Abstract
Tumor-initiating stem-like cells (TICs) are resistant to chemotherapy and associated with hepatocellular carcinoma (HCC) caused by HCV and/or alcohol-related chronic liver injury. Using HCV Tg mouse models and patients with HCC, we isolated CD133+ TICs and identified the pluripotency marker NANOG as a direct target of TLR4, which drives the tumor-initiating activity of TICs. These TLR4/NANOG–dependent TICs were defective in the TGF-β tumor suppressor pathway. Functional oncogene screening of a TIC cDNA library identified Yap1 and Igf2bp3 as NANOG-dependent genes that inactivate TGF-β signaling. Mechanistically, we determined that YAP1 mediates cytoplasmic retention of phosphorylated SMAD3 and suppresses SMAD3 phosphorylation/activation by the IGF2BP3/AKT/mTOR pathway. Silencing of both YAP1 and IGF2BP3 restored TGF-β signaling, inhibited pluripotency genes and tumorigenesis, and abrogated chemoresistance of TICs. Mice with defective TGF-β signaling (Spnb2+/– mice) exhibited enhanced liver TLR4 expression and developed HCC in a TLR4-dependent manner. Taken together, these results suggest that the activated TLR4/NANOG oncogenic pathway is linked to suppression of cytostatic TGF-β signaling and could potentially serve as a therapeutic target for HCV-related HCC.
Introduction
Tumor-initiating stem-like cells (TICs) represent a major factor in chemotherapy resistance (1) in the treatment of hepatocellular carcinoma (HCC), the fifth most common cancer in men and the seventh most common cancer in women in the world (2). In liver tissue, regeneration plays a vital role in a homeostatic response to injury. However, the occurrence of mutations during chronic liver injury is likely to prompt an expansion of altered stem cells, leading to the genesis of TICs for tumor development and progression. Because roughly 40% of HCC is considered clonal, early tumor initiation may be stem-like in origin. Therefore, it is important to understand the key functional pathways of these TICs for identification of new therapeutic targets for HCC.
Chronic inflammation is a major risk factor for cancer development (3), which may involve activation of NF-κB and STAT3 (4). Viral infection (HBV or HCV) and environmental factors (alcohol, obesity) give rise to chronic liver inflammation and increase the risk for HCC. The link between inflammation and cancer is supported by the fact that TLR4, the pathogen-associated molecular pattern, which mediates an inflammatory response to endotoxin and other ligands, is implicated in lung (5), colon (6), and skin carcinomas (7). Although we usually consider macrophages and lymphocytes to be the primary cell types that express TLR4 to mediate immune and inflammatory response, an increasing body of evidence points to the role of TLR4 ectopically expressed in epithelial parenchymal cells in oncogenesis (8, 9). To this end, we have recently identified the pluripotency marker Nanog, as a novel gene downstream of TLR4 in CD133+/CD49f+ TICs, which contributes to liver oncogenesis in HCV nonstructural 5A (Ns5a) Tg alcohol-fed mice (9). These Tg mice show upregulation of TLR4 expression in hepatocytes and develop liver tumors, when TLR4 signaling is chronically activated by endotoxemia associated with alcohol intake (9), serving as a model for synergistic HCC in alcoholic patients infected with HCV (10, 11).
The TGF-β pathway is instrumental in mammalian development and tumor suppression through inhibition of proliferation and induction of apoptosis. A defective TGF-β tumor suppressor pathway is implicated in a broad array of malignancies, including HCC (12). Paradoxically, TGF-β may also promote tumorigenesis — promoting tumor cell invasiveness and metastasis through modulation of the immune system and enhancement of the epithelial-mesenchymal transition (13, 14) in more progressed disease stages. Nevertheless, the roles and clinical relevance of TGF-β in its tumor suppressor signaling arm are becoming increasingly evident. This association is experimentally supported by development of spontaneous HCC in mice with impaired TGF-β signalling due to β2-spectrin deficiency (15). Loss or reduced expression of TGF-β receptor (TβRI/TβRII) or signalling molecules (e.g., SMAD4) also enhances malignant progression in various human tumor types, cancer xenografts, and Tg mice (16–22). This is due at least in part to activation of mitogenic and oncogenic pathways involving CDK4, PRAJA, β-catenin, TERT, and c-MYC that occur when the TGF-β pathway is inactivated. Suppressed TGF-β pathway may also contribute to the resistance of HCC to chemotherapy through induction of IL-6 (23) and downstream activation of STAT3 (24), and this chemoresistance presents an important clinical challenge for HCC.
In innate immunity, heightened TLR4 signaling is known to suppress the TGF-β pathway (25). Conversely, lack of a functional TGF-βR1 or TGF-βR2 (TβRI or TβRII) (26, 27) or SMAD3 (28) results in extensive inflammation due to increased TLR4 expression and LPS hyperresponsiveness (26). This reciprocal regulation of TLR4 and TGF-β pathways may also be important in liver oncogenesis and could present a new functional target for therapeutics of lethal cancers that evade cytostatic TGF-β control (29–32). Here, we report that the newly identified TLR4/NANOG oncogenic signaling pathway impairs the TGF-β pathway to confer TICs chemoresistance in HCV-associated HCC. Analysis of TICs isolated from HCC in alcohol-fed HCV Ns5a or Core mouse models and alcoholic HCV patients led to the identification of YAP1 and IGF2BP3 as novel TLR4/NANOG–dependent molecules responsible for suppression of the TGF-β pathway and chemoresistance. Further, HCC that developed due to a defective TGF-β pathway in β2-spectrin–deficient mice was shown to be similarly caused by reciprocally upregulated TLR4 signaling.
Results
Isolation of TLR4/NANOG–dependent CD133+/CD49f+ TICs from HCC mouse models and patients with HCC.
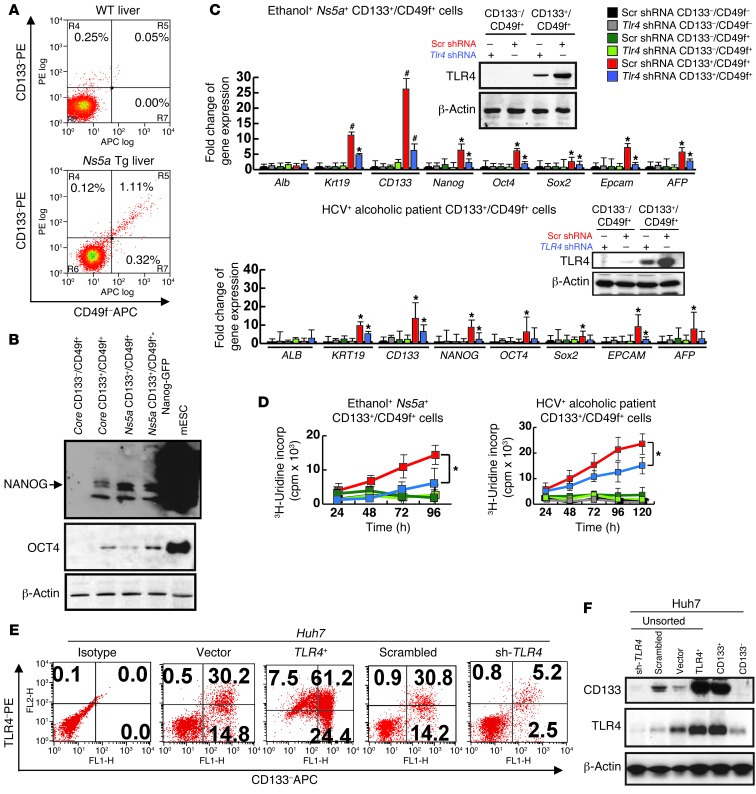
We first isolated TICs by CD133 and CD49f FACS sorting from liver tumors of ethanol-fed Ns5a Tg mice (Figure 1A) in which we had previously shown colocalization of NANOG and CD133 or CD49f in cells with a high nucleus/cytoplasm ratio (9). The liver tumors from the model had substantially increased percentages of both CD133–/CD49f+ and CD133+/CD49f+ populations, as compared with the WT mice (0.00% vs. 0.32% and 0.05% vs. 1.11%, respectively, Figure 1A). Quantitative PCR (qPCR) analysis of these populations demonstrates consistent inductions in stem cell markers, such as Nanog, Oct4, Sox2, Epcam, α-fetoprotein (AFP), and cytokeratin-19 (Krt19), in the CD133+/CD49f+ population as compared with CD133–/CD49f– or CD133–/CD49f+ population (Figure 1C). Based on this result, we designated and used CD133+/CD49f+ cells as TICs and CD133–/CD49f+ cells as control cells in subsequent studies. The Nanog induction in TICs was confirmed by immunoblot analysis (Figure 1B) and colocalization of NANOG immunofluorescence staining and GFP expressed under Nanog promoter of a transfected Nanog-GFP reporter plasmid (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI65859DS1). TLR4, which we proposed as the putative oncogenic factor (9), was conspicuously upregulated in TICs (inset of Figure 1C), and more importantly, knockdown of TLR4 with shRNA attenuated the induction of the stem cell genes (red vs. blue in Figure 1C). We performed microarray analysis on TICs vs. control cells and identified differentially regulated genes, including genes downstream of TLR4 (RNase L, Ifi44, Il10) and those implicated in invasion (Tiam-1) (33) and stemness (Fzd3 and Hhip3) (Supplemental Table 1). We then sought to validate the occurrence of these TICs in a different animal model as well as human HCC. For this purpose, we isolated the double-positive cells from HCC developed in alcohol-fed HCV Core Tg mice and from human HCC tissues surgically resected from HCV-infected alcoholic patients (representative histology for both shown in Supplemental Figure 1B). TICs from both groups showed TLR4 protein upregulation and similar inductions of the stemness genes in a manner dependent on TLR4 (Supplemental Figure 1C and Figure 1C). Next, we tested anchorage-independent growth and spheroid formation of TICs. The TICs but not control cells from the mouse model and from patients grew in soft agar (Supplemental Figure 1D) and formed spheroids after serial passages (Supplemental Figure 1E) in a manner dependent on Tlr4 and Nanog. Similarly, enhanced DNA synthesis of TICs was significantly attenuated 50%–80% by Tlr4 shRNA (Figure 1D and Supplemental Figure 1C, right panel). This inhibition was incomplete most likely because TLR4 was not silenced completely with shRNA, as shown by immunoblots in insets of Figure 1C and Supplemental Figure 1C. Invasiveness of the TICs was also 4-fold higher than that of the control cells, as shown for the cells from the Ns5a model in Supplemental Figure 1F. The human HCC cell line Huh7 expressed CD49f constitutively but CD133 in about 40%–50% of the cells. As CD133 positivity allowed isolation of self-renewing cells, we tested TLR4 expression in this cell line. Approximately two-thirds of CD133+ Huh7 cells were positive for TLR4 (Figure 1E). CD133 positivity increased to 85% when TLR4 was overexpressed (Tlr4+ vs. vector) and was abrogated when TLR4 was silenced with shRNA (shRNA for TLR4 vs. scrambled) (Figure 1E), suggesting a link between TLR4 and the stemness marker CD133. These results were confirmed by immunoblot analysis (Figure 1F). Furthermore, TLR4-transduced Huh7 cells showed increased proliferation as compared with vector-transduced cells, whereas TLR4 shRNA reduced proliferation of CD133+ cells but not CD133– Huh7 cells (Supplemental Figure 1G). Collectively, these results demonstrate TLR4-mediated proliferation of CD133+ TICs and Huh7 cells.
Figure 1. TLR4/NANOG–dependent TICs from liver tumors in alcohol-fed Ns5a Tg mice and patients.
