Abstract
Changes in global DNA linking number can be accommodated by localized changes in helical structure. We have used single-molecule torque measurements to investigate sequence-specific strand separation and Z-DNA formation. By controlling the boundary conditions at the edges of sequences of interest, we have confirmed theoretical predictions of distinctive boundary-dependent backbending patterns in torque-twist relationships. Abrupt torque jumps are associated with the formation and collapse of DNA bubbles, permitting direct observations of DNA breathing dynamics.
Force spectroscopy of biomolecules has stimulated general insights into the thermodynamics of small systems [1], including experimental tests of results from nonequilibrium statistical mechanics [2–4] and theoretical investigations of ensemble inequivalence [5–7]. To enable analogous contributions from torque spectroscopy, we have recently introduced a high-resolution approach [8] for measuring the torsional response of short (20–100 bp) sequences of interest (SOI), building on previous torque measurements that revealed sequence-averaged properties of long (2–15 kilobasepair) duplexes [8–10]. Local sequence-specific responses to supercoiling are relevant to the regulation of biological processes such as transcription [11] and replication [12, 13]. The associated torque signatures [8] illustrate general features of finite systems with constrained extensive properties, analogous to an Ising model at fixed magnetization (IMFM) [14].
In our initial study [8], we measured torque signatures of left-handed Z-DNA formation in d(pGpC)n sequences and disruption of strand-strand interactions in mismatched duplexes. For all the sequences we analyzed, we found good agreement with a 1D theoretical treatment of cooperative transitions, in which each basepair can adopt two available helical states, and junctions are disfavored by a domain wall penalty J. However, we performed only limited tests of theoretical predictions; we were unable to study the biologically important phenomenon of localized AT-rich strand separation [12]; and we did not investigate reversible dynamics.
For the current study, we designed a new set of molecular constructs and characterized their responses using the “static RBT” [8, 15] method. We previously described two distinct methods for measuring torque-twist relationships [8]. Both methods are based on rotor bead tracking (RBT), in which a DNA molecule is stretched using a micron-scale magnetic bead as a force handle, and rotation is monitored using a submicron non-magnetic bead attached to the side of the DNA. In “dynamic RBT”, torque is measured via the angular velocity of a calibrated viscous load. In static RBT, a reference DNA segment is used as a calibrated torsional spring (Fig. 1a). We have chosen static RBT here because it allows the use of smaller rotational probes, improving torque resolution [8, 15].
FIG. 1. Static RBT assay for high-resolution torque spectroscopy.
(a) A DNA tether is attached between the cover glass surface and a magnetic bead (MB, orange), with torsional constraints at both ends. A rotor bead (RB, green) is attached site-specifically to the side of the DNA molecule. The total twist θ is controlled by the angular position of the magnets. Transducer twist α is given by α = θ − ψ, where ψ is the angular position of the RB. The torque τ acting on the DNA can be calculated as τ = κTDα, where κTD is the torsional spring constant of the calibrated transducer segment. A sequence of interest (SOI, red) is inserted in the lower DNA segment. (b) Torque is plotted as a function of imposed twist for a DNA containing a 50 bp long d(pGpC)-repeat SOI (Z50). Data are shown for winding in the (+) direction only (see SFig. S1 for overlaid unwinding and rewinding curves). Raw 250 Hz data (gray) were averaged in 0.2 rotation bins (red) and fit to a model for cooperative structural transitions in polymers [8] with fixed boundary conditions (black line, Equation S7). The curve shows a transition between two linear elastic regions corresponding to B-DNA and Z-DNA (gray lines). Superhelical density σ = ΔLk / Lk0, where Lk0 is the linking number of the relaxed DNA (Lk0 = Ntotal / 10.4, where Ntotal is the length of the full DNA construct in basepairs) and ΔLk = θ for our experimental geometry.
