Abstract
Acute lymphoblastic leukemia (ALL) is the commonest childhood malignancy and is characterized by recurring structural genetic alterations. Previous studies of DNA methylation suggest epigenetic alterations may also be important, but an integrated genome-wide analysis of genetic and epigenetic alterations in ALL has not been performed. We analyzed 137 B-lineage and 30 T-lineage childhood ALL cases using microarray analysis of DNA copy number alterations and gene expression, and genome-wide cytosine methylation profiling using the HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) assay. We found that the different genetic subtypes of ALL are characterized by distinct DNA methylation signatures that exhibit significant correlation with gene expression profiles. We also identified an epigenetic signature common to all cases, with correlation to gene expression in 65% of these genes, suggesting that a core set of epigenetically deregulated genes is central to the initiation or maintenance of lymphoid transformation. Finally, we identified aberrant methylation in multiple genes also targeted by recurring DNA copy number alterations in ALL, suggesting that these genes are inactivated far more frequently than suggested by structural genomic analyses alone. Together, these results demonstrate subtype- and disease-specific alterations in cytosine methylation in ALL that influence transcriptional activity, and are likely to exert a key role in leukemogenesis.
Introduction
Acute lymphoblastic leukemia (ALL) is the commonest childhood malignancy (1); despite impressive advances in the success of therapy for ALL, it remains the leading cause of cancer-related death in young people. A detailed understanding of the genetic and epigenetic events contributing to leukemogenesis and treatment responsiveness is important in order to identify potential new avenues for therapy in this condition.
Pediatric ALL is characterized by a range of recurring numeric and structural chromosomal alterations. These include B-precursor ALL (B-ALL) with (a) high hyperdiploidy (>50 chromosomes), (b) hypodiploidy (<44 chromosomes), (c) t(12;21)(p13;q22) encoding ETV6-RUNX1 (also known as TEL-AML1), (d) t(1;19)(q23;p13.3) encoding TCF3-PBX1 (also known as E2A-PBX1), (e) t(9;22)(q34;q11) encoding BCR-ABL1, (f) cytokine receptor–like factor 2 (CRLF2) rearrangement (CRLF2r), and (g) MLL rearrangement at 11q23 (MLLr); MYC rearrangement in mature B cell leukemia/lymphoma; and TLX1 (also known as HOX11), TLX3 (also known as HOX11L2), LYL1, TAL1, and MLL rearrangement in T-lineage ALL (T-ALL) (2). These alterations are important events in leukemogenesis and are associated with responsiveness to therapy, treatment outcome, and distinct gene expression profiles (3). However, several of these translocations may be acquired years before development of overt leukemia (4), and mouse models have shown that these alterations are frequently insufficient to cause leukemia, which suggests that cooperating genetic events are required. Furthermore, recurring chromosomal alterations are absent in almost 20% of cases (5). Consequently, extensive genome-wide microarray profiling of DNA copy number alterations (deletions and gains) has identified multiple submicroscopic genetic changes in ALL that target critical cellular pathways, including lymphoid development, cell cycle regulation, tumor suppression, lymphoid signaling, and drug responsiveness (6–8). The nature and frequency of these alterations is associated with the cytogenetic subtype of ALL, and specific alterations have been shown to influence treatment outcome, such as deletion or mutation of IKZF1, which encodes the early lymphoid transcription factor IKAROS (8, 9).
However, somatic mutation is not the only mechanism through which gene expression is perturbed in tumors. Alteration of DNA methylation can also disrupt transcriptional regulation and is commonly perturbed in cancer. Genome-wide analysis of cytosine methylation has identified novel subtypes of acute myeloid leukemia (AML) and has provided important insights into the biology of this disease (10, 11). Previous studies in childhood ALL have examined cytosine methylation of the promoters of individual genes, investigated limited panels of genes (12–18), or performed genome-wide analyses of methylation in small patient cohorts (19) or specific subtypes of ALL, such as MLLr (20, 21), high-hyperdiploid, and ETV6-RUNX1–rearranged (22). These studies suggest that aberrant DNA methylation is a hallmark of childhood ALL and underscore the need for genome-wide studies in a large cohort of patients in order to better understand the contribution of DNA methylation to leukemogenesis. Few studies have analyzed large cohorts of ALL cases, and most have used candidate gene methods rather than genome-wide approaches (23). To date, no study has performed genome-wide analysis of methylation across the spectrum of B-ALL and T-ALL subtypes, correlating methylation status with changes in gene expression and concomitant structural DNA alterations.
Here we describe the first integrated genome-wide analysis of cytosine methylation, DNA copy number alterations, and gene expression in childhood ALL. We show that recurring cytogenetic alterations exhibited distinct methylation signatures and that methylation was an important determinant of the leukemic transcriptome. In addition, we identified a signature of aberrant methylation common to B-ALL cases and showed that genes targeted by recurring DNA copy number alterations frequently exhibited aberrant methylation.
Results
DNA methylation profiles in B-ALL are associated with genetic subtype.
We performed methylation profiling, using the HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) assay, of leukemic DNA samples obtained at diagnosis from 167 children with ALL (137 B-ALL and 30 T-ALL), 19 flow-sorted normal B cell progenitor subpopulations, and 8 CD3+ T cell samples purified from peripheral blood apheresis samples. The cohort was designed to encompass the range of known ALL subtypes, including several recently defined subtypes characterized by rearrangement and/or dysregulation of CRLF2 and the ETS family transcription factor ERG (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI66203DS1). DNA methylation status, defined by HELP, was significantly correlated with quantitative DNA methylation measurements of individual loci using MassARRAY EpiTYPER platform (r = 0.91; Supplemental Figure 1).