(A) FACS separation of CD133+/CD49f+ cells from liver tumors of a HCV Ns5a Tg mouse. (B) Immunoblot analysis confirms induction of NANOG and OCT4 in TICs from both Core and Ns5a mice. Mouse embryonic stem cells (mESC) serve as a positive control. (C) TICs isolated from liver tumors of Ns5a Tg mice or alcoholic HCV patients express Nanog, Oct4, Sox2, and Krt19 at higher levels than CD133–/CD49f+ or CD133–/CD49f– cells, as determined by qPCR. Immunoblots reveal increased TLR4 protein levels in TICs, which are effectively silenced by transduction of lentiviral shRNA (insets). This silencing abrogates stemness gene upregulation. *P < 0.05, #P < 0.01, compared with scrambled shRNA. (D) 3H-uridine incorporation assay in culture demonstrates enhanced, TLR4-dependent cell proliferation of TICs from Ns5a Tg model (left) and patients (right). *P < 0.05, compared with scrambled shRNA. (E) TLR4 expression correlates with the stemness marker CD133. Human HCC cell line Huh7 cells were transduced by a retroviral vector expressing TLR4 (TLR4+) or shRNA for TLR4 (sh-TLR4) and examined for surface expression of CD133 by FACS, as compared with those transduced with the control vector (Vector) or scrambled shRNA (Scrambled). (F) An immunoblot confirms induced CD133 expression in Huh7 cells transduced with Tlr4 (TLR4+) and a loss of this expression by transduction of shRNA against Tlr4 (sh-TLR4). A CD133+ population of Huh7 cells (CD133+) express abundant TLR4, and CD133– Huh7 cells (CD133–) lack this expression.
TICs give rise to tumor in vivo.
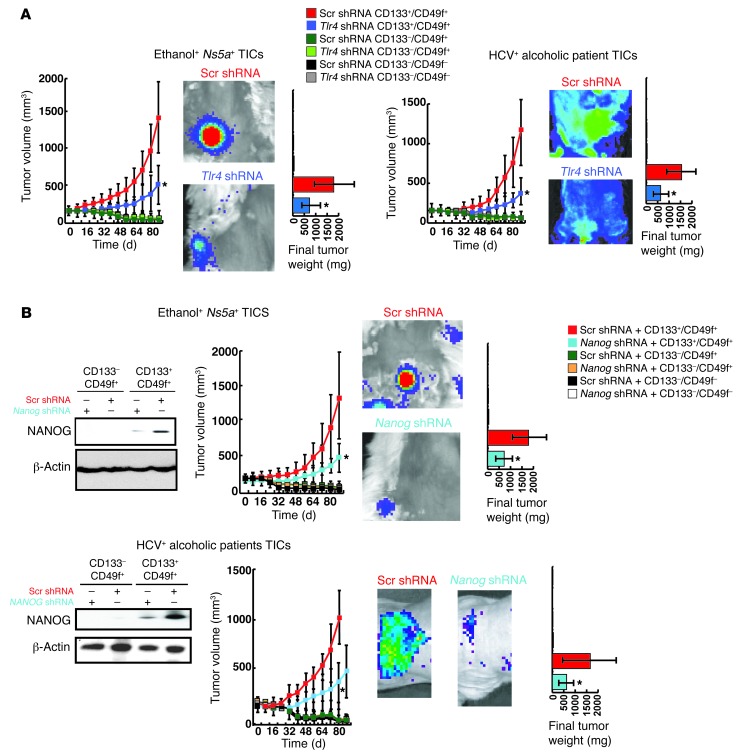
Next, we tested the tumor-initiating property of TICs from the Tg models and patients by infecting the TICs and control cells with a lentiviral GFP expression vector and transplanting them subcutaneously in NOD/Shi-scid/Il2r–/– (NOG) mice. Genesis and growth of tumors derived from transplanted cells were monitored by GFP imaging for 80 days. Growing tumors developed from TICs from all 3 groups, while corresponding control or CD133–/CD49f– cells failed to form tumors (Figure 2A and Supplemental Figure 2A). For mice bearing the control cells, we extended the observation up to 6 months but did not observe any tumor formation (data not shown). Both growth and weight of tumors generated by TICs from all 3 groups were significantly attenuated by infection of the cells with a lentivirus expressing Tlr4 shRNA prior to transplantation (Figure 2A and Supplemental Figure 2A). Histological analysis of tumors derived from Ns5a or Core Tg mouse and human patient TICs largely revealed dysplastic nodules or HCC (Supplemental Figure 2C), with occasional coexistence of cholangioma and fibrosarcoma. To test NANOG dependence of tumorigenesis from TICs, the cells were transduced with a Nanog shRNA lentiviral vector prior to transplantation. This manipulation reduced tumor growth by 60% to 75% (Figure 2B and Supplemental Figure 2B). As seen with Tlr4 shRNA, Nanog shRNA did not completely prevent the tumor growth, most likely due to incomplete silencing of NANOG expression with shRNA, as shown in accompanying immunoblots. One of the NANOG pseudogenes, NANOGP8, contributes to oncogenesis (34). To test the role of NANOGP8 vs. NANOG in tumorigenesis initiated by TICs, cells from human HCC were transduced with shRNA against each and used for a xenograft growth competition assay. Our results demonstrate suppressed tumorigenic activity by silencing of NANOG but not NANOGP8 (Supplemental Figure 3, A–D), suggesting a limited role of the latter in tumorigenesis caused by TICs.
Figure 2. TLR4/NANOG–dependent tumor-initiation property of TICs.
(A) GFP-transduced TICs, but not other populations from the Ns5a Tg mouse and patient HCC, give rise to growing tumors, as determined by whole-body imaging following subcutaneous transplantation in NOG mice. This growth is attenuated by Tlr4 shRNA transduction prior to transplantation (*P < 0.05). Final tumor weight is also reduced with Tlr4 shRNA. (B) Tumor growth by TICs from Ns5a Tg mouse or patient HCC is attenuated by transduction of Nanog shRNA. Final tumor weight is also reduced. Immunoblotting of cell lysates collected 10 days after the transplantation confirms expression of NANOG in the TICs and knockdown of this protein with specific shRNA. *P < 0.05.
To further validate the tumor-initiating property, the TICs and control cells from Ns5a mice were serially transplanted into NOG mice. The cells were derived from 3 different liver samples and subcutaneously injected with 3 different cell numbers (500; 2,500; and 10,000) into 6 NOG mice for each group (n = 54). In these mice, tumors arose exclusively from the TICs, and they were subsequently used for serial transplantation. Indeed, TICs repetitively formed tumors in the serial transplantation experiment (Supplemental Table 2). These results support the hypothesis that TICs have tumor-initiating and self-renewal properties. To address how TICs are maintained in tumors established after transplantation, the cells were isolated from the tumors generated by TICs from Ns5a and Core Tg mice and sorted to assess the CD133+/CD49f+ population. Interestingly, the double-positive TICs account for 1.1% to 1.4% of total cells (Supplemental Figure 4A), which is similar to the percentage observed for the double-positive cells from the original tumors, suggesting that TICs reside in a small population once a solid tumor is formed.
A recent study shows that CD24+ cells from patient HCC possess a tumor-initiating property in a manner dependent on STAT3 and NANOG (35). To address this possibility, we tested anchorage-independent growth and tumor formation by CD133+/CD24– vs. CD133+/CD24+ cells, and we found no difference in colony formation and tumor-initiation property in NOG mice (Supplemental Figure 4, B and C). These results suggest that CD24 positivity does not reflect tumor-initiating property in our TICs.
TLR4 induces Igf2bp3 and Yap1 through Nanog.
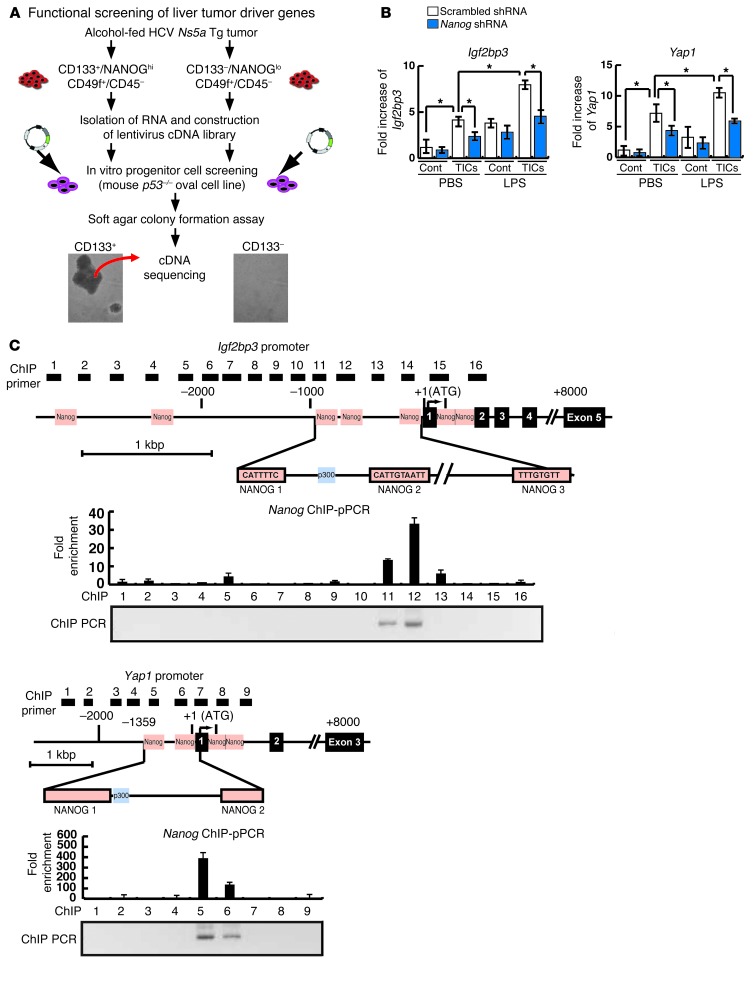
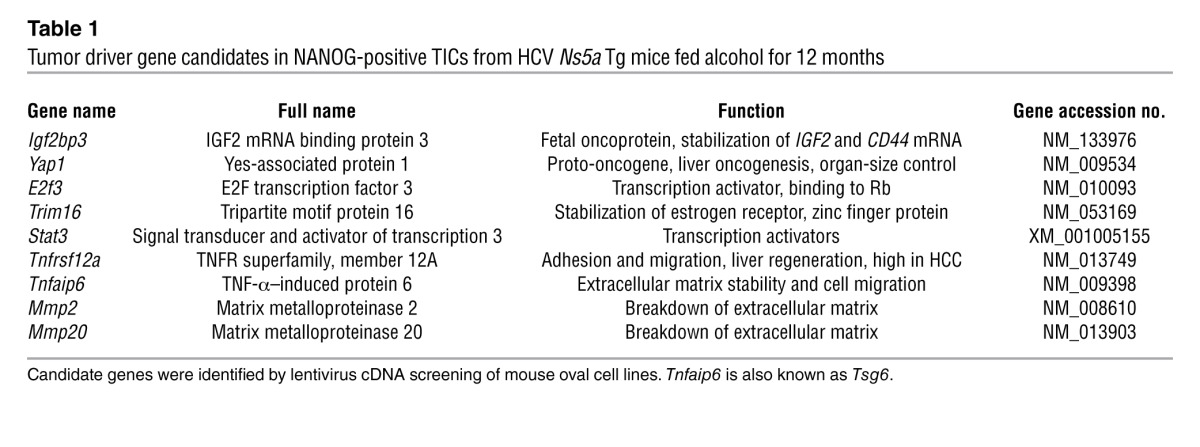
Having established tumorigenic activity of TICs, we next sought to identify oncogenes expressed by the putative TICs. This was achieved by generating a lentiviral cDNA library via reverse transcribing mRNA from the cells isolated from liver tumors of 5 independent Ns5a Tg mice, creating targeting vectors harboring cDNAs, and cloning them into lentivirus by the recombination method. Cells from an immortalized mouse liver progenitor p53-deficient cell line (PIL-4) were infected with the lentiviral library derived from either TICs or control cells and tested for colony formation in soft agar (Figure 3A). Following 2 to 3 weeks after infection with the lentiviral library from TICs, PIL-4 cells transformed and generated numerous colonies, but very few colonies were formed by the library from the control cells (Figure 3A). Colonies formed by infection with the TIC-derived library were individually isolated to screen for potential oncogenes. Isolation and sequencing of 45 cDNAs from 138 colonies have led to identification of 9 candidate genes, including Igf2bp3 (also known as IMP-3), Yap1, Tnfrsf12a, Trim16, Tnfaip6, E2f3, Stat3, Mmp2, and Mmp20 (Table 1). Most of these genes are known to have enriched NANOG binding motifs within 8 kb upstream of their transcription start site in embryonic stem cells (36). Among these genes, Yap1, E2f3, and Stat3 are reported to be oncogenic when overexpressed (37–39). YAP1 is a target of the organ-size control Hippo pathway (40). IGF2BP3 expression enhances tumor growth and invasion at least in part by promoting translation of IGF-II (41) and stabilization of CD44 mRNA (42), and its expression predicts early recurrence and poor prognosis in HCC (43, 44). We confirmed by qRT-PCR that Yap1 and Igf2bp3 were induced in TICs and were further upregulated by LPS in an at least partially NANOG-dependent manner (Figure 3B). Only 2 genes were isolated from a few colonies generated by the cDNA library from the control cells (Supplemental Figure 4D).