Figure 1b shows the response of a 50 bp long GC-repeat (Z50), a representative SOI that we previously investigated extensively using dynamic RBT [8]. For small twisting deformations, the molecule shows linear torsional elasticity characteristic of standard right-handed B-form DNA. At high levels of underwinding, the torsional response switches to a second linear region; the negative shift along the twist axis is consistent with a transformation to left-handed Z-DNA. The intervening region shows a characteristic abrupt torque jump associated with formation of a Z-DNA domain, and a B-Z coexistence region in which the torque is approximately constant. As seen in our dynamic RBT experiments [8], the torque-twist relationship is well described by an Ising-type model (Supplementary Equations) whose parameters include the changes in twist Δθ0, free energy ΔG0, and torsional compliance Δct associated with the state transition for a single basepair. The best-fit values for these parameters and for the domain wall penalty J are in good agreement with our dynamic RBT results (SFig. S1 and STab. S1).
We analyzed boundary effects in order to design rigorous challenges of our previous model. In the Z50 construct described above, the SOI is embedded in a continuous sequence with flanking regions of low Z-propensity. Our corresponding model calculations make use of “fixed boundary conditions”: we assume that the bases adjacent to the edges of the SOI remain in B-form, and a domain wall penalty is assessed if a basepair at the edge of the SOI adopts the Z-DNA state. In an alternative scenario, under “free boundary conditions”, the SOI is considered to be uncoupled from its surroundings, and there is never a domain wall penalty assessed at either edge of the SOI. For a given parameter set, these two conditions can predict very different responses (Fig. 2).
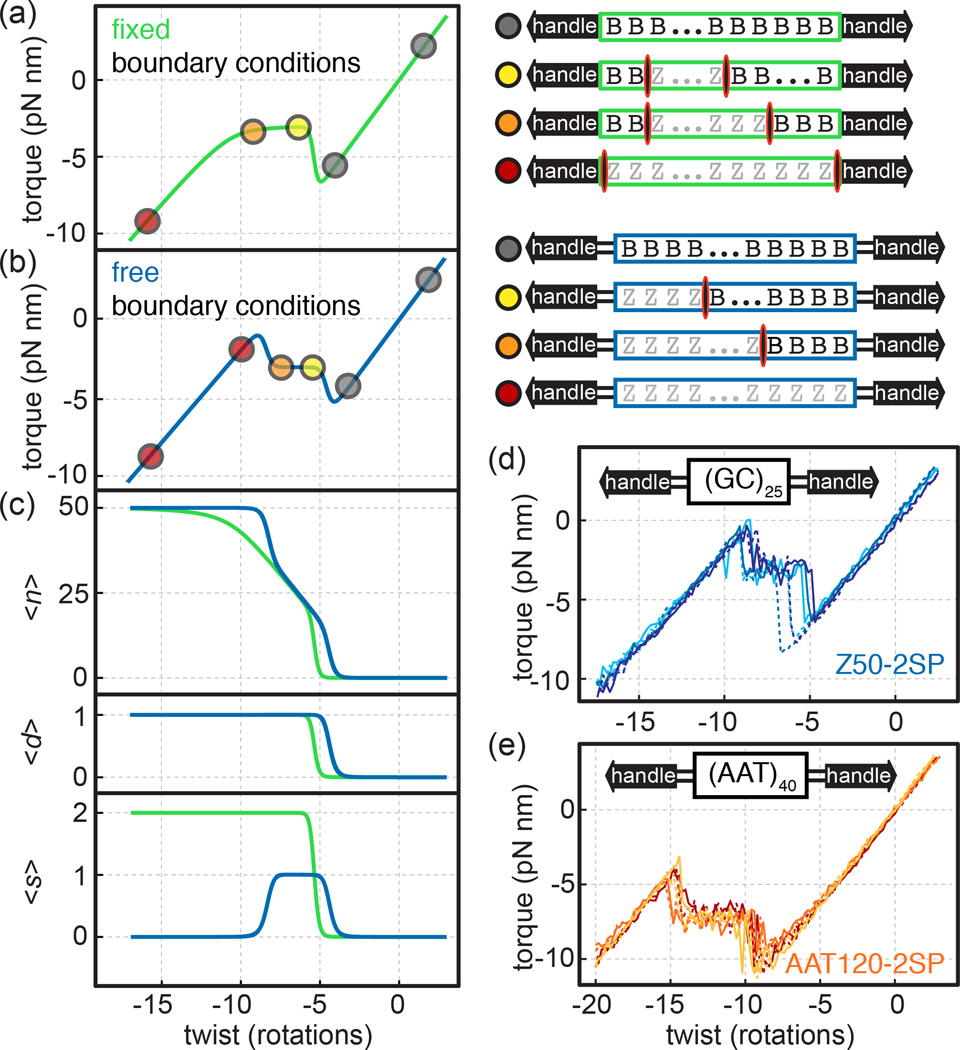
FIG. 2. Torque-twist signatures of free boundary conditions and the behavior of a 100% AT sequence.