To identify subgroups of ALL cases defined by methylation signatures, we performed unsupervised hierarchical clustering of high variance (SD >1 across the cohort; n = 4,037 probe sets), followed by determination of association between cluster membership and cytogenetic subtype by calculation of the gap statistic (24, 25) and Rand index (26). This procedure identified 8 robust clusters of leukemic samples using both indices. There was remarkable correlation between methylation cluster and ALL cytogenetic subtype (Figure 1A). As expected, distinct clusters of known ALL subtypes based on gene expression profiling data were observed (Figure 1B). In analysis of methylation data, all 29 T-ALL cases and 8 normal CD3+ T cell samples clustered separately from B-ALL cases (Figure 1A and Supplemental Figure 2). 7 clusters of B-ALL cases were identified. MLLr, BCR-ABL1, ETV6-RUNX1, and normal B cell progenitors (regardless of maturational stage) formed distinct clusters. The 4 cases with intrachromosomal amplification of chromosome 21 (iAMP21) clustered with ETV6-RUNX1 cases and B-ALL cases of hyperdiploid and miscellaneous karyotype. The MLLr cluster was 100% specific, containing 17 MLLr cases, but failed to include 3 cases positive for MLLr. No subclustering of MLLr cases was observed according to MLL fusion partner (MLLT3 [also known as AF9], 5 cases; AFF1 [also known as AF4], 7 cases; MLLT1 [also known as ENL], 2 cases; MLLT10 [also known as AF10], 2 cases; unknown, 2 cases; data not shown). Likewise, the BCR-ABL1 cluster was also 100% specific (13 cases), but 6 BCR-ABL1 cases failed to cluster with this group. Another cluster contained 24 ETV6-RUNX1 cases, but did not include 3 additional cases harboring the same fusion gene. Cases harboring high-hyperdiploid karyotypes or CRLF2r clustered together, whereas another cluster included cases with the TCF3-PBX1 fusion gene or cases with ERG alterations. Normal B and T cells formed specific clusters with 100% membership and no misclassifications. The concordance between DNA methylation and genetic subtypes was confirmed by repeating the hierarchical clustering using different subsets of probe sets (cross-patient SD 0.8–1.2; Supplemental Figure 3). These data indicate that DNA methylation profiles in ALL are highly associated with specific genetic subtypes and suggest that aberrant epigenetic patterning may have a central role in leukemogenesis and disease phenotype.
Figure 1. Hierarchical clustering of DNA methylation profiles and gene expression profiles of 167 childhood ALL cases and 19 normal B cells.
(A) Dendrogram of unsupervised hierarchical clustering of 4,037 methylation probe sets with SD >1. (B) Dendrogram of unsupervised hierarchical clustering of 842 probe sets of SD >1.2 of 154 ALL cases with Affymetrix gene expression profiling data. H50, high hyperdiploidy; Ph, BCR-ABL1; T-P, TCF3-PBX1.
Unique DNA methylation profiles distinguish ALL subtypes.
We next defined the DNA methylation signatures associated with each genetic subtype of childhood ALL. We performed supervised comparisons of methylation data between each B-ALL subtype and all other B-ALL cases and normal B cells, as well as comparing B-ALL with T-ALL cases and T-ALL cases with normal T cells. Differentially methylated regions (DMRs) were identified using limma (27) with a false discovery rate (FDR) of <0.1 and a log2 methylation delta of >1 (corresponding to a methylation difference of approximately 20% or greater). This analysis revealed that specific DNA methylation abnormalities were highly variable for the different subtypes, both in the magnitude of the signature identified and in the degree of hyper- and hypomethylation observed (Table 1 and Figure 2). For example, B-ALL cases with high hyperdiploidy exhibited the strongest signature, consisting of 719 DMRs, 635 of which were hypomethylated compared with other B-ALL cases. Given that the MspI fraction in the HELP assay acts as an internal control for copy number variations, the difference in log2(HpaII/MspI) ratios in the high-hyperdiploid B-ALL cases compared with the remaining B-ALL subtypes can only be explained by a detectable difference in methylation between the 2 groups. ALL cases harboring ERG alterations had a robust DNA methylation signature of 707 DMRs; however, this signature presented a balance between differential hyper- and hypomethylation relative to non–ERG-deleted B-ALL cases. On the other hand, cases with CRLF2r (many of which have activating JAK1/2 mutations; refs. 28, 29) were notable for their weak signature, with only 52 DMRs identified. Supplemental Table 2 lists all DMRs and their annotation for each signature.
Table 1.
Number of DMRs constituting the DNA methylation signatures of each ALL subtype

Figure 2. Subtype-specific DNA methylation signatures.
Heatmap representation of the DNA methylation profiles identified through supervised analysis distinguishing each ALL subtype from the other ALL cases. Rows represent probe sets; columns represent patients. In each case, the group of interest is represented at the far left of the heatmap (black bars). Red, low methylation; blue, high methylation.
Gene networks affected by aberrant DNA methylation in ALL.
We next performed Ingenuity Pathway Analysis (Ingenuity Systems) on each of the identified DNA methylation signatures. Analysis of the top 5 scoring networks demonstrated that, despite each ALL subtype presenting with a unique DNA methylation signature, functional analysis of the affected genes revealed some common biological themes (Supplemental Table 3). Networks involved in cell death and cell cycle were among the top 5 scoring networks for all subtypes, with the exception of the ERG signature, for which the top scoring network was involved in cellular growth, proliferation, and cancer. Another top network was cell-to-cell signaling, which was observed for most subtypes except ETV6-RUNX1 and BCR-ABL1 ALL. These 2 subtypes exhibited high-scoring networks involved in lipid metabolism.
Specific DNA methylation profiles correlate with gene expression patterns in genetic subtypes of ALL.