Figure 3. Identification of TLR4/NANOG–dependent oncogenes, Igf2bp3 and Yap1, from functional screening of a lentiviral cDNA library.
(A) Strategy of in vitro identification of liver oncogenes, with oval cells infected with the TIC library but not with the control cell library forming colonies. (B) Increased expression of Igf2bp3 and Yap1 in the TICs as confirmed by qPCR is further upregulated by LPS treatment. Nanog shRNA significantly reduces these inductions. (C) Schematic diagrams of Igf2bp3 and Yap1 promoters depicting the locations of NANOG consensus binding sequences (orange boxes) and the areas analyzed by ChIP (1–16 black boxes for Igf2bp3 and 1–9 black boxes for Yap1). NANOG ChIP-qPCR data for TICs from the Ns5a Tg model are shown below, with NANOG enrichment in the regions 11 and 12 within NANOG 1 and 2 sites for the Igf2bp3 promoter and in the region 5 encompassing the NANOG 1 site of the Yap1 promoter.
Table 1.
Tumor driver gene candidates in NANOG-positive TICs from HCV Ns5a Tg mice fed alcohol for 12 months
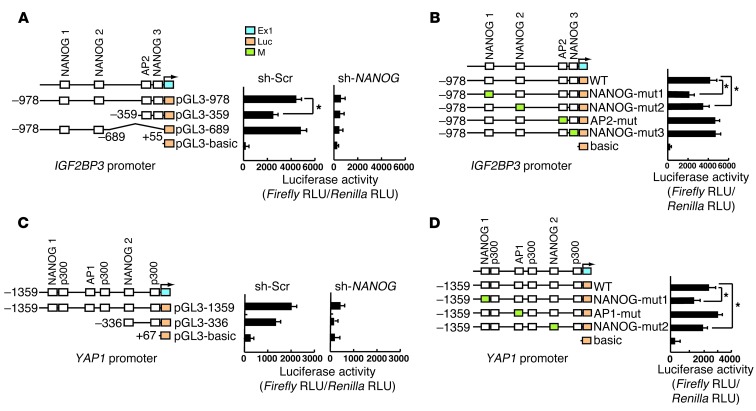
To determine how Igf2bp3 and Yap1 inductions are mediated by NANOG, the NANOG binding consensus sequences within their promoters were examined for recruitment of NANOG in LPS-stimulated TICs by ChIP-qPCR assays. NANOG recruitment is enriched in a region encompassing 3 NANOG binding sites in the IGF2BP3 promoter (–869, –746, and –147 nt) (Figure 3C) and 2 NANOG binding sites in the Yap1 promoter (–1294 and –323 nt) (Figure 3C). IGF2BP3 promoter analysis with deletion constructs in Huh7 cells suggests the importance of the segment (–978/–359 nt) containing 2 distal NANOG sites (NANOG 1 and 2, Figure 4A). NANOG dependence of this promoter activity was demonstrated by abrogating effects of NANOG shRNA (Figure 4A). To further confirm the functional importance of the NANOG sites within –978/+55 nt, we mutated each of the 3 sites (NANOG 1–NANOG 3). Mutations of NANOG 1 and 2 sites reduced LPS-induced IGF2BP3 promoter activity, while mutation of NANOG 3 site had no effect (Figure 4B), indicating that IGF2BP3 is a target of TLR4 signaling primarily via NANOG binding to the distal NANOG 1 and 2 sites. Similar results were obtained for a YAP1 proximal promoter region, with 2 NANOG sites (NANOG 1 and 2) being functional (Figure 4, C and D). These results demonstrate that TLR4 induces Igf2bp3 and Yap1 through NANOG.
Figure 4. Identification of TLR4/NANOG–dependent oncogenes, Igf2bp3 and Yap1, from functional screening of a lentiviral cDNA library.
(A) Igf2bp3 promoter analysis with deletion constructs demonstrates the importance of the proximal segment (–978/–359 nt) in LPS-induced promoter activity in TLR4-transduced Huh7 cells. NANOG dependence of this promoter activity is also demonstrated by the abrogating effects of NANOG shRNA. (B) Mutations of the NANOG consensus sites within –978/+55 nt (NANOG binding sites 1 and 2 but not 3) reduce LPS/TLR4–induced IGF2BP3 promoter activity. (C) YAP1 promoter analysis with deletion constructs demonstrates the importance of the proximal segment (–1359/–336 nt) in LPS-induced promoter activity in TLR4-transduced Huh7 cells. NANOG shRNA similarly abrogates the promoter activity. (D) Mutation of one of the NANOG consensus sites (NANOG binding site 2) reduces LPS/TLR4–induced YAP1 promoter activity. *P < 0.05.
Yap1 and Igf2bp3 are TIC oncogenes.
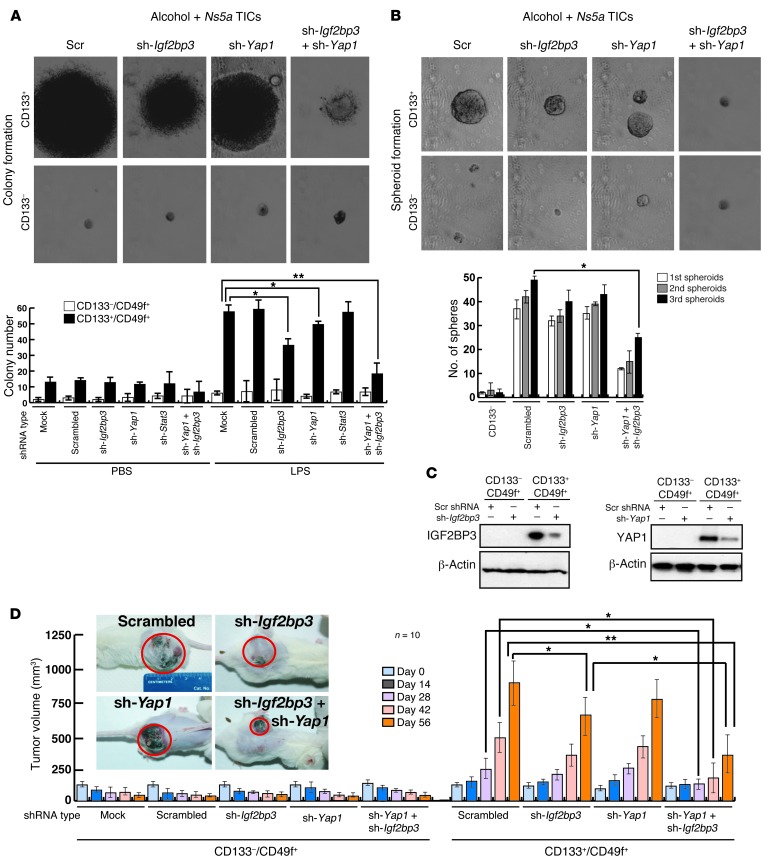
To validate the oncogenic activity of Yap1 and Igf2bp3 in vitro, TICs from Ns5a mice were infected with lentivirus expressing shRNA for each of the 2 genes prior to performing a colony formation assay. Colony formation was moderately inhibited by either Yap1 shRNA or Igf2bp3 shRNA alone. However, dual knockdown of Yap1 and Igf2bp3 almost completely abrogated colony formation (Figure 5A). Furthermore, transduction of shRNA against both Yap1 and Igf2bp3 significantly reduced the number of spheroids formed with TICs during the first, second, and third passages (Figure 5B). Immunoblot of lysates from the cells confirmed enhanced expression of YAP1 or IGF2BP3 in the TICs and effective knockdown of these proteins with the specific shRNA (Figure 5C). We then tested the oncogenic activities of these 2 genes in vivo by transplanting the TICs with or without silencing of Yap1 and/or Igf2bp3 in NOG mice. Growing tumors arose in the mice grafted with TICs transduced with an empty vector or scrambled shRNA over a period of 14 to 56 days (Figure 5D). Transduction of Igf2bp3 shRNA, but not that of Yap1 shRNA, moderately decreased the tumor size at day 56 only. However, silencing of both genes reduced tumor growth conspicuously at all 3 later time points (i.e., day 28, 42, and 56), confirming the cooperative effects of the 2 genes in oncogenesis. These results suggest that Yap1 and Igf2bp3 function via different mechanisms and work cooperatively for the oncogenic effect.
Figure 5. Validation of Yap1 and Igf2bp3 as TIC oncogenes.
(A) The number of colonies formed by TICs in soft agar is moderately reduced by lentiviral transduction of shRNA against Yap1 or Igf2bp3 but largely abrogated when knocking down both. Stat3 silencing has no effects. *P < 0.05, **P < 0.01. (B) Self-renewal ability, as assessed by spheroid formation assay, is reduced in sh-Yap1/sh-Igf2bp3–transduced TICs. Serial spheroid-forming capacity is also reduced by dual transduction of sh-Yap1 and sh-Igf2bp3. *P < 0.05. (C) Immunoblot analysis demonstrates induced expression of IGF2BP3 and YAP1 in TICs and effective knockdown by respective shRNA. (D) Transduction of Ifg2bp3 shRNA, but not that of Yap1 shRNA, modestly reduces subcutaneous tumor growth by the TICs from the Ns5a Tg mice in NOG mice, while transduction of both shRNA clearly abrogates the growth at the later 3 time points. Representative photos of NOG mice carrying TIC-derived tumors are shown to depict an appreciable reduction of tumor size by Igf2bp3 and Yap1 silencing. *P < 0.05, **P < 0.01 versus scrambled shRNA group.
YAP1 and IGF2BP3 suppress the TGF-β pathway.
