar]
- 82.Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 83.Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 2008;15:422–425. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 86.Tchou J, Conejo-Garcia J. Targeting the tumor stroma as a novel treatment strategy for breast cancer: shifting from the neoplastic cell-centric to a stroma-centric paradigm. Adv Pharmacol. 2012;65:45–61. doi: 10.1016/B978-0-12-397927-8.00003-8. [DOI] [PubMed] [Google Scholar]
- 87.Lisanti MP, Martinez-Outschoorn UE, Chiavarina B, Pavlides S, Whitaker-Menezes D, Tsirigos A, Witkiewicz A, Lin Z, Balliet R, Howell A, Sotgia F. Understanding the “lethal” drivers of tumor-stroma co-evolution: emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Biol Ther. 2010;10:537–542. doi: 10.4161/cbt.10.6.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weiland A, Roswall P, Hatzihristidis TC, Pietras K, Ostman A, Strell C. Fibroblast-dependent regulation of the stem cell properties of cancer cells. Neoplasma. 2012;59:719–727. doi: 10.4149/neo_2012_091. [DOI] [PubMed] [Google Scholar]
- 89.Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, Whitaker-Menezes D, Daumer KM, Lin Z, Witkiewicz AK, Flomenberg N, Howell A, Pestell RG, Knudsen ES, Sotgia F, Lisanti MP. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, Wang C, Pavlides S, Martinez-Cantarin MP, Capozza F, Witkiewicz AK, Flomenberg N, Howell A, Pestell RG, Caro J, Lisanti MP, Sotgia F. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFkappaB activation in the tumor stromal microenvironment. Cell cycle. 2010;9:3515–3533. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Outschoorn UE, Whitaker-Menezes D, Pavlides S, Chiavarina B, Bonuccelli G, Casey T, Tsirigos A, Migneco G, Witkiewicz A, Balliet R, Mercier I, Wang C, Flomenberg N, Howell A, Lin Z, Caro J, Pestell RG, Sotgia F, Lisanti MP. The autophagic tumor stroma model of cancer or “battery-operated tumor growth”: A simple solution to the autophagy paradox. Cell cycle. 2010;9:4297–4306. doi: 10.4161/cc.9.21.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, Pestell RG, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Loss of stromal caveolin-1 leads to oxidative stress, mimics hypoxia and drives inflammation in the tumor microenvironment, conferring the “reverse Warburg effect”: a transcriptional informatics analysis with validation. Cell cycle. 2010;9:2201–2219. doi: 10.4161/cc.9.11.11848. [DOI] [PubMed] [Google Scholar]
- 93.Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Howell A, Lisanti MP, Sotgia F. Ketone bodies and two-compartment tumor metabolism: stromal ketone production fuels mitochondrial biogenesis in epithelial cancer cells. Cell cycle. 2012;11:3956–3963. doi: 10.4161/cc.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Ketone body utilization drives tumor growth and metastasis. Cell cycle. 2012;11:3964–3971. doi: 10.4161/cc.22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ko YH, Lin Z, Flomenberg N, Pestell RG, Howell A, Sotgia F, Lisanti MP, Martinez-Outschoorn UE. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol Ther. 2011;12:1085–1097. doi: 10.4161/cbt.12.12.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Ketones and lactate “fuel” tumor growth and metastasis: Evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell cycle. 2010;9:3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, Wang C, Pestell RG, Martinez-Outschoorn UE, Howell A, Sotgia F, Lisanti MP. Transcriptional evidence for the “Reverse Warburg Effect” in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer’s disease, and “Neuron-Glia Metabolic Coupling”. Aging (Albany NY) 2010;2:185–199. doi: 10.18632/aging.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scherer PE, Okamoto T, Chun M, Nishimoto I, Lodish HF, Lisanti MP. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci U S A. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Razani B, Schlegel A, Liu J, Lisanti MP. Caveolin-1, a putative tumour suppressor gene. Biochem Soc Trans. 2001;29:494–499. doi: 10.1042/bst0290494. [DOI] [PubMed] [Google Scholar]
