Abstract
Efficient isolation of specific, intact, living neurons from the adult brain is problematic due to the complex nature of the extracellular matrix consolidating the neuronal network. Here, we present significant improvements to the protocol for isolation of pure populations of neurons from mature postnatal mouse brain using fluorescence activated cell sorting (FACS). The 10-fold increase in cell yield enables cell-specific transcriptome analysis by protocols such as nano-CAGE and RNA seq.
Keywords: FACS, parvalbumin, pyramidal, nanoCAGE, RNA seq
The mammalian brain consists of a variety of projection neurons, local interneurons, glial cells and extracellular matrix factors entangled in a complex network. Mature neurons are severely damaged by enzymatic dissociation and mechanical trituration, causing extensive cell death thus hampering downstream analyses. Collection of pure neuronal subtypes for RNA extraction and expression profiling has, therefore, been challenging, especially in the case of adult brain tissue. Gene expression profiling of isolated neuronal sub-types has the power to provide a catalog of genes expressed in neuronal sub-populations. Such catalogs would not only provide the means to understand normal neuronal development and function, but also permit distinctions between healthy and diseased states, establish biomarkers for diagnostic and prognostic applications, and facilitate the design of targeted molecular therapies (1, 2).
The proliferation of mouse strains with fluorescently labeled neuronal subpopulations has provided a means of identifying individual neuron sub-types in the intact brain tissue (3–6). Current technologies that enable the collection of pure populations of individual fluorescent neurons, such as laser capture micro-dissection (LCM) (7, 8), manual sorting (9, 10) or automated fluorescence activated cell sorting (FACS) have considerable limitations. LCM, conducted on fixed tissues, collects few cells of partial purity, while manual sorting also yields low cell numbers, and both provide inadequate amounts of high quality RNA required for whole transcriptome analysis. Automated FACS yields adequate numbers of pure cells and has been used to purify post-mitotic neurons from embryonic brains (5, 6, 11), as well as adult neural stem cells (12, 13) and more recently astrocytes (14). FACS has also been used in the microarray profiling of purified post-mitotic embryonic 5HT neurons (15). Although FACS was considered inapplicable for the isolation of pure neuronal populations from adult brain (1) due to neuronal fragility and shearing forces during sorting (9), at least four reports demonstrate the applicability of FACS to isolate neuronal subtypes (16–19). Arlotta et al. analyzed the array profile of FACS sorted, retrograde labeled corticospinal motor neurons during development (from E18 to P14) and compared them with callosal projection neurons and corticotectal projection neurons (16). Lobo et al. purified genetically labeled medium spiny neuron subtypes of the basal ganglia from striatal slices of up to 2 month old mice and reported a recovery of 5000–10,000 neurons yielding 3–10 ng of RNA (19). Guez-Barber et al. successfully isolated, from fixed immunolabeled cells, 200,000 NeuN positive cell bodies without neuronal processes, from striata or midbrains of adult rats (17, 18). To obtain high numbers of intact adult neurons, current protocols require gradient separation and FACS to isolate neuronal cell bodies, followed by culturing with appropriate factors over a few weeks to regenerate neuronal processes (20). Thus sequence based expression profiling of specific neurons isolated from the adult mouse brain has not been conducted to date.
Here, we aimed to overcome the poor isolation rate of single intact neurons dissociated from adult mouse brain by supplementation with trehalose and essential modifications of existing methodology. Trehalose is a disaccharide and its role in maintaining cell viability during heat stress and cryopreservation is well documented (21). Trehalose helps maintain catalytic activity of proteins at high temperatures and is known to act as a chaperonin-like small molecule (22). Upon induction of stress in bacteria, yeast, fungi and invertebrate cells, trehalose synthesis is induced intracellularly (23, 24), but endogenous trehalose is absent in mammalian cells. Although trehalose has been shown to be essential on both sides of the plasma membrane to provide cryoprotection (25), extracellular trehalose alone can improve cell viability and membrane integrity of mammalian cells (26, 27). In Dauer larvae of C. elegans, intracellular trehalose is essential to confer protection against dessication (24). A role for trehalose has been proposed in barotolerance (28), however recent simulations on membrane integrity during mechanical stress did not reveal a protective role for 2 Molal Trehalose solution under the experimental conditions (29).