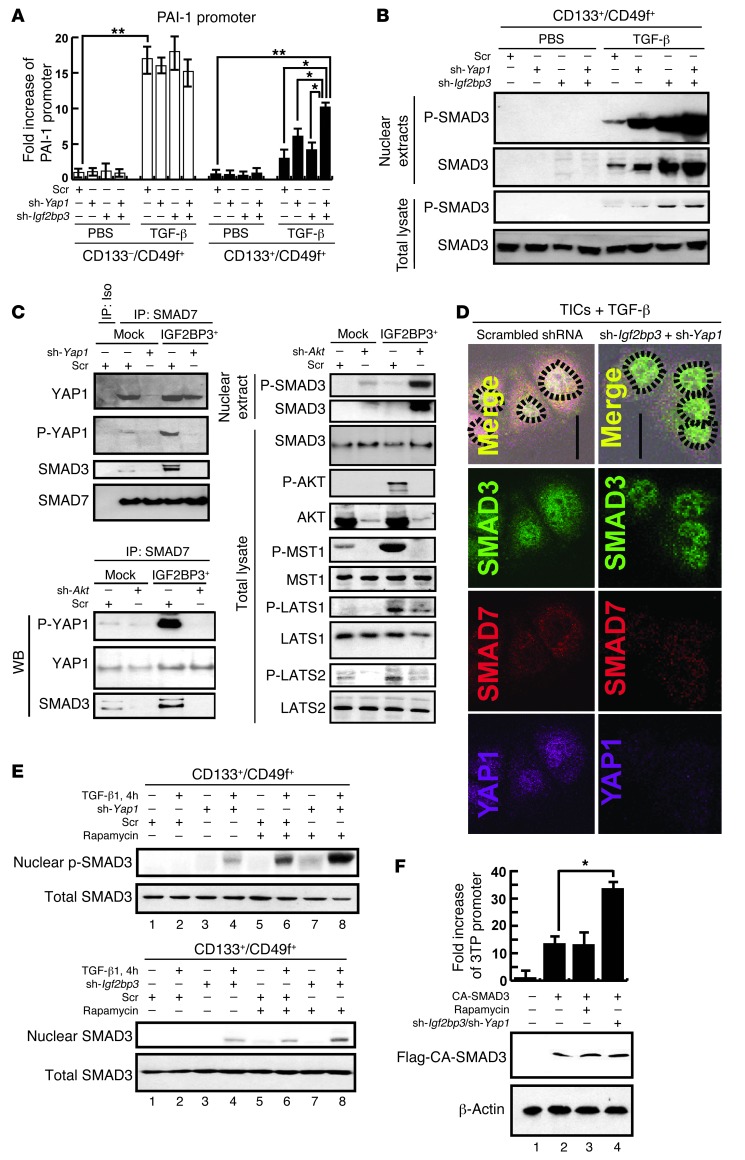
While TLR4/NANOG is identified as a novel oncogenic pathway in our TICs, a deficient TGF-β pathway caused by inactivation of at least one of the TGF-β signaling components is also a well-known risk factor for HCC in humans (45, 46) and a causal oncogenic mechanism in animal models (23, 47). Thus, we hypothesized that these 2 pathways may be reciprocally regulated and linked to the tumorigenic activity of TICs. Indeed, PAI-1 promoter activity, a parameter for TGF-β–mediated SMAD3/4 activity, was reduced in TICs under either LPS or TGF-β stimulation, as compared with that in the control cells (Supplemental Figure 5A), despite comparable expression of Tgfbr2 or Smad3 in the 2 populations (Supplemental Figure 5B). YAP1 may interact with SMAD7 and potentiates SMAD7’s inhibitory activity against TGF-β/SMAD3 signaling (48). Igf2bp3 encodes an mRNA binding protein, which promotes IGF-II translation by binding to the 5′UTR of Igf-II mRNA (41). IGF-II activates AKT and subsequently mTOR, which may suppress SMAD3 activation (49). Thus, we hypothesized that both YAP1 and IGF2BP3, induced in TICs via TLR4/NANOG, suppress TGF-β signaling and promote oncogenesis. To test this notion, we silenced Yap1 and Igf2bp3 individually or simultaneously with shRNA in TICs to determine their effects on TGF-β–induced PAI-1 promoter activity. The TICs showed again a lower TGF-β–induced promoter activity as compared with that of the control cells. Yap1 or Igf2bp3 silencing modestly increased the activity, but silencing both genes clearly upregulated the activity in the TICs but not in the control cells (Figure 6A). To specifically examine the canonical TGF-β pathway, we examined p-SMAD3 levels in the whole-cell and nuclear extracts from TGF-β–treated TICs with or without Yap1 and/or Igf2bp3 silencing. This analysis demonstrates that TGF-β–induced p-SMAD3 levels were increased by silencing, and more conspicuously by silencing both genes (Figure 6B), corroborating the PAI-1 promoter results.
Figure 6. TICs are defective in TGF-β signaling due to SMAD3 pathway interference by YAP1 and IGF2BP3.
(A) TGF-β–stimulated PAI-1 promoter activity is lower in TICs than in control cells. This defect is modestly improved with Yap1 or Igf2bp3 shRNA alone and synergistically corrected by both. *P < 0.05, **P < 0.01. (B) Nuclear levels of p-SMAD3 or SMAD3 in TGF-β–treated TICs are increased by Yap1 or Igf2bp3 silencing with respective shRNA and conspicuously by silencing of both genes. (C) Interactions of SMAD7 with YAP1, p-YAP1, and SMAD3 are increased by Igf2bp3 transduction in PIL4 cells expressing YAP1 (PIL4-YAP1 cells), and these interactions are attenuated with Yap1 shRNA (top left). In the same PIL4-YAP1 cells with Igf2bp3 transduction, increased SMAD7 interactions with p-YAP1 and SMAD3 are abrogated by Akt silencing. Conversely, reduced nuclear p-SMAD3 level is increased. Phosphorylation of MST1 and LATS1/2 is increased by overexpression of IGF2BP3 but reduced by Akt silencing. (D) Fluorescent microscopy depicting increased nuclear staining of SMAD3 in TGF-β–treated TICs by silencing Igf2bp3 and Yap1. Dotted circles indicate the outline of nuclei. Original magnification, ×200. (E) Rapa (200 nM) further increases TGF-β–induced nuclear p-SMAD3 level, which is already augmented by silencing Yap1 or Igf2bp3. (F) Igf2bp3/Yap1 silencing but not rapa augments 3TP-Luc promoter activity induced by CA-SMAD3. *P < 0.05.
Next, we examined whether YAP1 interacts with SMAD7/SMAD3 to suppress TGF-β signaling and Igf2bp3 affects this interaction. For this analysis, we used the PIL-4 cells transduced with Yap1 cDNA (PIL-4-YAP1). Immunoprecipitation with SMAD7 antibody and subsequent immunoblotting with anti-YAP1, anti-p-YAP1 (p-Ser127), or anti-SMAD3 revealed the interactions of SMAD7 with YAP1, p-YAP1, and SMAD3, all of which were abrogated by Yap1 silencing but increased by IGF2BP3 expression (Figure 6C, top left panel). The YAP1/SMAD7/SMAD3 interaction stimulated with IGF2BP3 was also reduced with Yap1 shRNA, suggesting that YAP1 and IGF2BP3 suppress the TGF-β signal via increased YAP1/SMAD7/SMAD3 interaction. We further examined the role of AKT in regulating these interactions, since AKT is known to suppress TGF-β signaling (50). Akt silencing with specific shRNA reduced the SMAD7 interaction with YAP1, p-YAP1, and SMAD3 while increasing nuclear p-SMAD3 levels in PIL4-YAP1 cells, and these effects are more conspicuous in Igf2bp3-transduced cells (Figure 6C, bottom left). As AKT phosphorylates MST1, which in turn phosphorylates Ser127 of YAP1 to promote cell survival (51), p-MST1 levels were also examined by immunoblot analysis. IGF2BP3 expression increased both p-AKT and p-MST1 in a manner dependent on AKT (Figure 6C, right), concomitant with increased interaction of p-YAP1 with SMAD7 (Figure 6C, bottom left), supporting potential involvement of the AKT/MST1/YAP1 pathway. LATS1 and LATS2 are other kinases also shown to phosphorylate YAP1 either directly or downstream of MST1 (52, 53). Indeed, both p-LATS1 and p-LATS2 levels were increased by IGF2BP3 expression and reduced by Akt silencing (Figure 6C, right). To attain direct evidence for association of the YAP1/SMAD7/SMAD3 interaction in the cytosol with reduced nuclear p-SMAD3, immunofluorescence microscopy of TGF-β–treated TICs was performed, demonstrating colocalization of SMAD7, YAP1, and SMAD3 mainly in the cytosol of TICs transduced with scrambled shRNA. However, in TICs with Yap1 and Igf2bp3 silencing, SMAD3 nuclear localization became more evident (Figure 6D), supporting the notion that YAP1/SMAD7/SMAD3 interaction prevents SMAD3 activation and nuclear translocation in our TICs. Conversely, overexpression of both YAP1 and IGF2BP3 inhibited nuclear translocation of SMAD3 in PIL-4 cells (Supplemental Figure 5C).
AKT-activated mTOR inhibits SMAD3 phosphorylation (54, 55), and YAP1 phosphorylation at Ser127 prevents nuclear translocation and mitogenic activity of YAP1 (56). Thus, we further tested whether rapamycin (Rapa), the mTOR inhibitor, or the YAP1 S127A mutant reduces the YAP1/SMAD7 interaction stimulated by the constitutively active myristoylated AKT (AKTMyr). PIL-4 cells transduced with Yap1 and AktMyr show a strong interaction between YAP1 and SMAD7, which is not affected by Rapa, suggesting that mTOR plays no role in the YAP1/SMAD7 interaction (Supplemental Figure 5D). In contrast, when the YAP1-S127A mutant was expressed, it reduced the interaction of YAP1 and SMAD7, even in the presence of AKTMyr, suggesting the importance of Ser127 phosphorylation by AKT in the YAP1/SMAD7 interaction (Supplemental Figure 5D).
We next examined whether mTOR regulates TGF-β–induced SMAD3 phosphoactivation in PIL-4 cells. TGF-β–induced p-SMAD3 levels were reduced by IGF2BP3 overexpression but augmented by Rapa (Supplemental Figure 5E, lane 2 vs. lane 4 or lane 6), and this Rapa-augmented SMAD3 activation was not inhibited by IGF2BP3 expression (Supplemental Figure 5E, last lane), suggesting that mTOR is downstream of IGF2BP3 in suppressing SMAD3 activation. In our TICs, Rapa or Yap1 shRNA augmented TGF-β–induced p-SMAD3 levels (Figure 6E, top, lane 2 vs. lane 4 or lane 6), and both treatments result in a synergistic effect (Figure 6E, last lane), suggesting that YAP1 and mTOR inhibit activation of SMAD3 at different levels. With Igf2bp3 silencing, we also observed an augmenting effect of Rapa on nuclear p-SMAD3 (Figure 6E, bottom). To confirm the role of mTOR, this kinase was silenced with shRNA. This manipulation also augmented TGF-β–induced p-SMAD3 levels in the TICs (Supplemental Figure 5F), confirming the negative regulatory role of mTOR in SMAD3 activation. Next, we tested the effects of Rapa plus silencing of both Yap1 and Igf2bp3. Indeed, Rapa, together with Yap1 and Igf2bp3 silencing, had a synergistic effect on TGF-β–induced p-SMAD3 levels (Supplemental Figure 5G, last lane), suggesting that mTOR has an independent, negative regulation on SMAD3 activation from that achieved by YAP1 and IGF2BP3. Rapa also abrogated suppressed p-SMAD3 levels caused by the constitutively active AKTMyr (Supplemental Figure 5G, lane 5 vs. lane 8).