- 100.Witkiewicz AK, Dasgupta A, Sotgia F, Mercier I, Pestell RG, Sabel M, Kleer CG, Brody JR, Lisanti MP. An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am J Pathol. 2009;174:2023–2034. doi: 10.2353/ajpath.2009.080873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Witkiewicz AK, Dasgupta A, Sammons S, Er O, Potoczek MB, Guiles F, Sotgia F, Brody JR, Mitchell EP, Lisanti MP. Loss of stromal caveolin-1 expression predicts poor clinical outcome in triple negative and basal-like breast cancers. Cancer Biol Ther. 2010;10:135–143. doi: 10.4161/cbt.10.2.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.El-Gendi SM, Mostafa MF, El-Gendi AM. Stromal caveolin-1 expression in breast carcinoma. Correlation with early tumor recurrence and clinical outcome. Pathol Oncol Res. 2012;18:459–469. doi: 10.1007/s12253-011-9469-5. [DOI] [PubMed] [Google Scholar]
- 103.Thomas NB, Hutcheson IR, Campbell L, Gee J, Taylor KM, Nicholson RI, Gumbleton M. Growth of hormone-dependent MCF-7 breast cancer cells is promoted by constitutive caveolin-1 whose expression is lost in an EGF-R-mediated manner during development of tamoxifen resistance. Breast Cancer Res Treat. 2010;119:575–591. doi: 10.1007/s10549-009-0355-8. [DOI] [PubMed] [Google Scholar]
- 104.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar DH, Kutty MK. Review of stem cell deregulation and breast cancer: an emerging hypothesis. Indian J Pathol Microbiol. 2012;55:147–153. doi: 10.4103/0377-4929.97842. [DOI] [PubMed] [Google Scholar]
- 107.Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell cycle. 2011;10:3871–3885. doi: 10.4161/cc.10.22.17976. [DOI] [PubMed] [Google Scholar]
- 108.Sanchez CG, Penfornis P, Oskowitz AZ, Boonjindasup AG, Cai DZ, Dhule SS, Rowan BG, Kelekar A, Krause DS, Pochampally RR. Activation of autophagy in mesenchymal stem cells provides tumor stromal support. Carcinogenesis. 2011;32:964–972. doi: 10.1093/carcin/bgr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 110.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4:351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 111.Debnath J. Detachment-induced autophagy in three-dimensional epithelial cell cultures. Methods Enzymol. 2009;452:423–439. doi: 10.1016/S0076-6879(08)03625-2. [DOI] [PubMed] [Google Scholar]
- 112.Taddei ML, Giannoni E, Fiaschi T, Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J Pathol. 2012;226:380–393. doi: 10.1002/path.3000. [DOI] [PubMed] [Google Scholar]
- 113.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc Natl Acad Sci U S A. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):vii89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, Yan J, Lv X, Chen X, Hu ZW. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238–3250. doi: 10.1158/0008-5472.CAN-11-3832. [DOI] [PubMed] [Google Scholar]
- 117.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 118.Alderton GK. Metastasis: Epithelial to mesenchymal and back again. Nat Rev Cancer. 2013;13:3. doi: 10.1038/nrc3428. [DOI] [PubMed] [Google Scholar]
- 119.Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer. 2010;10:871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- 120.Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, Liao WS, Bast RC Jr. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 122.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, Rabinowich H. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–19677. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martinet W, Agostinis P, Vanhoecke B, Dewaele M, De Meyer GR. Autophagy in disease: a double-edged sword with therapeutic potential. Clin Sci (Lond) 2009;116:697–712. doi: 10.1042/CS20080508. [DOI] [PubMed] [Google Scholar]