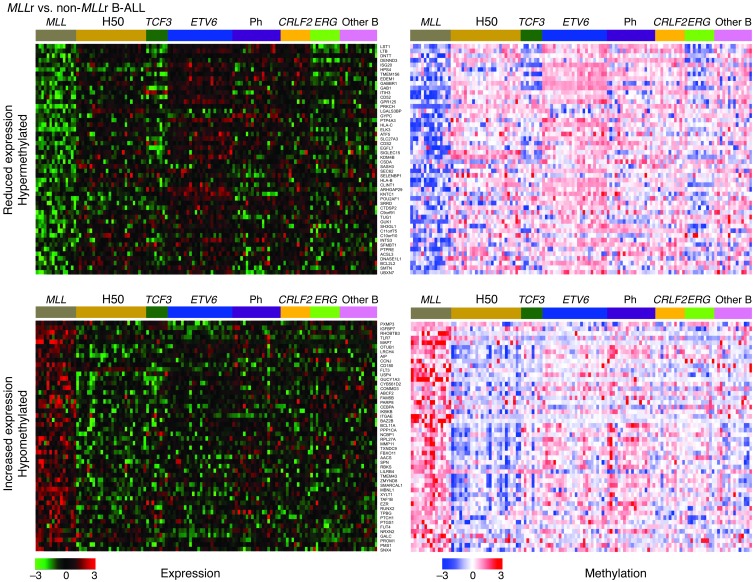
We next used 2 approaches to examine correlations between methylation and gene expression in 154 cases with both methylation and gene expression data available. In a “Refseq-centric” analysis, Pearson correlation coefficients were calculated between all possible pairs of methylation probe sets and gene expression probe sets, and the methylation-expression probe set pair with the maximum correlation coefficient was used for downstream analysis. These coefficients were used to rank genes of interest and generate heatmaps with highly correlated genes. Genes with both gene expression and methylation FDR of <0.1, and for which gene expression and methylation had a positive Pearson correlation (which, given the nature of the HELP assay, translates to an inverse biological correlation) with P < 0.05, were considered highly correlated. This demonstrated that a substantial fraction of differentially methylated Refseq genes were differentially expressed. Figures 3 and 4 show examples of gene expression and methylation heatmaps for MLLr B-ALL and for T-ALL. Supplemental Table 4 shows the results of this Refseq analysis, and Supplemental Table 5 summarizes the number and proportion of correlated genes.
Figure 3. Expression and methylation heatmaps for genes exhibiting differential methylation and expression in MLLr B-ALL.
Heatmaps of expression (left; green, reduced expression; red, increased expression) and methylation (right; blue, hypermethylation; red, hypomethylation) show the top 50 correlated genes for each subtype from Refseq-centric correlation analysis. The total number of correlated probe sets for each subtype is summarized in Supplemental Table 4.
Figure 4. Expression and methylation heatmaps for genes exhibiting differential methylation and expression in T-ALL.
Heatmaps of expression (left; green, reduced expression; red, increased expression) and methylation (right; blue, hypermethylation; red, hypomethylation) show the top 50 correlated genes for each subtype from Refseq-centric correlation analysis. The total number of correlated probe sets for each subtype is summarized in Supplemental Table 4.
We next used gene set enrichment analysis (GSEA; ref. 30) to gain insight into the relationship between methylation and gene expression profiles. For this analysis, we defined hyper- and hypomethylated gene sets for each subtype of ALL, which were then tested for enrichment in the gene expression profiles of each ALL subtype (31). In all but 2 of the genetic subtypes, we found significant enrichment of the hyper- and/or hypomethylated gene sets in the corresponding gene expression profile (Figure 5 and Supplemental Table 6), which demonstrated that changes in DNA methylation were accompanied by matching changes in gene expression. This indicates that aberrant DNA methylation in childhood ALL is an important determinant of gene expression in genetically defined disease subtypes. We did not find significant enrichment for the CRLF2r B-ALL methylation signature in the CRLF2r gene expression profile, consistent with our observation that this subtype of ALL had relatively weak methylation and gene expression signatures.
Figure 5. GSEA of DNA methylation signatures in gene expression profiles.
Shown are enrichment scores and leading edge plots for each of the DNA methylation gene sets that showed significant enrichment in the corresponding gene expression profile.
High-hyperdiploid ALL is characterized by the gain of at least 5 additional chromosomes. As many, but not all, genes on the aneuploid chromosomes exhibit increased expression, we examined the relationship between methylation status and gene expression in this subtype of leukemia. With the exception of chromosome 21, for which 27.5% of HELP probe sets were methylated, we did not observe preferential hypomethylation of trisomic chromosomes in hyperdiploid ALL (Supplemental Table 7). As expected, the proportion of genes exhibiting reduced expression was lower in aneuploid (3.5%) than diploid (20.8%) chromosomes; however, almost half of such genes exhibited concomitant hypermethylation (Supplemental Table 8). Thus, methylation is an important determinant of gene silencing on aneuploid chromosomes in ALL.
DNA methylation plays a role in the differential regulation of lineage-specific pathways.
Using a similar supervised analysis approach (limma FDR <0.1 and methylation change >20%), we compared methylation profiles of T-ALL cases with B-ALL cases, and normal T cells with normal B cells, and subtracted the normal from the leukemic methylation signatures to determine the extent to which the T-ALL versus B-ALL signature was influenced by lineage rather than leukemia-specific factors. Although the extent of overlap of the normal and leukemic signatures may be influenced by the number of samples in each comparison group, we observed that the extent of overlap (∼40%) remained remarkably consistent despite varying the delta threshold used to define the signatures (Supplemental Table 8). As expected, the T-ALL versus B-ALL signature consisted of a large number of DMRs (n = 1,319). Notably, three-quarters of the signature consisted of DMRs that were hypermethylated in T-ALL versus B-ALL. GSEA showed significant enrichment of both the hypermethylated and the hypomethylated components of the methylation signature in the corresponding gene expression profile (Figure 5). Strikingly, we found that the mediators of T cell receptor (TCR) signaling and differentiation were silenced by methylation in B-ALL cases; conversely, the B cell receptor (BCR) signaling and differentiation program was silenced in T-ALL (Figure 6). Key components of both pathways were found among these epigenetically regulated genes, including CD79A, CD79B, BLNK, BTK, and SYK in the BCR signaling pathway and CD3G, CD3D, LCK, LAT, and ZAP70 in the TCR signaling pathway (Figure 6). Notably, the “residual” leukemic T versus B cell methylation signature derived after subtraction of the signature of normal T versus B cells exhibited differential methylation of lymphocyte signaling pathways (BCR signaling, DAVID P = 0.008; TCR signaling, DAVID P = 0.12). Moreover, several key lymphoid signaling genes, including CD3 and CD79B, exhibited differential methylation between normal and leukemic cells (Supplemental Figure 4). Together, these findings suggest that DNA methylation is a critical determinant of lineage specification in ALL by its regulation of the expression levels of lineage-specific signaling pathways.