We then asked whether mTOR, YAP1, and IGF2BP3 affect the TGF-β pathway downstream of SMAD3 activation (nuclear translocation and promoter activation) by testing these negative regulators against constitutively active SMAD3 (CA-SMAD3) in a TGF-β–specific 3TP-luciferase reporter assay in TICs. Rapa failed to augment the promoter activity induced by CA-SMAD3 (Figure 6F, bar 2 vs. bar 3), suggesting that mTOR’s inhibitory effect is primarily at the level of phosphoactivation of SMAD3. In contrast, CA-SMAD3–mediated promoter activity was still augmented by silencing of Yap1 and Igf2bp3 (Figure 6F, bar 2 vs. bar 4), suggesting that these molecules also have a negative effect downstream of SMAD3 activation, most likely by blocking p-SMAD3 nuclear translocation via augmented interactions of p-Yap1, p-SMAD3, and SMAD7, as demonstrated above. Last, we determined the effects of Yap1 and Igf2bp3 silencing on expression of stemness genes (Nanog, CD133, Oct4, and Sox2) and differentiation-related genes (Alb and Krt19). qRT-PCR results demonstrate suppression of all stemness genes and Krt19 by this manipulation, while inducing albumin expression (Supplemental Figure 5H). Further, Yap1 and Igf2bp3 silencing abrogated Nanog promoter activity in TICs to the level similar to the activity in the control cells (Supplemental Figure 5I). Collectively, these results demonstrate the mechanisms and functional importance of suppressed TGF-β canonical pathway by NANOG-induced YAP1 and IGF2BP3 in cell fate regulation of TICs.
Suppressed TGF-β signaling activates the TLR4/NANOG oncogenic pathway.
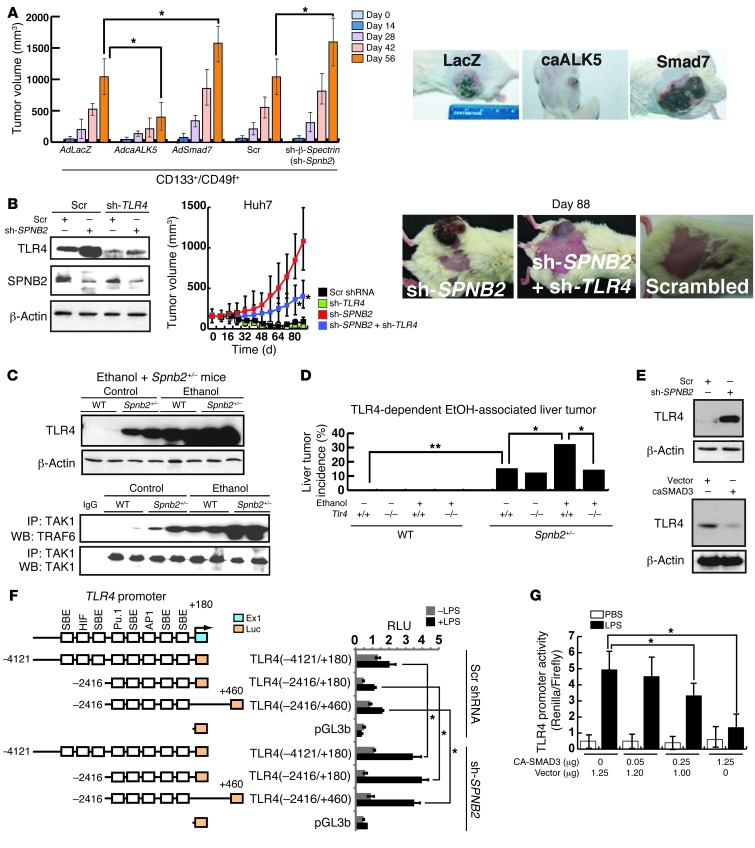
Our results point to the critical role of an inhibited TGF-β tumor suppressor pathway for activation of oncogenic TLR4 signaling and TIC generation. We next asked a question as to whether direct genetic manipulations of the TGF-β pathway in TICs influence their tumorigenic activity. A gain-of-function approach for TGF-β signaling was achieved by adenoviral transduction of the constitutively active TβRI (57) in TICs, and we tested its effects on subcutaneous tumor growth in NOG mice. This manipulation indeed reduces tumor growth as compared with the cells infected with the virus expressing LacZ as a control (Figure 7A). Conversely, overexpression of the inhibitory SMAD7 or expression of shRNA against the chaperone protein β2-spectrin (Spnb2), which recruits p-SMAD3/SMAD4 into the nucleus, promoted tumor growth (Figure 7A). These results confirm the importance of an inhibited TGF-β tumor suppressor pathway in the oncogenic activity of the TICs.
Figure 7. Inhibition of the TGF-β pathway promotes TLR4-mediated oncogenesis.
(A) Subcutaneous tumor growth by TICs from Ns5a Tg mouse tumors in NOG mice is attenuated by a gain-of-function approach for TGF-β signaling using adenoviral transduction of the constitutively active TβRI (caALK5), while it is promoted by overexpression of Smad7 or shRNA against Spnb2. (B) SPNB2 knockdown induces TLR4 and tumor-initiating property of Huh7 cells, which do not cause spontaneous xenograft growth. This growth by SPNB2 knockdown is largely prevented by knockdown of TLR4 with shRNA. Representative pictures of NOG mice bearing tumors at day 88 are shown. (C) Heterozygosity of Spnb2 induces TLR4 expression and downstream signaling (TAK1/TRAF6 association), and these changes are accentuated by alcohol feeding for 12 months. (D) The spontaneous liver tumor incidence in the Spnb2+/– mice, but not in Spnb2+/–Tlr4 mice, is increased by alcohol feeding. (E) Knockdown of SPNB2 with shRNA induces TLR4, while overexpression of caSMAD reduces TLR4 in Huh7 cells. (F) Knockdown of SPNB2 in Huh7 cells equally promotes LPS-induced activity of the TLR4 promoter containing 3 proximal SMAD-responsive elements (SRE). (G) Expression of CA-SMAD3 inhibits LPS-mediated TLR4 promoter (–4121/+180 nt) activity in Huh7 cells. *P < 0.05, **P < 0.01.
To attain the relevance of this finding in humans, we knocked down SPNB2 to suppress the TGF-β pathway in the human HCC cell line Huh7 and determined its effect on tumor growth in NOD mice. Interestingly, SPNB2 knockdown itself induces TLR4 in Huh7 cells (Figure 7B). Although Huh7 cells transduced with scrambled shRNA did not grow, SPNB2 knockdown allowed the cells to form rapidly growing tumors composed of cells with increased TLR4 immunoreactivity (Supplemental Figure 6). This growth was attenuated with shRNA against TLR4 (Figure 7B).
To further test this notion in a whole animal model, we studied Spnb2+/– heterozygous mice. Spnb2+/– mice with reduced TGF-β signaling spontaneously develop HCC (15). As shown in Huh7 cells with SPNB2 knockdown, TLR4 was induced in the liver of this genetic mouse model as compared with that in WT mice (Figure 7C). To activate TLR4 signaling, Spnb2+/– mice were fed alcohol for 12 months. TLR4 activation, as assessed by TAK1/TRAF6 interaction, was evident in the liver of alcohol-fed Spnb2+/– mice (Figure 7C). More importantly, this treatment increased liver tumor incidence in Spnb2+/– mice but not in Spnb2+/–Tlr4–/– compound mice (Figure 7D), indicating that activation of the TLR4 pathway by alcohol feeding promotes tumorigenesis in Spnb2+/– mice with reduced TGF-β signaling and reciprocally upregulated TLR4.
We then asked how TLR4 is negatively regulated by TGF-β. Interaction between SMAD3 and AP1 is known to inhibit SMAD3 DNA binding ability (58). As there are several SMAD-binding elements in the TLR4 promoter, we tested whether SMAD3 inhibits LPS-mediated TLR4 promoter activity in Huh7 cells (Figure 7F). Silencing of SPNB2 in Huh7 cells (Figure 7E) significantly enhanced LPS-induced TLR4 promoter activation (Figure 7F). In contrast, CA-SMAD3 overexpression markedly reduced TLR4 protein expression (Figure 7E) and promoter activity (Figure 7G). These results demonstrate that TGF-β signaling directly inhibits the TLR4 promoter and that defective TGF-β signaling promotes LPS-induced TLR4 transcription. These results, together with our YAP1/IGF2BP3 data, support the reciprocal antagonistic regulation of TLR4 and TGF-β and its implication in liver oncogenesis.
Human HCCs have accentuated expression of TLR4/NANOG/YAP1/IGF2BP3 pathway components.
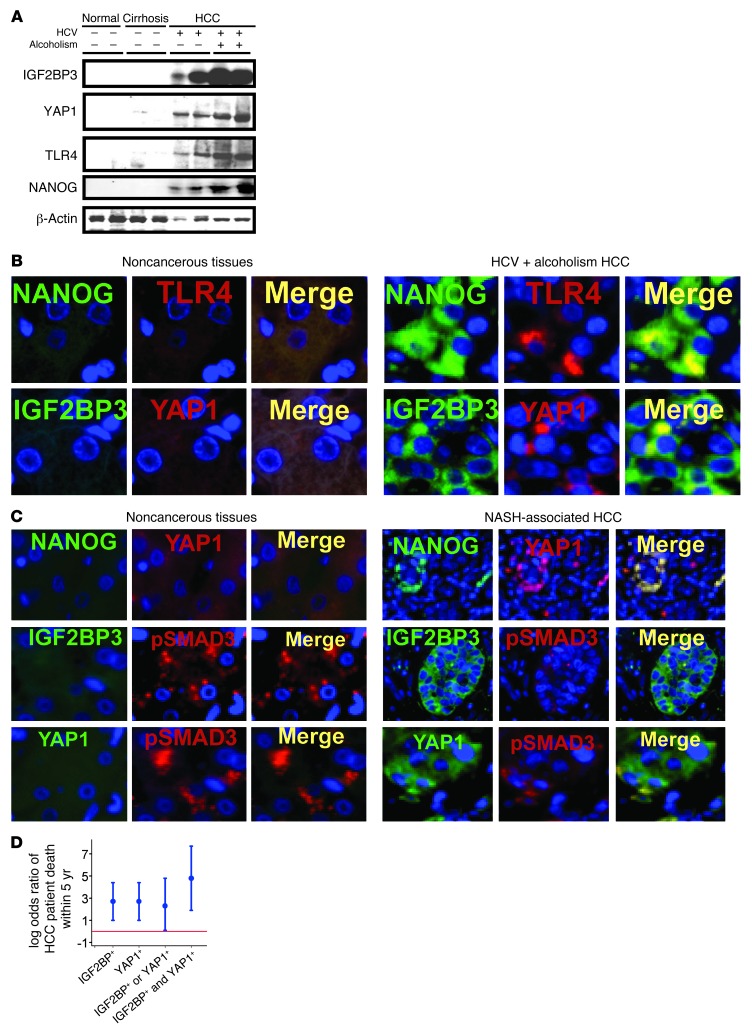
To attain clinical relevance of our findings, human HCC specimens (clinicopathological features described in Supplemental Tables 3 and 4) were analyzed. Immunoblotting shows dramatically higher levels of YAP1, IGF2BP3, TLR4, and NANOG in HCC from patients with HCV with or without alcoholism as compared with nontumorous cirrhosis or normal liver tissues (Figure 8A). Furthermore, HCC sections show increased TLR4, NANOG, YAP1, and IGF2BP3 staining compared with that in nontumorous liver tissues (Figure 8B and Supplemental Figure 7, A and B). To broaden the relevance, we also examined HCC in patients with nonalcoholic steatohepatitis without HCV infection. Similarly, NANOG, YAP1, and IGF2BP3 staining was increased while p-SMAD3 staining was reduced in HCC of this etiology, as compared with noncancerous liver tissues (Figure 8C and Supplemental Figure 7C). Of 34 patients with HCC, 18 died of the disease within 5 years of the date of diagnosis, and the remaining 16 survived over 5 years. In this patient cohort, the odds of dying within 5 years was significantly higher among patients with positive IGF2BP3 immunoreactivity as compared with patients with negative IGF2BP3 immunoreactivity (odds ratio [OR] = 15.2, 95% CI = 2.8–81; P = 0.001) (Figure 8D). Similarly, the odds of 5-year mortality were significantly higher among patients with positive YAP1 immunoreactivity as compared with patients with no immunoreactivity (OR = 15, 95% CI = 2.8–80; P = 0.002) (Figure 8D). Patients with both IGF2BP3 and YAP1 positivity in liver tumors have conspicuously elevated odds to die within 5 years as compared with patients for whom only 1 out of the 2 factors were positive (Figure 8D, P < 0.001). These results indicate that the TLR4/NANOG/YAP1/IGF2BP3 pathway is activated in human HCC and is associated with poor patient outcome.