- 126.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berardi DE, Campodonico PB, Diaz Bessone MI, Urtreger AJ, Todaro LB. Autophagy: friend or foe in breast cancer development, progression, and treatment. Int J Breast Cancer. 2011;2011:595092. doi: 10.4061/2011/595092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Notte A, Leclere L, Michiels C. Autophagy as a mediator of chemotherapy-induced cell death in cancer. Biochem Pharmacol. 2011;82:427–434. doi: 10.1016/j.bcp.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 129.Crazzolara R, Cisterne A, Thien M, Hewson J, Baraz R, Bradstock KF, Bendall LJ. Potentiating effects of RAD001 (Everolimus) on vincristine therapy in childhood acute lymphoblastic leukemia. Blood. 2009;113:3297–3306. doi: 10.1182/blood-2008-02-137752. [DOI] [PubMed] [Google Scholar]
- 130.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 131.Li DD, Guo JF, Huang JJ, Wang LL, Deng R, Liu JN, Feng GK, Xiao DJ, Deng SZ, Zhang XS, Zhu XF. Rhabdastrellic acid-A induced autophagy-associated cell death through blocking Akt pathway in human cancer cells. PloS One. 2010;5:e12176. doi: 10.1371/journal.pone.0012176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi WY, Xiao D, Wang L, Dong LH, Yan ZX, Shen ZX, Chen SJ, Chen Y, Zhao WL. Therapeutic metformin/AMPK activation blocked lymphoma cell growth via inhibition of mTOR pathway and induction of autophagy. Cell Death Dis. 2012;3:e275. doi: 10.1038/cddis.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bommareddy A, Hahm ER, Xiao D, Powolny AA, Fisher AL, Jiang Y, Singh SV. Atg5 regulates phenethyl isothiocyanate-induced autophagic and apoptotic cell death in human prostate cancer cells. Cancer Res. 2009;69:3704–3712. doi: 10.1158/0008-5472.CAN-08-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 135.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 137.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 138.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PloS One. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee KH, Hsu EC, Guh JH, Yang HC, Wang D, Kulp SK, Shapiro CL, Chen CS. Targeting energy metabolic and oncogenic signaling pathways in triple-negative breast cancer by a novel adenosine monophosphate-activated protein kinase (AMPK) activator. J Biol Chem. 2011;286:39247–39258. doi: 10.1074/jbc.M111.264598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rao R, Balusu R, Fiskus W, Mudunuru U, Venkannagari S, Chauhan L, Smith JE, Hembruff SL, Ha K, Atadja P, Bhalla KN. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012;11:973–983. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- 141.Thomas S, Sharma N, Golden EB, Cho H, Agarwal P, Gaffney KJ, Petasis NA, Chen TC, Hofman FM, Louie SG, Schonthal AH. Preferential killing of triple-negative breast cancer cells in vitro and in vivo when pharmacological aggravators of endoplasmic reticulum stress are combined with autophagy inhibitors. Cancer Lett. 2012;325:63–71. doi: 10.1016/j.canlet.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 142.Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, Rodemann HP. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol. 2011;99:287–292. doi: 10.1016/j.radonc.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 143.Bristol ML, Di X, Beckman MJ, Wilson EN, Henderson SC, Maiti A, Fan Z, Gewirtz DA. Dual functions of autophagy in the response of breast tumor cells to radiation: cytoprotective autophagy with radiation alone and cytotoxic autophagy in radiosensitization by vitamin D3. Autophagy. 2012;8:739–753. doi: 10.4161/auto.19313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sheng Y, Sun B, Guo WT, Zhang YH, Liu X, Xing Y, Dong DL. 3-Methyladenine induces cell death and its interaction with chemotherapeutic drugs is independent of autophagy. Biochem Biophys Res Commun. 2013;432:5–9. doi: 10.1016/j.bbrc.2013.01.106. [DOI] [PubMed] [Google Scholar]
- 145.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 146.Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–212. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]