We reasoned that trehalose might stabilize stressed cells and cell membranes during tissue digestion and dissociation. Accordingly, we supplemented trehalose at concentration of 0.132M in all solutions during tissue digestion and neuron disaggregation to maintain cell viability. Here, we present substantial improvements to the protocol for the isolation of fluorescent neurons from adult mouse cortex by FACS sorting and total RNA extraction for whole transcriptome sequencing applications. Our protocol consistently yields high numbers of viable cells in a single-cell suspension and good quality and quantity of RNA.
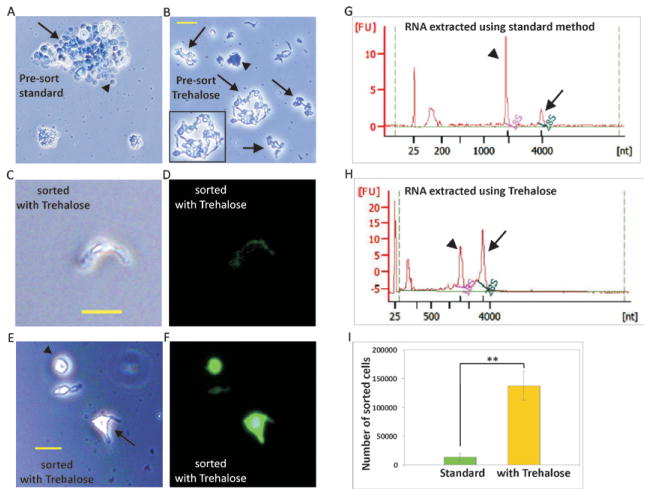
We used BAC transgenic mice expressing EGFP in Pvalb (4) or EYFP in y1-positive neurons (line M) on a C57BL/6 background (3). For digestion and dissociation, we chose to modify a standard method (protocol provided by the Neural Dissociation Kit from Worthington), which is generally successful for the isolation of neurons from neonatal brain. Each solution in the kit is supplemented with 10% v/v trehalose. After digestion each cortex is divided equally into 2 tubes and gentle, measured trituration is performed sequentially. Washed cells are triturated again and after a final wash, cells are ready for sorting. Before flow cytometry, cell viability and complete dissociation is confirmed by analyzing cells under a fluorescence microscope. In contrast to the standard method (Figure 1A), our optimized method produces viable intact isolated neurons, some dead cells as well as debris at the pre-sort stage (Figure 1B). The initial drops of sorted cells are visualized under a fluorescent microscope to adjust gating and ensure collection of viable and fluorescence positive cells alone (Figure 1C–F). To prevent RNA degradation, the sorted cells are collected directly in Trizol in the 25:75 sample: Trizol LS ratio. At this stage, RNA may be extracted using a method of choice.
Figure 1. Improved neuron and RNA acquisition using our optimized protocol.
Images taken before sorting (A–B) for standard protocol (A) and optimized protocol with Trehalose (B). Note the dead cells in undissociated tissue (arrow) as well as dead single cells without projections (arrowhead) in the standard protocol. Dissociated intact live neurons (arrows and inset) as well as dead dissociated neurons (arrowhead) are seen with the optimized protocol. (C–F) Images taken after sorting for optimized protocol. Note the intact neuron (C–D) and cells with small (arrow) or no projections (arrowhead) (E–F) in the after sort images from optimized protocol. Scale bar 20μm. Bioanalyzer profiles of RNA extracted from sorted pyramidal neurons isolated using the standard protocol (G) and optimized protocol with Trehalose (H). Note the size of the 28S peak (arrow) is smaller than the 18S peak (arrowhead) in the RNA extracted using the standard method, suggesting RNA degradation with the standard protocol. (I) Chart depicting the yields of fluorescent pyramidal cells using the standard method (n = 2) and our optimized method (n = 6), (Cell numbers shown in Tables 1 and 2). Cell numbers are increased 10-fold when trehalose is used. ** P < 0.001; Student’s t-test, error bars represent s.d.