Figure 8. Induction of TLR4/NANOG/YAP1/IGF2BP3 pathway components in human HCC.
(A) TLR4, NANOG, IGF2BP3, and YAP1 protein levels are increased in HCC specimens from patients with HCV infection without or with alcoholism, as compared with cirrhotic or healthy livers. (B) Immunofluorescent microscopy demonstrates higher expression of NANOG, TLR4, YAP1, and IGF2BP3, which are often colocalized in patient HCC specimens, as compared with normal liver. Original magnification, ×200. (C) Immunofluorescent microscopy demonstrates increased expression of NANOG, YAP1, and IGF2BP3, which are often colocalized in HCC specimens from patients with nonalcoholic steatohepatitis (NASH), as compared with normal liver. p-SMAD3 staining is conversely reduced in HCC. Original magnification, ×200. (D) The log OR (and 95% CI) of patient death within 5 years from initial diagnosis for HCC patients is increased for those with positive IGF2BP3 or/and YAP1 immunoreactivity, as compared with those with negative IGF2BP3 and YAP1 immunoreactivity (red line).
Silencing of Igf2bp3 and Yap1 sensitizes TICs to drug-induced cell death.
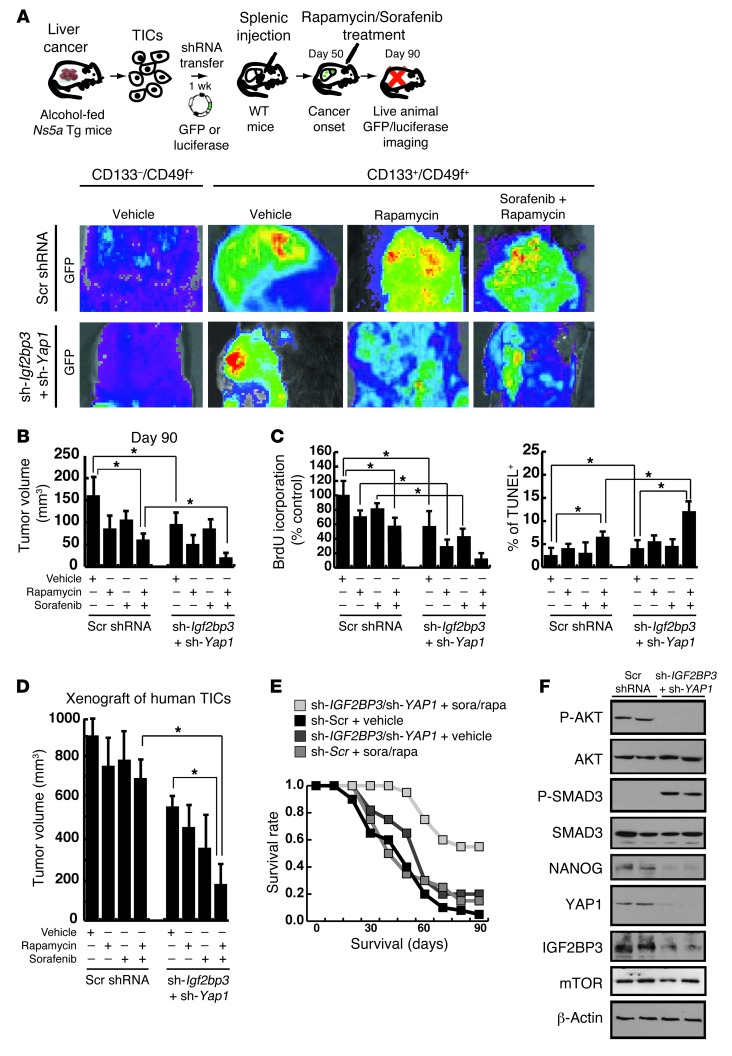
Next, we tested biological significance and therapeutic relevance of Yap1 and Igf2bp3 by determining the effects of silencing these genes on the efficacy of 2 common chemotherapeutic agents, Rapa and sorafenib (inhibitor of pan-tyrosine kinases targeting the MAPK pathway) (59), after assessing their IC50 in cultured TICs (Supplemental Figure 8, A and B). In vitro, silencing of Yap1 and Igf2bp3 made TICs susceptible to suppressed proliferation and increased apoptosis by the drugs (Supplemental Figure 8C). Then, we tested the effects of gene silencing in mice transplanted with TICs infected with lentivirus-GFP via splenic injection, as shown in Figure 9A. TIC liver engraftment was facilitated by retrorsine pretreatment and CCl4 injections following transplantation. Whole-body imaging, tumor volume, and BrdU incorporation measurements were performed at the terminal time point of day 90 after transplantation (Figure 9, B and C), demonstrating that Yap1 and Igf2bp3 silencing promotes retardation of tumor growth by the chemotherapy to the extent that the tumor size was reduced approximately 90% (representative gross photos shown in Supplemental Figure 8D). TUNEL staining of tumor tissues revealed increased incidence of apoptotic cell death by the combination therapy (Figure 9C).
Figure 9. Silencing of Igf2bp3 and Yap1 sensitizes TICs to drug-induced cell death.
(A) Silencing of Igf2bp3 and Yap1 in TICs from HCV Ns5a Tg mice promotes growth inhibition of tumor upon chemotherapeutic drug treatment (rapa and sorafenib) in immune-competent C57BL/6 mice, as monitored by GFP imaging after splenic injection and engraftment enrichment procedures with retrorsine and CCl4. (B) The tumor volume measurement performed at day 90 after transplantation shows a maximal growth retardation achieved by the combination of the drugs and Igf2bp3/Yap1 silencing (*P < 0.05). (C) TIC proliferation, as determined by BrdU incorporation in vivo at day 90, is most inhibited by the combination of the chemotherapeutic drugs and silencing of Igf2bp3 and Yap1. Concomitantly, the incidence of apoptotic cell death increases conspicuously by the combined treatment. *P < 0.05. (D) Xenograft tumor growth by TICs from patient HCC in NOG mice is most conspicuously suppressed by rapa and sorafanib treatment only in the presence of IGF2BP3 and/or YAP1 silencing (*P < 0.05). (E) Improved survival of mice injected with human TICs transduced with shRNA targeting YAP1 and IGF2BP3 and subjected to injection of the chemotherapeutic drugs, as compared with other groups with single therapies. (F) Immunoblot analysis demonstrates that silencing of both YAP1 and IGF2BP3 induces p-SMAD3 levels, while reducing NANOG expression in xenograft tissue of mice treated with rapa and sorafenib.
Finally, the chemosensitizing effect of IGF2BP3 and YAP1 silencing was tested by xenograft transplantation with the human HCC-derived TICs. These human TICs were very resistant to the growth inhibitory effect of Rapa and/or sorafenib (Figure 9D). However, silencing of IGF2BP3 and YAP1 abrogated this resistance and retarded the tumor growth by 70%, as compared with the drug treatment alone, and by 85%, as compared with no treatment (Figure 9D). This profound effect on drug sensitivity correlates with an improved survival of the mice with the combination therapy as opposed to chemotherapy or gene silencing alone (Figure 9E). The levels of p-SMAD3, p-AKT, and NANOG were also examined in the tumor tissues collected from the mice treated with only the drugs or with combination therapy. These analyses demonstrate that the combination therapy induced SMAD3 phosphorylation and reduced NANOG, IGF2BP3, p-AKT, and YAP-1 expression (Figure 9F).
Discussion
CD133+ TICs have previously been isolated from liver tumors of Pten- or Mat1a-deficient mice (60, 61). Using the same surface marker, we isolated TLR4/NANOG–dependent TICs from two mouse models of HCV-related liver tumors and patients with HCC. CD133+ TICs from Mat1a-deficient mice show heightened MAPK activation in response to TGF-β treatment, which promotes cell survival and antiapoptotic effects of TGF-β (60). This prosurvival effect may be caused by a shift in TGF-β signaling from the SMAD-mediated to the MAPK-dominant pathway. Indeed, our study demonstrates defects in the SMAD pathway in CD133+ TICs, which may underlie this shift of the signaling. Further, using the TIC-derived cDNA library, we identify Yap1 and Igf2bp3 as NANOG target genes, which exert their oncogenic activities via suppression of the SMAD3 pathway (Figure 10). IGF2BP3 and YAP1 render this effect at two different levels, SMAD3 phosphoactivation and p-SMAD3 nuclear translocation. The former regulation is dependent on mTOR activated by the IGF2BP3/AKT pathway, while the latter is achieved by the interaction of YAP1 with SMAD7 and SMAD3, which is enhanced by IGF2BP3 via Ser127 phosphorylation of YAP1 (Figure 10). More importantly, defective TGF-β signaling caused by YAP1 and IGF2BP3 promotes stemness, oncogenic activity, and chemoresistance of TICs, as silencing of both Yap1 and Igf2bp3 restores TGF-β signaling, reduces stemness gene expression, and abrogates resistance of both animal and human TICs to chemotherapy. This outcome has an important implication in new therapeutic developments targeting TICs and may potentially lead to improved patient survival and reduced medical costs, if these experimental modalities are translated successfully.
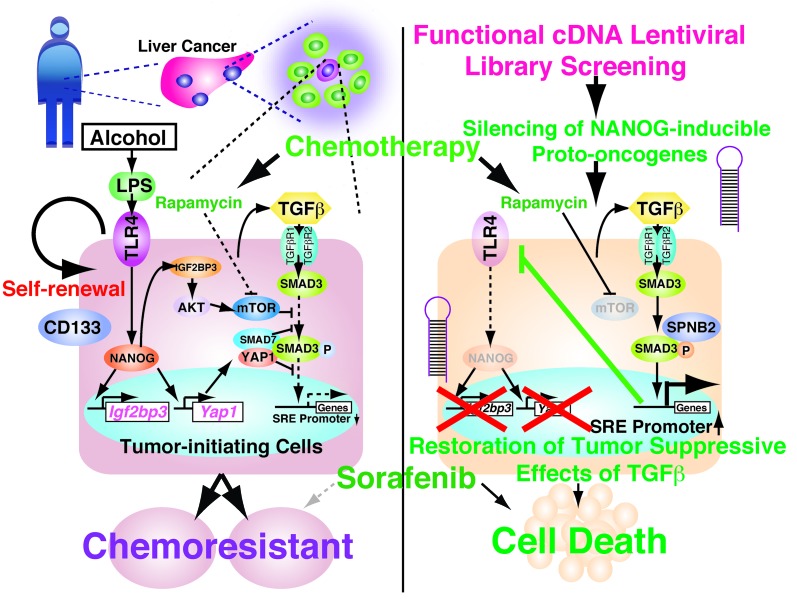
Figure 10. A schematic representation of the proposed link between oncogenic TLR4/NANOG signaling and a defective TGF-β tumor suppressor pathway in generating TICs.
Ectopic upregulation of TLR4 and its activation by LPS induce the pluripotency factor NANOG, other stem cell genes, and self-renewal of TICs. NANOG induces IGF2BP3 and YAP1, which in turn inhibit TGF-β signaling at the level of SMAD3 phosphoactivation and p-SMAD3 nuclear translocation. The former effect is dependent on the IGF2BP3/AKT/mTOR pathway, while the latter is caused by p-YAP1/SMAD7/SMAD3 interactions, which are enhanced by IGF2BP3/AKT–mediated YAP1 phosphorylation. Restoration of the TGF-β tumor suppressor pathway by silencing IGF2BP3 and YAP1 downregulates TLR4 via SMAD3-mediated transcriptional repression and inhibits TLR4/NANOG–mediated TIC self-renewal and oncogenic activity while chemosensitizing TICs. A defective canonical TGF-β pathway, as in Spnb+/– mice, upregulates TLR4 by transcriptional derepression and activates the TLR4/NANOG oncogenic pathway for liver tumorigenesis.