Figure 6. Representation of the BCR and TCR signaling pathways and their regulation through DNA methylation.
(A) Schematic representation of the BCR signaling pathway, which was significantly differentially methylated in B-ALL versus T-ALL (DAVID P = 0.00064). Genes that were hypomethylated or overexpressed in B-ALL are highlighted red; genes that were hypermethylated or silenced in B-ALL are highlighted green. (B) Schematic representation of the TCR signaling pathway (DAVID P = 0.029). Genes that were hypomethylated or overexpressed in T-ALL are highlighted red; genes that were hypermethylated or silenced in T-ALL are highlighted green.
Recurring epigenetic lesions are present across all genetic subtypes of ALL.
These analyses identified distinct methylation signatures for each ALL subtype. In order to determine whether a common epigenetic signature of ALL exists, we compared the methylation profiles of each ALL subtype with a cohort of normal B cells at different stages of differentiation. Unsupervised clustering showed minimal intersample variability between the normal B cells at different differentiation stages, which allowed us to use the entire cohort of normal B cells, including pro-B, pre-B, and mature B cells, as our normal reference for comparison. In order to do this, we performed ANOVA analysis across the different B-ALL subtypes followed by Dunnett’s post-hoc test, using the normal B cells as the reference, in order to identify genes aberrantly methylated in each B-ALL subtype. We next identified those genes that were aberrantly methylated in at least 7 of the 8 B-ALL subtypes. At this significance cutoff, we identified 75 DMRs annotated to 85 unique genes (Figure 7 and Supplemental Table 9). 66 of the 75 DMRs were hypermethylated in the leukemia samples, while only 9 were hypomethylated. 5 genes from this common signature were randomly selected, and their recurrent aberrant methylation was validated by MassARRAY EpiTYPER in a subset of randomly selected samples (Supplemental Figure 5). 36 of 55 aberrantly hypermethylated genes represented on gene expression arrays exhibited reduced expression, while 3 of 5 hypomethylated genes were overexpressed. This common methylation signature of ALL included genes involved in signaling (TIE1, MOS, CAMLG, and GPRC5C), genes involved in cell cycle regulation and proliferation (MCTS1 and DGKG), genes involved in RNA metabolism (PABPN1 and PABPC5), transcription factors and transcriptional regulators (PROP1, TAF3, H2AFY2, ELF5, ZBTB16, CNOT1, and TADA2A), and the homeobox genes HOXA5 and HOXA6. Importantly, since the analysis compared the ALL samples with a cohort of normal B cells from different stages of differentiation, this methylation profile cannot be interpreted as a reflection of the differentiation stage from the cell of origin, but rather represents a true leukemic signature.
Figure 7. Heatmap representation of genes frequently aberrantly methylated in B-ALL.
75 DMRs annotated to 84 unique genes were identified as aberrantly methylated in at least 7 of the 8 B-ALL subtypes compared with methylation status in normal B cells. Each row represents a DNA methylation probe set; each column represents a patient. Columns are arranged by genetic subtype.
Genes frequently affected by structural abnormalities are also targets for aberrant DNA methylation.
We next examined whether targets of structural genetic alteration in ALL are also dysregulated by aberrant methylation. We identified 76 genes (Supplemental Table 10) known to be mutated in ALL (7, 8), as well as genes in signaling pathways of which other components are frequently targeted by genetic mutations and structural abnormalities. DNA methylation status was available for 71 of the 76 genes. We observed that 26 of these 71 genes were aberrantly methylated in certain B-ALL cases; that is, their DNA methylation status was different than normal B cells. 15 of the 71 were aberrantly methylated in 5% or more of the cases, with some genes presenting aberrant methylation frequencies as high as 76% (Figure 8A and Table 2). An additional 11 genes were observed as aberrantly methylated in fewer than 5% of the ALL cases, while the remaining 45 genes were never targeted by abnormal methylation in our cohort. Aberrant methylation of these genes included both abnormal gains and losses of methylation compared with normal B cells, and it targeted both promoter and gene body regions. Table 2 shows the affected region and specific abnormality observed for each gene. In most cases, aberrant methylation of these genes occurred in patients who did not harbor a copy number abnormality as well. However, in a small number of cases, we detected concomitant occurrence of structural and epigenetic abnormalities (GAB1, 2 of 77; RAG2, 2 of 32; KRAS, 2 of 39; RB1, 8 of 106; ATP10A, 1 of 14). In the case of CDKN2A (also known as P16) and CDKN2B (also known as P15), we detected 5 of 24 and 7 of 28 cases, respectively, with both types of abnormalities, which indicates that aberrant hypermethylation of the promoter region of these genes could play a role in silencing the remaining allele in cases of heterozygous deletions. While the lack of gene expression data for the normal B cell cohort prevents a complete understanding of the changes in gene expression that accompany these lesions, we did observe that hypermethylation of the promoter regions of CDKN2A, CDKN2B, and PTEN was accompanied by low levels of expression of these genes, while expression levels above the median were observed for KRAS, which displayed hypomethylation of its promoter region (Figure 8B). Unlike KRAS mutations, which are common in hyperdiploid B-ALL, aberrant hypomethylation of this gene was observed more frequently in ETV6-RUNX1 (n = 22), BCR-ABL1 (n = 12), and CRLF2r (n = 8) cases.
Figure 8. Gene expression of genes mutated in ALL that also exhibit aberrant methylation.
(A) Proportion of cases showing aberrant methylation of genes also involved by somatic mutation. (B) Relative gene expression levels for PTEN, CDKN2A, CDKN2B, and KRAS in patients with aberrant methylation. Shown are box plot representations of gene expression for a subset of genes frequently affected by mutations in patients that instead presented aberrant methylation of these genes. PTEN, CDKN2A, and CDKN2B were expressed at low levels in the affected patients, whereas KRAS was expressed at high levels. Bottom bound, middle line, and top bound of boxes denote 25th, 50th, and 75th percentiles, respectively; whiskers denote minimum and maximum non-outlier values; symbols denote outliers.