(a) Average values for torque and other variables were calculated as a function of imposed twist for a transition with fixed boundary conditions using parameters measured for Z50 (STab. S1). Schematic depictions show representative microstates for portions of the torque-twist curves marked with colored dots. The SOI is outlined as a green rectangle. Domain walls are shown as red/black ovals. (b) Calculations and illustrations for the transition of Z50 under free boundary conditions (blue), shown as in (a). (c) calculations of n, number of nucleotides in the high-energy state; d, number of domains in the high-energy state; s, number of domain walls for Z50 as function of imposed twist under fixed (green) and free (blue) boundary conditions. (d)–(e) Measured torque-twist plots. For each construct, three independent experiments are shown; dashed and solid lines indicate (+) and (−) winding directions, respectively. (d) Z50 SOI flanked by atomic spacers (Z50-2SP). (e) 120 bp AAT-repeat SOI flanked by atomic spacers (AAT120-2SP).
In our treatment of Z50 and in related physical models, domain wall penalties lead to “overshoot” features in plots of mean torque as a function of imposed twist. Under fixed boundary conditions, there is an abrupt torque jump at the beginning of the transition as an initial domain is formed (Fig. 1b, 2a and SFig. S2). This domain then grows, and domain walls persist in the final all-Z configuration. In contrast, under free boundary conditions (Fig. 2b, c and SFig. S2) there are no domain wall penalties in the final configuration, and as a result there are two abrupt transitions: a torque “overshoot” as an initial nucleus is formed, followed later by an “undershoot” as the molecule abruptly transitions from partially converted to fully converted, ridding itself of domain walls. We previously presented the partition functions for both fixed and free boundary conditions [8]. We also described an experimental realization proposed to approximate free boundary conditions, in which atomic spacers placed at the edges of the SOI isolate the sequence from the surrounding DNA, while preserving a torsional constraint [8] (Tab. S3). However, we tested these atomic spacers only with short sequences that show two-state behavior, and did not discuss or investigate the “overshoot-undershoot” behavior described above.
AAT- and GC-repeats show torque “undershoots” predicted for free boundary conditions
We measured torque as a function of imposed twist for two new constructs in which a long SOI is flanked by atomic spacers. In Z50-2SP, the SOI is the 50 bp Z-forming GC-repeat studied earlier. In AAT120-2SP, we used a 120 bp AAT-repeat that we predicted would readily undergo strand separation. Both constructs show the predicted overshoot and undershoot features surrounding the torque plateau (Fig. 2d,e). Torque-twist plots collected for AAT120-2SP show no perceptible hysteresis under our conditions, and we have used averaged unwinding and rewinding curves for subsequent equilibrium analysis. For Z50-2SP, we observe some hysteresis, and have used rewinding curves as an approximation for equilibrium curves in subsequent analysis, as before [8].