Our finding on the cytosolic activity of p-Ser127 YAP1 inhibiting the TGF-β tumor suppressor pathway contrasts with oncogenic nuclear activity of YAP1, which is inhibited by MST1/2-mediated phosphorylation of the protein (53). This study did not examine the nuclear role of YAP1 in TICs. However, it is possible that YAP1 may serve as an oncogene in both cytosol and nucleus, and p-Ser127 YAP1 still exerts an indirect oncogenic effect via suppression of SMAD3 nuclear translocation, as also previously suggested (48). Our results on coordinated increases in p-MST1 and p-LATS1/2 with p-AKT and p-YAP1 (Figure 6C) further suggest that MST1 and LATS1/2 are kinases potentially responsible for p-Ser127 of YAP1 downstream of p-AKT (62, 63).
While activated TLR4 inhibits the TGF-β pathway, a suppressed TGF-β pathway induces TLR4 in the liver. The importance of this reciprocal regulation in liver tumorigenesis is supported by our findings in mice with SPNB2 haploinsufficiency. These mice with defective canonical TGF-β signaling ectopically upregulated TLR4 in the liver and developed liver tumors at a higher incidence when fed alcohol in a manner dependent on TLR4 (Figure 7, C and D). We also demonstrated that TLR4 upregulation by a suppressed TGF-β pathway involves at least in part transcriptional derepression by reduced SMAD3-mediated repression (Figure 7, E–G).
Multiple cancers derived from meso-endodermally derived epithelium are associated with TGF-β/BMP pathway inactivation, in which it may regulate progenitor cell fate. Indeed, the functions of TGF-β are more complex than simply inhibiting cell growth, as TGF-β can induce the growth of mesenchymal cells involved in cell invasion (13, 64). TGF-β signals also modulate the immune response to tumors and are thought to play a role in tumor angiogenesis (65). TGF-β is also commonly overproduced by cancer cells. Besides breast cancer, hepatocellular and lung carcinomas overproduce TGF-β1 in vivo, and the higher the level of this cytokine, the higher their chance for metastasis (66, 67). Yet, in vivo models have not yielded robust protumorigenic roles for the cytokine, and it is possibly that its levels reflect the loss of the linear SMAD signaling pathway and a negative feedback increase in the cytokine. In TICs of epithelial tissues, such as pancreas and liver (23, 60, 68, 69), a defective TGF-β pathway may promote tumorigenesis via stemness gene induction, while in nonepithelial tumor cells, such as glioma cells, an active TGF-β pathway may promote the oncogenic potential.
NANOG is per se a well-established negative transcriptional regulator of the tumor suppressor p53 (70), and this mechanism may certainly contribute to the genesis of TLR4-dependent TICs. Intriguingly, a recent high-throughput screening identified an inhibitor of TGF-β signaling, which replaces a role of SOX2 and induces NANOG to complete reprogramming for generation of iPS cells (71). Indeed, our findings of TLR4-mediated oncogenic activity in the Spnb2+/– mice and SPNB2-knocked down Huh7 cells support the notion that inhibition of the TGF-β pathway is required for stemness gene induction in the genesis of TICs. We also noticed that knockdown of Nanog leads to attenuation of Tlr4 expression in TICs (data not shown), implying a feedback loop. Currently, the mechanism underling this loop is unknown but may be critical in fully understanding the TLR4/NANOG pathway in regulation of pluripotency. Similarly, YAP1 and IGF2BP3 may have a positive feedback regulation on NANOG, as indicated by reduced NANOG expression upon silencing of these 2 factors (Supplemental Figure 5H). This regulation may be due to restored TGF-β signaling and reciprocally suppressed TLR4 activation, which is upstream of NANOG.
Our study extends the TLR4/NANOG oncogenic mechanism identified in the TICs from the animal models to HCC in patients via (a) demonstration of a similar population of TICs from patient HCC, (b) immunohistochemical evidence of increased TLR4 and NANOG as well as their downstream targets YAP1 and IGF2BP3 in tumor sections from patients with HCC, and (c) correlation of YAP1 and IGF2BP3 immunoreactivity with outcome of patients with HCC. Further, immunohistochemical results on nonalcoholic steatohepatitis–associated HCC suggest that the TLR4/NANOG pathway may have a broad implication in HCC, regardless of etiology. A recent study demonstrates that TLR4 is expressed in embryonic stem cells and adult progenitor cells from mammary and intestine and its activation leads to proliferation and stem cell expansion (72). Based on these data, combined with our findings, we propose that TLR4 is a proto-oncogene or a tumor-promoter gene whose aberrant expression and activation leads to induction of pluripotency genes and genesis of TICs via activation of the TLR4/NANOG pathway and consequent inhibition of the TGF-β/SMAD pathway.
Methods
More information is available in the Supplemental Methods.
Isolation of TICs.
CD133+/CD49f+ TICs were separated from CD133–/CD49f+ or CD133–/CD49f– cells by FACS from a CD45– fraction of tumor tissue cell suspension (61, 73) from the animal models and patients, as described in detail in Supplemental Methods.
Mice.
HCV Ns5a Tg mice were provided by Ratna Ray (Saint Louis University, Saint Louis, Missouri, USA).
Functional cDNA screening for oncogenes.
A lentiviral cDNA library, which expresses GFP driven by EMCV IRES, was established from NANOGhi/TICs and NANOGlo/CD133/CD49f+ control cells by the lentiviral RL recombination method based on the Gateway system (Invitrogen) using HEK293T cells. The mouse oval cell line (progenitor cell), deficient in p53 (immortalized, nontransformed PIL-4, 1 × 106 cells) (74), was infected with each lentiviral cDNA library (approximately 10 million copies = MOI of 10). This viral cDNA library technique has previously been used to clone and test cancer driver genes in vitro (75–77) and in vivo (37, 78). Following 7 days after infection, GFP-positive PIL-4 cells were sorted by FACS to eliminate nontransduced cells. The GFP-positive cells were then seeded into soft agar plates for colony formation assay as described previously (76, 79). In brief, a base layer was made by mixing 1% soft agar with an equivalent 2× medium in 6-well plates. The transduced PIL-4 cells were harvested, suspended in medium containing 0.35% soft agar, and seeded on the base layer at different densities, starting from 5,000 cells per well, using a 24-well plate. Plates were maintained at 37°C and fed every 3 days with 0.1 ml complete Williams E medium (30 ng/ml IGF2, 20 ng/ml EGF, 10 μg/ml insulin, and 10% fetal bovine serum). After 2 weeks, the number of colonies that formed was counted, and the colonies were isolated by ring cloning and propagated in the same media for subsequent assays under a microscope as previously described (80, 81). As a control, a lentiviral vector expressing scrambled shRNA was used. Oncogenic parameters (colony size, proliferative and invasive activity) were assessed using 24-well plates to determine the priority of the candidate genes to be tested by this assay, and we knocked down the highest priority genes on the list as described previously (75–77). Cell cultures infected with these libraries developed many distinctly transformed colonies, with a variety of morphology and growth rates, while uninfected cell cultures or those infected with viruses derived from the CD133–/CD49f+ control cells had lower frequencies of occurrence of transformed colonies, equivalent to the low spontaneous rate of transformation of the PIL-4 cells used in these experiments. The cell proliferation rate was assessed by BrdU staining in 96-well plates.
In vitro and in vivo oncogenic activities.
The TICs and control CD133–/CD49f– cells were tested for colony formation using 24-well plates with soft agar or spheroid formation in ultra-low adhesion plates. Cell proliferation rate was assessed by incorporation of 3H-uridine in 96-well plates. Invasive activity was determined by a Biocoat Matrigel Invasion Chamber (BD Biosciences Labware). Vectors (pCR2.1-NANOGP8 and pPyCAG-NANOGP8) were provided by Dean G. Tang (The University of Texas MD Anderson Cancer Center). For xenograft transplantation, the cells were infected with lentiviral vectors expressing GFP and shRNA targeting Yap1 or Igf2bp3 or scrambled shRNA. Infected cells (1 × 104) were injected with Matrigel into nude, NOG, or immune-competent mice. The tumor tissues were divided for snap freezing for mRNA and protein analysis for TLR4 and TGF-β signaling components and oncogenic factors, fixation with 3% paraformaldehyde for subsequent immunostaining of the TIC markers, or H&E staining and histological evaluation of neoplastic cells.
Liver TIC engraftment via splenic injection.
To test the effects of the chemotherapeutic drugs and Igf2bp3/Yap1 silencing on liver tumor growth derived from mouse TICs, TICs or CD133–/CD49f+ control cells were prelabeled with lentiviral GFP and transduced with shRNA for Igf2bp3 and Yap1, followed by splenic injection of C57BL/6 mice as previously described (37). The lentiviral transduction efficiency is monitored by FACS analysis for GFP positivity. More than 95% of TICs were routinely labeled by lentiviral transduction. The recipient mice were pretreated before surgery with 2 doses of retrorsine (70 mg/kg, i.p.), an alkaloid that exerts a strong and persistent block of native hepatocyte proliferation, thereby increasing the competitive advantage of transplanted cells. For intrasplenic injection of cells, anesthesia was induced with an i.p. mixture of ketamine and xylazine (Abbott Laboratories) at a strength of 100 mg/ml, given as a dose of 0.1 ml per 20 g of body weight. Under direct vision, cells were injected into the spleen through a mini incision on left flank, using a 27-gauge needle. The spleen was retracted, and the lower pole of the spleen was looped with a 4-0 vicryl ligation. The indicated number of cells in 0.2 ml media was injected into the lower pole of the spleen within 2 minutes. The abdomen was sutured using 4-0 vicryl. Transplanted cells were allowed to migrate to the recipient liver and engraft the organ. To further facilitate expansion of the transplanted cells, recipient mice were treated with CCl4 (dose: 0.5 ml/kg i.p., 1-week interval, 3 cycles during 30 days at day 14, 21, and 28 after splenic injection), which induces liver injury and regeneration. Acute liver damage was induced by subcutaneous injection of CCl4 dissolved in olive oil 2 days before transplantation. To determine whether the cells colonize recipient livers, we monitored the growth of liver tumors via GFP imaging at the Animal Imaging Core of University of Southern California Radiology Department. Approximately one percent of the host liver consisted of “seeded” GFP-positive cells embedded within the normal liver architecture. Images were taken in at least 2 (typically perpendicular) levels and underwent processing for contrast and brightness, with subsequent analysis by the Image Pro Plus 3.1 software (Media Cybernetics). Images were captured directly on a microcomputer or continuously through video output on a high-resolution VCR (SLV-R1000, Sony Corp.). Imaging at lower magnification that visualized the entire animal was carried out in a light box illuminated by blue light fiber optics (Lightools Research Inc.) and performed using the thermoelectrically cooled color CCD camera.
Statistics.
The data in bar and line graphs are shown as mean + SD. P values of less than 0.05 are considered significant. Linear mixed effect models were used to examine whether there was a difference in tumor growth speed among different groups of mice, e.g., mice grafted with TICs transduced with shRNA targeting a cellular gene vs. scrambled shRNA. Difference in mean tumor size across different groups of mice at a certain time point was assessed with ANOVA. Logistic regression was used to compare the odds of having positive IGF2BP3 or YAP1 immunoreactivity among patients who died within 5 years vs. the odds of having positive IGF2BP3 or YAP1 immunoreactivity among those who survived over 5 years. YAP1 and IGF2BP3 immunoreactivity was considered as negative when the percentage of stained cells with immunofluorescence microscopy was 0 and positive if the percentage was greater than 0. These statistical computations were performed using Stata 11.0 (StataCorp, Stata Statistical Software: Release 11. 2009, StataCorp LP). All P values reported are 2 sided.