Table 2.
Epigenetic alterations of genes known to be targeted by genetic lesions

Discussion
Aberrant DNA methylation is a hallmark of many neoplasms, including hematological malignancies. Genome-wide abnormalities in DNA methylation can be found in chronic and acute leukemias, both lymphoid (21, 32–34) and myeloid (10, 35, 36), as well as in preleukemic conditions, such as myelodysplastic syndromes (37, 38). Furthermore, DNA methylation of certain genes has been shown to have prognostic significance in AML and ALL (10, 36, 39). DNA methyltransferase 1–deficient (Dnmt1-deficient) murine models have demonstrated that DNA methylation is required for tumorigenesis, both in solid tumors and in leukemias, even when using a potent leukemia oncogene such as MLLT3 (40–42). Therefore, the aberrant DNA methylation patterns observed in malignancies cannot be interpreted as simple secondary events passively acquired during transformation, but rather must play an active role in determining the malignant phenotype. In order to explore the relationship between genetic and epigenetic lesions in childhood ALL, we carried out a comprehensive integrative analysis of DNA methylation, structural abnormalities, copy number alterations, and gene expression in a large cohort of primary ALL. In this study, not only did we observe that epigenetic subtypes identified through unsupervised analysis approaches revealed a remarkably high degree of concordance with known genetic subtypes, but furthermore, these epigenetic abnormalities associated with each of the ALL genetic subtypes were highly correlated with their respective transcriptional profiles. These findings suggest that changes in DNA methylation patterns in ALL may play an important role in determining the transcriptional program and therefore the phenotype of the leukemic blasts.
Recent studies of the hematopoietic system have revealed marked epigenetic plasticity during hematopoietic differentiation (43). These epigenetic changes are in fact required for lineage determination, and interference with DNA methylation — either genetically or chemically — results in skewing of the hematopoietic differentiation toward the myeloid lineage (40, 43). However, the specific regulation of lineage-specific signaling pathways through epigenetic regulation in acute leukemias has not been previously reported. Our study revealed extensive differential DNA methylation of multiple components of the BCR and TCR signaling pathways in B-ALL and T-ALL, respectively. Moreover, these epigenetic changes correlated with changes in gene expression, which indicates that lineage-specific signaling pathways in lymphoid leukemic cells may be regulated at the epigenetic level.
Our observation that a set of recurrent epigenetic alterations is observed across all ALL subtypes, regardless of genetic or epigenetic subtype, lends further support to a model in which certain epigenetic events are required to take place during malignant transformation. The near-universal nature of these alterations in methylation suggests that they represent a set of genes whose silencing is likely to constitute a necessary event in order to complete the transformation process. This finding is in concordance with prior studies in both ALL and AML that have demonstrated that most recurrent genetic events observed in these leukemias are insufficient to fully transform a normal hematopoietic progenitor (44–50). A similar scenario has been previously described for adult AML (10), and the set of genes targeted in that form of leukemia, while almost identical to those observed in myelodysplastic syndromes, is distinct from the set of genes affected in ALL. This leads us to conclude that these common epigenetic lesions that need to be established during transformation are lineage specific and may play a central role in determining the final phenotype of the malignant clone. However, 9 of these recurrent epigenetic hits are observed in to both ALL and AML cases, which indicates that they may play an even more universal role in leukemic transformation, independent of lineage. Among these are DHRS12, which has been shown to be frequently deleted and underexpressed in some solid tumors (51); MCART1, which functions as a mitochondrial carrier and is therefore likely to play an important role in cellular metabolism; and SPNS2 (a homolog of HSpin1), which has been implicated in regulating caspase-independent cell death (52). Functional studies in murine models will be required in the future to determine the precise mechanism and timing by which these lesions contribute to the leukemogenic process. Another observation of our current study was that genes that are well-known to be frequently affected by sequence and copy number alterations in childhood ALL may also be targeted by aberrant DNA methylation. We compared the DNA methylation profile in ALL blasts of 71 of these genes with their status in normal B cells and found that approximately one-third was also susceptible to aberrant DNA methylation. Among these, we observed hypermethylation of the promoter regions of CDKN2A, CDKN2B, and PTEN and hypomethylation of the promoter region of KRAS. The finding that certain ALL cases presented with genetic abnormalities of these well-established oncogenes and tumor suppressors, while others instead presented with epigenetic abnormalities, raises the possibility that during malignant transformation, the cell can resort to both genetic and epigenetic mechanisms to ensure deregulation of key biological pathways that would favor expansion of the malignant clone. Aberrant methylation of these genes has been previously reported in the context of leukemias as well as solid tumors (17, 53, 54), which indicates that epigenetic deregulation of these key regulatory genes is indeed a frequent event in tumorigenesis.
A better understanding of the mechanisms through which malignant cells deregulate central pathways to ensure their proliferation and survival advantage is central to the design of appropriate therapeutic targeting strategies. By combining several high-throughput platforms and integrating genetic and epigenetic information, we demonstrated that these 2 mechanisms of regulation both played a key role in regulating the transcriptional program in ALL, thereby contributing to the determination of disease phenotype.
Methods
Patient samples.
The cohort, composed of 167 children with ALL treated at St. Jude Children’s Research Hospital between 1988 and 2007 (primarily on the St. Jude Total XIII and Total XV protocols; refs. 55, 56), included 123 B-ALL cases, with high hyperdiploidy (>50 chromosomes; n = 26), TCF3-PBX1 (n = 9), ETV6-RUNX1 (n = 27; 2 with iAMP21; ref. 57), BCR-ABL1 (n = 19), CRLF2r (n = 11; 2 with iAMP21; refs. 29, 58), a recently identified group with a distinct gene expression profile and alteration of ERG (n = 11; ref. 59), and MLLr (n = 20); 14 cases with normal, pseudodiploid, or miscellaneous karyotype; and 30 T-ALL cases, 4 of which had MLLr, and 7 of which had early thymic progenitor immunophenotype (refs. 60, 61, and Supplemental Table 1). We also studied 19 normal B cell samples flow sorted from the bone marrow of healthy children as controls, including pro-B cells (CD34+CD19– surface Ig negative [sIg–]), pre-B cells (CD34+CD19+sIg–), and mature B cells (CD34–CD19+sIg+); and 8 CD3+ T cell samples purified from peripheral blood apheresis samples of healthy donors by MACS separation (Miltenyi Biotech).