To evaluate viability, isolated neurons were stained with trypan blue, by adding a 1:1 volume of 0.4% trypan blue to a small aliquot of the cell suspension. After 5 min incubation at room temperature, the number of viable (unstained) and dead (stained) cells were counted on a glass slide. Trypan blue staining revealed that trehalose treatment boosted cell viability from 58.1% to 81.0%. Although during FACS, the gating is adjusted to collect viable cells based on forward scatter, we also find that dead cells lose their fluorescence and are sorted out of the collection. Our direct comparison to the standard kit protocol reveals an overall improvement of 10-fold in cell yield (Figure 1 and Table 1). The poor cell numbers obtained with the standard protocol using gradient centrifugation appear to result from incomplete dissociation of tissue and greater cell death.
Table 1.
Direct comparison of the standard method with our optimized protocol.
| Parallel comparison between standard and optimized protocols | |||||
|---|---|---|---|---|---|
| THY1-YFP mouse id | Method | Cell numbers | RNA amount ng | RNA pg/cell | RIN |
| 1216a | standard | 8,500 | 2.05 | 0.24 | 6.8 |
| 1216b | standard | 17,770 | 1.35 | 0.08 | 7.7 |
| 1216c | optimized | 137,200 | 185.4 | 1.41 | 7.2 |
Data shown for standard protocol (n=2) and optimized protocol (n=1) obtained from each mouse cerebral cortex (P68) processed in parallel on the same day.
With our protocol we routinely collect an average of 120,842 pyramidal neurons (n = 24 mice) and 20,000 parvalbumin neurons (n = 24 mice) from each whole cortex, with an average RIN score of 7.9 as tested on the Bioanalyzer pico kit (Figure 1H and Table 2). Our protocol produces an average of 1.21 pg of RNA per pyramidal neuron, and 3.7 pg of RNA per parvalbumin neuron. Using trehalose we processed parvalbumin neurons from adult mice and prepared libraries for expression profiling using cDNA arrays (30). More recently, using this protocol, we prepared libraries from parvalbumin and pyramidal neurons with the nanoCAGE protocol (31, 32), and the data from these libraries is being prepared for submission. Using our method, profiling can also be performed using RNA seq (33), or a single cell can be plated per well to perform single cell transcriptome sequencing (34).
Table 2.
Data from adult mouse cortex processed using our optimized protocol.
| THY1-YFP mouse id | Age (days) | Cell number | RNA amount (ng) | pg/cell | RIN | Tag Count | Map rate(%) |
|---|---|---|---|---|---|---|---|
| 0824a | 68 | 140,000 | 274 | 1.96 | 7.9 | 7,362,128 | 99.8 |
| 0824b | 68 | 100,000 | 208 | 2.08 | 7.1 | 7,319,088 | 99.9 |
| 0904 | 60 | 168,500 | 270 | 1.60 | 8.0 | 6,388,969 | 99.9 |
| 0907 | 58 | 160,000 | 320 | 2.00 | 8.3 | 7,306,675 | 99.9 |
| 0828 | 70 | 120,000 | 262 | 2.18 | 8.2 | 6,711,929 | 99.9 |
Data shown for optimized protocol on cerebral cortex processed by the same operator on different days.