Study approval.
The present studies in animals and humans were reviewed and approved by Institutional Animal Care and Use Committee and Institutional Review Board of University of Southern California.
Microarray data.
Microarray data have been deposited in a MIAME-compliant public database (GEO accession no. GSE45646, GSM1111249, and GSM1111250).
Supplementary Material
Acknowledgments
We thank Akiko Ueno and Raul Lazaro, the animal core personnel for performing mouse experiments; Rajeshwar Nityanandan, Dinesh Babu Uthaya Kumar, and Naomi Anderson (University of Southern California) for technical support and critical reading; Ratna Ray (Saint Louis University) for providing HCV Ns5a Tg mice; Dean G. Tang (The University of Texas MD Anderson Cancer Center) for providing vectors (pCR2.1-NANOGP8 and pPyCAG-NANOGP8); Michael Karin (UCSD) for suggestions and reagents; and Steve Weinman (University of Kansas) for critiques and discussions. This project was supported by NIH grants 1R01AA018857, 5RC2AA019392, P50AA11999 (Animal Core, Morphology Core, and Pilot Project Program), 5P30DK048522-13 (pilot project funding), R24AA012885 (Non-Parenchymal Liver Cell Core), AI83025U19, CA123328, CA108302, R01 CA042857 (to L. Mishra), P01 CA130821 (to L. Mishra), P30 CA016672 (to R. DePinho), and P30 DK56338 (to M. Estes) as well as U19AI83025, Zumberge Foundation. This research is also supported by a Research Scholar Grant (RSG MPC122545 (to K. Machida) and pilot funding (IRG-58-007-48) from American Cancer Society and the SFB-TRR77 “Liver Cancer” from the German Research Foundation (to S. Dooley). Microscopy was performed by the Cell and Tissue Imaging Core of the University of Southern California Research Center for Liver Diseases (P30 DK048522). Statistical analysis was performed by Susan Groshen and Lingyun Ji in Norris Comprehensive Cancer Center Biostatistics Core (supported by NIH/NCI P30 CA 014089). Animal imaging was performed by the University of Southern California Molecular Imaging Center (supported by NIH/NVRR S10). Tissue pathological slide preparation was performed by Moli Chen in Translational Pathology Core of Norris Comprehensive Cancer Center. Liver tissues were obtained from The Liver Tissue Cell Distribution System at University of Minnesota.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2013;123(7):2832–2849. doi:10.1172/JCI65859.
References
- 1.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer AK, et al. Toll-like receptor 4 in butylated hydroxytoluene-induced mouse pulmonary inflammation and tumorigenesis. J Natl Cancer Inst. 2005;97(23):1778–1781. doi: 10.1093/jnci/dji403. [DOI] [PubMed] [Google Scholar]
- 6.Fukata M, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133(6):1869–1881. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal D, Saccheri F, Venereau E, Pusterla T, Bianchi ME, Rescigno M. TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J. 2010;29(13):2242–2252. doi: 10.1038/emboj.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukata M, et al. Constitutive activation of epithelial TLR4 augments inflammatory responses to mucosal injury and drives colitis-associated tumorigenesis. Inflamm Bowel Dis. 2011;17(7):1464–1473. doi: 10.1002/ibd.21527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machida K, et al. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A. 2009;106(5):1548–1553. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan JM, Govindarajan S, Arakawa K, Yu MC. Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the US. Cancer. 2004;101(5):1009–1017. doi: 10.1002/cncr.20427. [DOI] [PubMed] [Google Scholar]
- 11.Hassan MM, et al. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36(5):1206–1213. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- 12.Mishra L, et al. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49(1):318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez S, et al. Hepatitis C virus compartmentalization and infection recurrence after liver transplantation. Am J Transplant. 2009;9(7):1591–1601. doi: 10.1111/j.1600-6143.2009.02666.x. [DOI] [PubMed] [Google Scholar]
- 14.Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24(37):5775–5789. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299(5606):574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, et al. Expression of transforming growth factor beta type II receptor leads to reduced malignancy in human breast cancer MCF-7 cells. J Biol Chem. 1994;269(42):26449–26455. [PubMed] [Google Scholar]
- 17.Wang J, et al. Reduced expression of transforming growth factor beta type I receptor contributes to the malignancy of human colon carcinoma cells. J Biol Chem. 1996;271(29):17366–17371. doi: 10.1074/jbc.271.29.17366. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, et al. Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J Biol Chem. 1995;270(37):22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- 19.Markowitz S, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 20.Papageorgis P, et al. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borczuk AC, Kim HK, Yegen HA, Friedman RA, Powell CA. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med. 2005;172(6):729–737. doi: 10.1164/rccm.200504-615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gobbi H, et al. Loss of expression of transforming growth factor beta type II receptor correlates with high tumour grade in human breast in-situ and invasive carcinomas. Histopathology. 2000;36(2):168–177. doi: 10.1046/j.1365-2559.2000.00841.x. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105(7):2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28(7):961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Zhang Y, Yu Y, Yang X, Cao X. SOCS3 promotes TLR4 response in macrophages by feedback inhibiting TGF-beta1/Smad3 signaling. Mol Immunol. 2008;45(5):1405–1413. doi: 10.1016/j.molimm.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 26.McCartney-Francis N, Jin W, Wahl SM. Aberrant Toll receptor expression and endotoxin hypersensitivity in mice lacking a functional TGF-beta 1 signaling pathway. J Immunol. 2004;172(6):3814–3821. doi: 10.4049/jimmunol.172.6.3814. [DOI] [PubMed] [Google Scholar]
- 27.Hahm KB, et al. Loss of transforming growth factor beta signalling in the intestine contributes to tissue injury in inflammatory bowel disease. Gut. 2001;49(2):190–198. doi: 10.1136/gut.49.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18(5):1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snippert HJ, et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136(7):2187–2194.e1. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White LK, Wright WE, Shay JW. Telomerase inhibitors. Trends Biotechnol. 2001;19(3):114–120. doi: 10.1016/S0167-7799(00)01541-9. [DOI] [PubMed] [Google Scholar]
- 32.Sotillo R, et al. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 2001;20(23):6637–6647. doi: 10.1093/emboj/20.23.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habets GG, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77(4):537–549. doi: 10.1016/0092-8674(94)90216-X. [DOI] [PubMed] [Google Scholar]
- 34.Jeter CR, et al. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells. 2009;27(5):993–1005. doi: 10.1002/stem.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9(1):50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125(7):1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281(47):36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 39.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 40.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17(23):2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 41.Liao B, Hu Y, Herrick DJ, Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem. 2005;280(18):18517–18524. doi: 10.1074/jbc.M500270200. [DOI] [PubMed] [Google Scholar]
- 42.Vikesaa J, et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 2006;25(7):1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeng YM, et al. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008;48(4):1118–1127. doi: 10.1002/hep.22459. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19(2):1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park YN, Chae KJ, Oh BK, Choi J, Choi KS, Park C. Expression of Smad7 in hepatocellular carcinoma and dysplastic nodules: resistance mechanism to transforming growth factor-beta. Hepatogastroenterology. 2004;51(56):396–400. [PubMed] [Google Scholar]
- 46.Kitisin K, et al. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007;26(50):7103–7110. doi: 10.1038/sj.onc.1210513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang B, et al. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4(7):802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 48.Ferrigno O, et al. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21(32):4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 49.Song K, Wang H, Krebs TL, Danielpour D. Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. EMBO J. 2006;25(1):58–69. doi: 10.1038/sj.emboj.7600917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen RH, Su YH, Chuang RL, Chang TY. Suppression of transforming growth factor-beta-induced apoptosis through a phosphatidylinositol 3-kinase/Akt-dependent pathway. Oncogene. 1998;17(15):1959–1968. doi: 10.1038/sj.onc.1202111. [DOI] [PubMed] [Google Scholar]
- 51.Jang SW, Yang SJ, Srinivasan S, Ye K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J Biol Chem. 2007;282(42):30836–30844. doi: 10.1074/jbc.M704542200. [DOI] [PubMed] [Google Scholar]
- 52.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J Biol Chem. 2008;283(41):27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 53.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16(5):425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song K, Cornelius SC, Reiss M, Danielpour D. Insulin-like growth factor-I inhibits transcriptional responses of transforming growth factor-beta by phosphatidylinositol 3-kinase/Akt-dependent suppression of the activation of Smad3 but not Smad2. J Biol Chem. 2003;278(40):38342–38351. doi: 10.1074/jbc.M304583200. [DOI] [PubMed] [Google Scholar]
- 55.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 56.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130(6):1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dooley S, et al. Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 2001;502(1–2):4–10. doi: 10.1016/S0014-5793(01)02656-4. [DOI] [PubMed] [Google Scholar]
- 58.Dennler S, Prunier C, Ferrand N, Gauthier JM, Atfi A. c-Jun inhibits transforming growth factor beta-mediated transcription by repressing Smad3 transcriptional activity. J Biol Chem. 2000;275(37):28858–28865. doi: 10.1074/jbc.M910358199. [DOI] [PubMed] [Google Scholar]
- 59.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48(4):1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding W, et al. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49(4):1277–1286. doi: 10.1002/hep.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rountree CB, Ding W, He L, Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cells. 2009;27(2):290–299. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11(1):11–23. doi: 10.1016/S1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 64.Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 65.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134(3):392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang M, et al. Whole-body optical imaging of green fluorescent protein-expressing tumors and metastases. Proc Natl Acad Sci U S A. 2000;97(3):1206–1211. doi: 10.1073/pnas.97.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito N, et al. Positive correlation of plasma transforming growth factor-beta 1 levels with tumor vascularity in hepatocellular carcinoma. Cancer Lett. 1995;89(1):45–48. doi: 10.1016/0304-3835(95)90156-6. [DOI] [PubMed] [Google Scholar]
- 68.Ijichi H, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20(22):3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek HJ, et al. Hepatocellular cancer arises from loss of transforming growth factor beta signaling adaptor protein embryonic liver fodrin through abnormal angiogenesis. Hepatology. 2008;48(4):1128–1137. doi: 10.1002/hep.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 71.Ichida JK, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5(5):491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SH, Hong B, Sharabi A, Huang XF, Chen SY. Embryonic stem cells and mammary luminal progenitors directly sense and respond to microbial products. Stem Cells. 2009;27(7):1604–1615. doi: 10.1002/stem.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47(4):1288–1297. doi: 10.1002/hep.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jellicoe MM, et al. Bioenergetic differences selectively sensitize tumorigenic liver progenitor cells to a new gold(I) compound. Carcinogenesis. 2008;29(6):1124–1133. doi: 10.1093/carcin/bgn093. [DOI] [PubMed] [Google Scholar]
- 75.Kisanuki H, et al. Retroviral expression screening of oncogenes in pancreatic ductal carcinoma. Eur J Cancer. 2005;41(14):2170–2175. doi: 10.1016/j.ejca.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 76.Whitehead I, Kirk H, Kay R. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol Cell Biol. 1995;15(2):704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi YL, et al. Retroviral expression screening of oncogenes in natural killer cell leukemia. Leuk Res. 2005;29(8):943–949. doi: 10.1016/j.leukres.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turkson J, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276(48):45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 80.Berquin IM, Dziubinski ML, Nolan GP, Ethier SP. A functional screen for genes inducing epidermal growth factor autonomy of human mammary epithelial cells confirms the role of amphiregulin. Oncogene. 2001;20(30):4019–4028. doi: 10.1038/sj.onc.1204537. [DOI] [PubMed] [Google Scholar]
- 81.Ding W, et al. CD133(+) liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49(4):1277–1286. doi: 10.1002/hep.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.