DNA methylation microarrays.
All samples had >80% leukemic cells, or were flow sorted to >90% purity prior to DNA extraction, as previously described (62). High–molecular weight DNA was isolated from leukemic cells using phenol-chloroform extraction. The HELP assay was carried out as previously described (63, 64), and samples were hybridized onto a custom human promoter array covering 25,626 HpaII amplifiable fragments (HAFs; representing approximately 50,000 CpGs), annotated to approximately 14,000 genes (design ID 4802; Roche NimbleGen). HAFs are defined as genomic regions contained between 2 flanking HpaII restriction sites found 200–2,000 bp apart. HAFs were first realigned to the hg19 build of the human genome, then annotated to the nearest transcription start site (TSS), allowing for a maximum distance of 5 kb from the TSS. Hybridization and scanning were performed as previously described (65, 66). Quality control for regional artifacts and dye bias was performed on all hybridizations following standard procedures for HELP microarrays (67), and any arrays that failed were repeated. After summarization of probe-level data into probe sets, background subtraction, and log2 transformation, simple quantile normalization was performed on each channel separately, and the log ratio was then calculated for each sample.
Microarray gene expression profiling.
RNA was extracted from leukemic cells using the TRIzol method, and gene expression profiling was performed using Affymetrix U133A arrays (Affymetrix), in accordance with the manufacturer’s protocols and as previously described (31), for 154 patients (Supplemental Table 1). Gene expression signals were scaled to a median value of 500 using the Affymetrix MAS 5.0 algorithm. Probe sets with absent calls for all samples were excluded, and probe set signals were variance stabilized by adding the number 32 and subsequently log2-transformed.
Quantitative DNA methylation sequencing by MassARRAY EpiTYPER.
Validation of HELP data was performed on 21 randomly selected samples. MALDI-TOF mass spectrometry using EpiTYPER by MassARRAY (Sequenom) on bisulfite-converted DNA was performed as previously described (68). MassARRAY primers (Supplemental Table 11) were designed to cover the flanking HpaII sites of 22 randomly selected HAFs, as well as all other HpaII sites found up to 2,000 bp upstream of the downstream site and up to 2,000 bp downstream of the upstream site, in order to cover all possible alternative sites of digestion.
Clustering analysis.
All microarray data analyses were performed using R version 2.9.0. ( http://www.r-project.org) and Bioconductor version 2.4 ( http://www.bioconductor.org/; ref. 69). Unsupervised hierarchical clustering of HELP data was performed, with 1 – Pearson correlation distance, followed by a Lingoes transformation of the distance matrix to a Euclidean matrix (70) and subsequent clustering using Ward’s method. Initial analysis was performed on probe sets with across-patient SD >1 (n = 4,037) in order to filter out uninformative probe sets. Gap statistic analysis (24, 25) was used in order to identify the optimal cutoff of the tree, and the adjusted Rand index (26) was used to compare the extent of concurrence between the clustering results and the underlying cytogenetic subtypes. To further determine the stability of these clustering results, the hierarchical clustering was repeated at different SD cutoffs, and the gap statistic and Rand index was determined for each.
Definition of DNA methylation signatures.
2 approaches were taken to define the methylation signatures. Pairwise comparisons were performed using limma (27) on HELP methylation log2(Hpall/MspI) ratios at a Benjamini-Hochberg FDR (71) of <10%. A second criterion of an absolute log2 methylation delta of >1 between sample means, corresponding to a methylation difference of 20% or greater, was required in order to ensure the capturing of most likely biologically relevant methylation changes. We compared each B-ALL subtype with all other B-ALL cases (excluding the normal B cell cohort), B-ALL versus T-ALL, and T-ALL and B-ALL versus normal T and B cells. To identify epigenetic lesions common to all ALL subtypes, ANOVA analysis was performed across the normal B cells and the different B-ALL subtypes, followed by adjustment of the P value using the Benjamini-Hochberg method (71) and subsequent Dunnett’s post-hoc test using normal B cells as the reference. Aberrantly methylated genes were identified for each of the B-ALL subtypes compared with normal B cells, at P < 0.005 and absolute methylation delta of >1.5 (corresponding to ≥25% methylation change). Recurrently occurring epigenetic lesions in childhood ALL were defined as genes that were aberrantly methylated in at least 7 of the 8 B-ALL subtypes. To determine the methylation status of a set of 76 genes with frequent structural abnormalities in B-ALL (7, 8) relative to their status in normal B cells, HELP values were categorized into the following 2 groups: methylated, log2(HpaII/MspI) < 0.5; unmethylated, log2(HpaII/MspI) > 1.5. log2(HpaII/MspI) values between 0.5 and 1.5 were considered uncertain. For each gene, the methylation status in the normal B cells was determined. Abnormal methylation in B-ALL was defined as any case in which the methylation status of a given gene fell in the opposite category as that of the normal B cells for that gene.
Ingenuity and DAVID Pathway Analysis.
For each ALL subtype, we performed Ingenuity Pathway Analysis and The Database for Annotation, Visualization, and Integrated Discovery (DAVID; refs. 72, 73, and see below) Pathway Analysis using the entire HELP array as a user-defined reference background and the observed DNA methylation signature at FDR 0.1 and absolute log2 methylation delta of >1. In Ingenuity analysis, both direct and indirect interactions were allowed.
Integration of methylation and gene expression profiling data.
2 approaches were used. In a Refseq-centric correlation analysis to identify cis regulation, Pearson correlation coefficients for each Refseq annotated gene (74) were first calculated among all possible pairs of methylation probe sets and gene expression probe sets across the cohort or subgroups of interest (e.g., all patients or B-ALL or T-ALL patients). The methylation-expression probe set pair with the maximum absolute correlation coefficient was then chosen for each Refseq gene. These coefficients were used to rank genes of interest and generate heatmaps with highly correlated genes. Genes with both gene expression and methylation FDR of <0.1, and for which gene expression and methylation had a positive Pearson correlation (which, given the nature of the HELP assay, translates to an inverse biological correlation) with P < 0.05 were considered highly correlated.