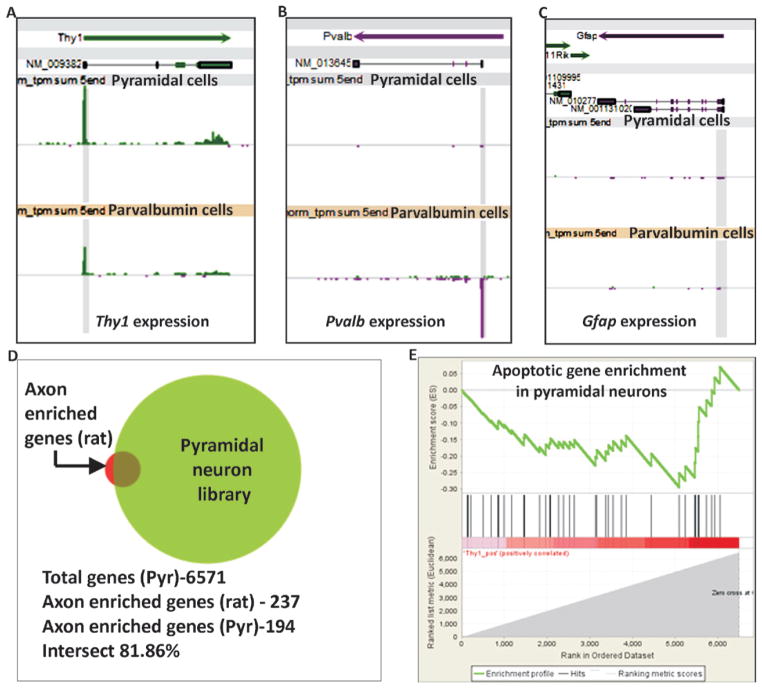
To investigate the purity of collected cells, we analyzed the nanoCAGE libraries prepared from sorted pyramidal neurons for the expression of Pvalb (parvalbumin neuron-specific marker) or Gfap (glial cell-specific marker). We found that the average expression of y1 promoter is almost 306-fold higher (306 tags/million/library) than Pvalb (1.24 tags/million/library) (Figure 2A and B) and 100 fold higher than Gfap (3.09 tags/million/library) (Figure 2C) in our wildtype pyramidal neuron libraries (n = 12), indicating successful isolation of a pure population of y1 expressing neurons alone. Similar analyses of libraries prepared from wildtype parvalbumin neurons (n = 6) reveal that the average expression of Pvalb (427.52 tags/million/library) is higher than Gfap (0.26 tags/million/library) (Figure 2B and 2C). Interestingly, in our parvalbumin neuron libraries we find endogenous expression of y1 (114 tags/million/library) (Figure 2A) suggesting y1 may be co-expressed with Pvalb in some cells, as has been reported previously for the somatosensory cortex (10).
Figure 2. Quality of libraries prepared using RNA obtained with our optimized protocol.
(A–C) Expression levels of Thy1, Pvalb and Gfap in libraries prepared using our optimized protocol. Each snapshot shows Refseq transcripts in the top section and tags per million (tpm) in libraries in the middle and bottom sections. Fixed expression scale of 1000 is shown for all panels. Pyramidal cell libraries (n = 12) and parvalbumin cell libraries (n = 6) as labeled. Tags at the transcript start site (TSS) are shaded in gray. Thy1 expression is found in both pyramidal and parvalbumin cells. Pvalb expression is seen in Parvalbumin libraries alone and Gfap expression is not seen in either of the libraries. (D) Graphical representation of the intersection between axon-enriched mRNAs cataloged in rat (237 genes) (38) and our adult mouse pyramidal neuron nanoCAGE libraries (6571 genes), showing the presence of 194 axons enriched genes in our pyramidal libraries. (E) GSEA plot depicting the enrichment score of genes up-regulated in programmed cell death, Enrichment score (ES) −0.29 and Normalized Enrichment Score (NES) −0.96. The analysis reveals that our pyramidal neuron libraries are not enriched in genes up-regulated during programmed cell death.
To confirm that the FACs sorted cells were viable, we investigated the pyramidal neuron library for genes reported to be up-regulated during programmed cell death (PCD) of cortical neuronal cells (35). Yang et al. cultured mouse cortical cells from E14 brains and identified 69 genes highly expressed during apoptosis induced by serum starvation (35). For our analysis, we first created a list of over 6000 genes expressed in our pyramidal neuron library and ranked them by expression level. Gene set enrichment analysis (GSEA) (36, 37) of genes expressed in pyramidal neurons revealed that, of the 69 genes reported to be up-regulated in PCD, only 43 genes were found expressed in our pyramidal neurons and that these genes were not enriched in our libraries (Figure 2E). We further conducted GO analysis on the top 700 expressed genes in our Pyramidal neuron libraries. Our data reveal that of the 49 genes listed in the KEGG apoptosis pathway (mmu04210), only 2 genes (Nfkbia and Cycs) were in the top 10% genes expressed in our libraries, thus indicating that the collected neurons were viable.