In a complementary approach, GSEA (30) implemented in R ( www.r-project.org) was used to examine the enrichment of the hypomethylated and hypermethylated gene sets in gene expression signatures of each ALL subtype. Hyper- and hypomethylated gene lists for each methylation signature were generated, as defined by FDR of <0.1 and absolute methylation delta of >1.0. Gene sets with <10 or >500 genes were excluded, and significantly enriched gene sets after 1,000 permutations at FDR of <0.25 were reported.
To examine the association between gene expression and methylation in high-hyperdiploid B-ALL cases, Affymetrix gene expression probe sets and HELP methylation probe sets corresponding to the same Refseq genes were ordered by chromosomal location; those below a limma FDR threshold of 0.1 (10%) were considered differentially expressed/methylated.
Data access.
HELP microarray data are available at GEO (ref. 75; accession no. GSE44862). Gene expression microarray data are available at GEO (accession no. GSE26281). A comprehensive archive of all tables and figures generated by the analyses described above can be found at http://stjuderesearch.org/site/data/ALL-Methylation.
Statistics.
Gap statistic analysis (24, 25) was used in order to identify the optimal cutoff of hierarchical clustering trees, and the adjusted Rand index (26) was used to examine correlation between the clustering results and the underlying cytogenetic subtypes. Limma (27) with FDR correction (typically 10%) and ANOVA with FDR and Dunnett’s post-hoc test were used for supervised analysis of methylation signatures. Pearson’s correlation was used to examine correlation of methylation and expression status. A P value less than 0.05 was considered significant.
Study approval.
The study fulfilled requirements of the Helsinki declaration and was approved by the IRBs of St. Jude Children’s Research Hospital and Weill Cornell Medical College. Written informed consent and assent was obtained from patients and their legal guardians.
Supplementary Material
Acknowledgments
The authors thank the Tissue Resources Core Facility of St. Jude Children’s Research Hospital for providing leukemia samples; R. Cross for performing flow sorting; R. Iyengar for performing CD3+ lymphocyte purification; D. Payne-Turner for DNA extraction; K. Spelshouse for figure preparation; the Clinical Application of Core Technology Laboratory of the Hartwell Center for Bioinformatics and Biotechnology of St. Jude for performing SNP and gene expression microarrays; and the Weill Cornell Medical Center Epigenetics Core Facility for performing the HELP microarrays. This work was supported by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, an American Society of Hematology Scholar Award (to C.G. Mullighan), and an American Association for Cancer Research/Aflac Career Development Award (to C.G. Mullighan). C.G. Mullighan is a Pew Scholar in the Biomedical Sciences and is a St. Baldrick’s Scholar. A.K. Andersson was supported by the Swedish Childhood Cancer Foundation. M.E. Figueroa is supported by a Leukemia and Lymphoma Society Special Fellow award and a Clinical Scientist Development Award from the Doris Duke Charitable Foundation. A. Melnick is funded by a Leukemia and Lymphoma Society SCOR and TRP award and is a Burroughs Wellcome Clinical Translational Scholar and Scholar of the Leukemia and Lymphoma Society. A. Melnick is also supported by the Sackler Center for Biomedical and Physical Sciences.
Footnotes
Conflict of interest: Ari Melnick has consulting agreements with BioReference Laboratories and Celgene and is on the speakers bureau for Genentech.
Citation for this article:J Clin Invest. 2013;123(7):3099–3111. doi:10.1172/JCI66203.
Maria E. Figueroa’s present address is: Department of Pathology, University of Michigan, Ann Arbor, Michigan, USA.
Anna K. Andersson’s present address is: Department of Clinical Genetics, Lund University Hospital, Lund, Sweden.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeoh EJ, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. doi: 10.1016/S1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 4.Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood. 1999;94(3):1057–1062. [PubMed] [Google Scholar]
- 5.Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009;144(2):147–156. doi: 10.1111/j.1365-2141.2008.07417.x. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan CG, Downing JR. Global genomic characterization of acute lymphoblastic leukemia. Semin Hematol. 2009;46(1):3–15. doi: 10.1053/j.seminhematol.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 8.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinelli G, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27(31):5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- 10.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batova A, et al. Frequent and selective methylation of p15 and deletion of both p15 and p16 in T-cell acute lymphoblastic leukemia. Cancer Res. 1997;57(5):832–836. [PubMed] [Google Scholar]
- 13.Agirre X, Vizmanos JL, Calasanz MJ, Garcia-Delgado M, Larrayoz MJ, Novo FJ. Methylation of CpG dinucleotides and/or CCWGG motifs at the promoter of TP53 correlates with decreased gene expression in a subset of acute lymphoblastic leukemia patients. Oncogene. 2003;22(7):1070–1072. doi: 10.1038/sj.onc.1206236. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Aberrant methylation in promoter-associated CpG islands of multiple genes in acute lymphoblastic leukemia. Leuk Res. 2006;30(1):98–102. doi: 10.1016/j.leukres.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Zheng S, et al. Hypermethylation of the 5′ CpG island of the FHIT gene is associated with hyperdiploid and translocation-negative subtypes of pediatric leukemia. Cancer Res. 2004;64(6):2000–2006. doi: 10.1158/0008-5472.CAN-03-2387. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez MI, et al. Concurrent methylation of multiple genes in childhood ALL: Correlation with phenotype and molecular subgroup. Leukemia. 2003;17(9):1845–1850. doi: 10.1038/sj.leu.2403060. [DOI] [PubMed] [Google Scholar]
- 17.Roman-Gomez J, et al. CpG island methylator phenotype redefines the prognostic effect of t(12;21) in childhood acute lymphoblastic leukemia. Clin Cancer Res. 2006;12(16):4845–4850. doi: 10.1158/1078-0432.CCR-05-2592. [DOI] [PubMed] [Google Scholar]
- 18.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Prosper F, Heiniger A, Torres A. Lack of CpG island methylator phenotype defines a clinical subtype of T-cell acute lymphoblastic leukemia associated with good prognosis. J Clin Oncol. 2005;23(28):7043–7049. doi: 10.1200/JCO.2005.01.4944. [DOI] [PubMed] [Google Scholar]
- 19.Taylor KH, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67(18):8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 20.Stumpel DJ, et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood. 2009;114(27):5490–5498. doi: 10.1182/blood-2009-06-227660. [DOI] [PubMed] [Google Scholar]