To confirm whether our protocol retained RNA transcripts localized to neurites, we examined the nanoCAGE libraries for mRNAs reported to be enriched in axons. Since a list of axon-enriched mRNAs for mouse is not available, we compared the axonal mRNA list for mature 13 day-old cultured E18 rat cortical neurons created by Taylor et al. (38) with level 3 clusters in our nanoCAGE libraries made from adult (>P55) mouse pyramidal neurons with an average of 7.5 million tags per multiplexed library on the Illumina GA IIx. At this depth of sequencing, we found an 81% intersection of mRNAs enriched in rat cortical axons indicating that our protocol preserves axon enriched mRNAs (Figure 2D). The missing 19% of mRNAs from our libraries might reflect differences due to species (rat vs mouse), age (embryonic vs >P55) or sequencing depth.
In conclusion, we show that the use of trehalose greatly enhances cell viability, thus preventing RNA degradation during cell processing. Our technique of purification of labeled neurons can likely be applied to the adult brain of other mammals as well as complex heterogeneous tissues, where individual cell types are too fragile and therefore difficult to isolate/dissociate. Intact cells from the pre-sorting stage may be plated or sorted in individual wells for single cell profiling, as has been done more readily with peripheral neurons (39,40). Our method has the potential to facilitate the unprecedented creation of transcriptome catalogs from neuronal sub-populations from a single adult mouse brain.
Supplementary Material
Acknowledgments
We thank H. Monyer (Heidelberg Univ) for kindly providing Pvalb-EGFP mouse breeding pairs and P. Arlotta (Harvard Univ) for valuable advice. This work is supported by a JSPS Grant-in-Aid and postdoctoral fellowship to A.S.; Grant-in-aid for Scientific Research on Priority Areas (‘Integrative Brain Research’) No.17021047 from the Japanese Ministry of Education, Culture, Sports, Science & Technology (MEXT) and Human Frontiers Science Program grant (HFSP PGP 0018/2007-C) to T.K.H.; a 7th Framework grant (Dopaminet), MEXT Grant-in-Aid for Scientific Research (A) No.20241047 to P.C., Funding Program for Next Generation World-Leading Researchers by MEXT to P.C, and the National Human Genome Research Institute grant U54 HG004557 to P.C.; and MEXT funding to the RIKEN Brain Science and Omics Science Center.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/113878
Competing interests
The authors declare no competing interests.
References
- 1.Dougherty JD, Geschwind DH. Progress in realizing the promise of microarrays in systems neurobiology. Neuron. 2005;45:183–185. doi: 10.1016/j.neuron.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Saxena A, Carninci P. Whole transcriptome analysis: what are we still missing? Wiley Interdiscip Rev Syst Biol Med. 2011;3:527–543. doi: 10.1002/wsbm.135. [DOI] [PubMed] [Google Scholar]
- 3.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 4.Meyer AH, et al. In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J Neurosci. 2002;22:7055–7064. doi: 10.1523/JNEUROSCI.22-16-07055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedlund E, et al. Embryonic stem cell-derived Pitx3-enhanced green fluorescent protein midbrain dopamine neurons survive enrichment by fluorescence-activated cell sorting and function in an animal model of Parkinson’s disease. Stem Cells. 2008;26:1526–1536. doi: 10.1634/stemcells.2007-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou W, et al. Embryonic stem cells with GFP knocked into the dopamine transporter yield purified dopamine neurons in vitro and from knock-in mice. Stem Cells. 2009;27:2952–2961. doi: 10.1002/stem.216. [DOI] [PubMed] [Google Scholar]
- 7.Gurok U, et al. Laser capture microdis-section and microarray analysis of dividing neural progenitor cells from the adult rat hippocampus. Eur J Neurosci. 2007;26:1079–1090. doi: 10.1111/j.1460-9568.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 8.Khodosevich K, et al. Gene expression analysis of in vivo fluorescent cells. PLoS One. 2007;2:e1151. doi: 10.1371/journal.pone.0001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protocols. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- 10.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 11.Sawamoto K, et al. Direct isolation of committed neuronal progenitor cells from transgenic mice coexpressing spectrally distinct fluorescent proteins regulated by stage-specific neural promoters. J Neurosci Res. 2001;65:220–227. doi: 10.1002/jnr.1145. [DOI] [PubMed] [Google Scholar]