- 21.Schafer E, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood. 2010;115(23):4798–4809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidsson J, et al. The DNA methylome of pediatric acute lymphoblastic leukemia. Hum Mol Genet. 2009;18(21):4054–4065. doi: 10.1093/hmg/ddp354. [DOI] [PubMed] [Google Scholar]
- 23.Milani L, et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood. 2010;115(6):1214–1225. doi: 10.1182/blood-2009-04-214668. [DOI] [PubMed] [Google Scholar]
- 24.Tibshirani R, Walther G, Hastie T. Estimating the number of data clusters via the gap statistic. J R Statist Soc. B. 2001;63(2):411–423. doi: 10.1111/1467-9868.00293. [DOI] [Google Scholar]
- 25.Yan M, Ye K. Determining the number of clusters using the weighted gap statistic. Biometrics. 2007;63(4):1031–1037. doi: 10.1111/j.1541-0420.2007.00784.x. [DOI] [PubMed] [Google Scholar]
- 26.Hubert L, Arabie P. Comparing partitions. J Classif. 1985;2(1):193–218. doi: 10.1007/BF01908075. [DOI] [Google Scholar]
- 27.Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 28.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106(23):9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell LJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114(13):2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross ME, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102(8):2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 32.Kuang SQ, et al. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22(8):1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 33.Arthur GL, et al. Large-scale analysis of DNA methylation in chronic lymphocytic leukemia. Epigenomics. 2009;1(1):39–61. doi: 10.2217/epi.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong W-G, et al. Genome-wide DNA methylation profiling of chronic lymphocytic leukemia allows identification of epigenetically repressed molecular pathways with clinical impact. Epigenetics. 2010;5(6):499–508. doi: 10.4161/epi.5.6.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez S, et al. DNA methylation profiles and their relationship with cytogenetic status in adult acute myeloid leukemia. PLoS One. 2010;5(8):e12197. doi: 10.1371/journal.pone.0012197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullinger L, et al. Quantitative DNA-methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115(3):636–642. doi: 10.1182/blood-2009-03-211003. [DOI] [PubMed] [Google Scholar]
- 37.Figueroa ME, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2008;113(6):1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, et al. Residual DNA methylation at remission is prognostic in adult Philadelphia chromosome-negative acute lymphocytic leukemia. Blood. 2009;113(9):1892–1898. doi: 10.1182/blood-2008-02-141002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broske A-M, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 41.Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic mice. Cancer Res. 2002;62(5):1296–1299. [PubMed] [Google Scholar]
- 42.Trinh BN, Long TI, Nickel AE, Shibata D, Laird PW. DNA Methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair. Mol Cell Biol. 2002;22(9):2906–2917. doi: 10.1128/MCB.22.9.2906-2917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castilla LH, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 45.Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1(1):63–74. doi: 10.1016/S1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 46.Cheng K, et al. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood. 2010;115(16):3341–3345. doi: 10.1182/blood-2009-03-208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrow M, Horton S, Kioussis D, Brady HJ, Williams O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood. 2004;103(10):3890–3896. doi: 10.1182/blood-2003-10-3695. [DOI] [PubMed] [Google Scholar]
- 48.Tsuzuki S, Seto M, Greaves M, Enver T. Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci U S A. 2004;101(22):8443–8448. doi: 10.1073/pnas.0402063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dedera DA, et al. Chimeric homeobox gene E2A-PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell. 1993;74(5):833–843. doi: 10.1016/0092-8674(93)90463-Z. [DOI] [PubMed] [Google Scholar]
- 50.Kamps MP, Baltimore D. E2A-Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol Cell Biol. 1993;13(1):351–357. doi: 10.1128/mcb.13.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartuma H, et al. Gene expression and single nucleotide polymorphism array analyses of spindle cell lipomas and conventional lipomas with 13q14 deletion. Genes Chromosomes Cancer. 2011;50(8):619–632. doi: 10.1002/gcc.20884. [DOI] [PubMed] [Google Scholar]
- 52.Yanagisawa H, Miyashita T, Nakano Y, Yamamoto D. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-xL, induces a caspase-independent autophagic cell death. Cell Death Differ. 2003;10(7):798–807. doi: 10.1038/sj.cdd.4401246. [DOI] [PubMed] [Google Scholar]
- 53.Raj K, et al. CDKN2B methylation status and isolated chromosome 7 abnormalities predict responses to treatment with 5-azacytidine. Leukemia. 2007;21(9):1937–1944. doi: 10.1038/sj.leu.2404796. [DOI] [PubMed] [Google Scholar]
- 54.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111(1):47–54. doi: 10.1016/S0006-291X(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 55.Pui CH, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24(2):371–382. doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pui CH, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730–2741. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strefford JC, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci U S A. 2006;103(21):8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mullighan CG, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nature Genetics. 2009;41(11):1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullighan CG, et al. ERG deletions define a novel subtype of B-progenitor acute lymphoblastic leukemia. Blood. 2007;110:691. [Google Scholar]
- 60.Coustan-Smith E, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 63.Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- 65.Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44(3):305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 66.Figueroa ME, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS One. 2008;3(3):e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson RF RM. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24(9):1161–1167. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gentleman R, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chessel D, Dufour AB, Thioulouse J. The ade4 package - I: One-table methods. R News. 2004;4:5–10. [Google Scholar]
- 71.Benjamini Y HY. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 72.Huang DW, Sherman BT, Lempicki RA. Systemic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 73.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37(Database issue):D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.