- 12.Murayama A, et al. Flow cytometric analysis of neural stem cells in the developing and adult mouse brain. J Neurosci Res. 2002;69:837–847. doi: 10.1002/jnr.10339. [DOI] [PubMed] [Google Scholar]
- 13.Rietze RL, et al. Purification of a pluri-potent neural stem cell from the adult mouse brain. Nature. 2001;412:736–739. doi: 10.1038/35089085. [DOI] [PubMed] [Google Scholar]
- 14.Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci USA. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wylie CJ, et al. Distinct transcriptomes define rostral and caudal serotonin neurons. J Neurosci. 2010;30:670–684. doi: 10.1523/JNEUROSCI.4656-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlotta P, et al. Neuronal subtype-specific genes that control corti-cospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 17.Guez-Barber D, et al. FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci. 2011;31:4251–4259. doi: 10.1523/JNEUROSCI.6195-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guez-Barber D, et al. FACS purification of immunolabeled cell types from adult rat brain. J Neurosci Methods. 2012;203:10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobo MK, et al. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 20.Brewer GJ, Torricelli JR. Isolation and culture of adult neurons and neurospheres. Nat Protocols. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- 21.Eroglu A, et al. Intracellular trehalose improves the survival of cryopreserved mammalian cells. Nat Biotechnol. 2000;18:163–167. doi: 10.1038/72608. [DOI] [PubMed] [Google Scholar]
- 22.Carninci P, et al. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci USA. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez-Peral FJ, et al. Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology. 2002;148:2599–2606. doi: 10.1099/00221287-148-8-2599. [DOI] [PubMed] [Google Scholar]
- 24.Erkut C, et al. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol. 2011;21:1331–1336. doi: 10.1016/j.cub.2011.06.064. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan SS, et al. Cryopreservation of stem cells using trehalose: evaluation of the method using a human hematopoietic cell line. Stem Cells Dev. 2004;13:295–305. doi: 10.1089/154732804323099226. [DOI] [PubMed] [Google Scholar]
- 26.Erdag G, et al. Cryopreservation of fetal skin is improved by extra-cellular trehalose. Cryobiology. 2002;44:218–228. doi: 10.1016/s0011-2240(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 28.Iwahashi H, et al. Barotolerance is dependent on both trehalose and heat shock protein 104 but is essentially different from thermotolerance in Saccharomyces cerevisiae. Lett Appl Microbiol. 1997;25:43–47. doi: 10.1046/j.1472-765x.1997.t01-1-00069.x. [DOI] [PubMed] [Google Scholar]
- 29.Pereira CS, Hunenberger PH. Effect of trehalose on a phospholipid membrane under mechanical stress. Biophys J. 2008;95:3525–3534. doi: 10.1529/biophysj.108.131656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plessy C, et al. A resource for transcriptomic analysis in the mouse brain. PLoS One. 2008;3:e3012. doi: 10.1371/journal.pone.0003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plessy C, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salimullah M, et al. NanoCAGE: A High-Resolution Technique to Disco er and Interrogate Cell Transcriptomes. Cold Spring Harb Protoc. 2011;2010:pdb prot5559. doi: 10.1101/pdb.prot5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta S, et al. Highly consistent, fully representative mRNA-Seq libraries from ten nanograms of total RNA. BioTechniques. 2010;49:898–904. doi: 10.2144/000113556. [DOI] [PubMed] [Google Scholar]
- 34.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 35.Yang MH, et al. The gene expression profiling in murine cortical cells undergoing programmed cell death (PCD) induced by serum deprivation. J Biochem Mol Biol. 2007;40:277–285. doi: 10.5483/bmbrep.2007.40.2.277. [DOI] [PubMed] [Google Scholar]
- 36.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor AM, et al. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tietjen I, et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38(2):161–75. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 40.Gustincich S, et al. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc Natl Acad Sci U S A. 2004;101(14):5069–74. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.