To allow quantitative fits to torque-twist curves containing atomic spacers, we introduce a generalized model that includes “free” and “fixed” boundary conditions as special cases (Equation S13). We account for variable coupling with the surrounding DNA by including a parameter J’ that represents a penalty assessed for a domain wall that occurs at the edge of the SOI. This model reduces to fixed boundary conditions for J’ = J and to free boundary conditions for J’ = 0 (Fig. 3a, inset). By fitting the response of Z50-2SP using this model, we measure ΔG = 0.5 kcal/(mol bp), J = 5.6 kcal/mol, and Δθ0 = −1.1 rad/bp, agreeing well with fits to spacer-free Z50 (Fig. 3a and STab. S1). As expected, the edge-specific domain wall penalty was found to be low, J’ = 1 kcal/mol, supporting our proposal that atomic spacers create a close approximation to free boundary conditions. To further test this result and challenge our model, we constructed DNA tethers with a spacer on only one side of the SOI (Z50-1SP, Fig. 3b), and fit to a one-sided generalized model (Equation S14). Measurements closely match the predicted shape of the torque-twist relationship, and the parameters extracted from Z50-1SP data are in good agreement with Z50-2SP measurements, including J’ ≈ 1 kcal/mol (Fig. 3b and STab. S1).
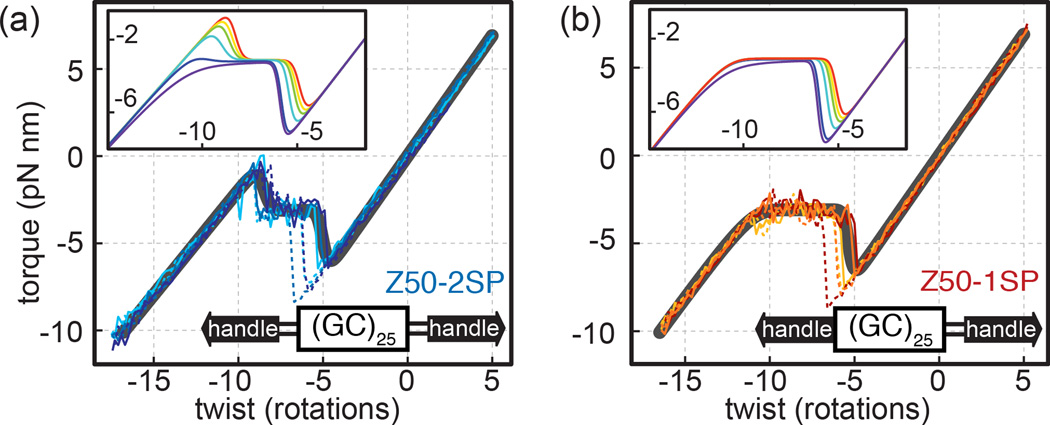
FIG. 3. The effect of boundary conditions on transitions in GC-repeats.
(a) Torque-twist plots for Z50-2SP acquired during unwinding (dashed blue lines) and rewinding (solid blue lines) together with a fit (grey) to a generalized model (Supplementary Equations) which includes a variable boundary penalty J’ for domain walls at the edges of the SOI. Inset, calculated plots for J’ = 0, 0.5, 1, 2, 4 and J’ = J = 5.6 kcal/mol are in red, yellow, green, light blue, dark blue and violet, respectively. All other parameters have been kept constant (STab. S1, Z50-2SP). The generalized model reduces to free boundary conditions for J’ = 0 and to fixed boundary conditions for J’ = J. (b) Torque-twist plots for Z50-1SP, which contains an atomic spacer on only one side of the SOI, are shown together with a fit to a one-sided model. Inset shows the effect of varying J’ on the one-sided model, with color-coding as in (a) ranging from J’ = 0 to J’ = J = 6.3 kcal/mol (all other parameters taken from STab. S1, Z50-1SP).
AAT-repeats undergo strand-separation when underwound
We used the generalized model to extract thermodynamic and structural parameters from the AAT120-2SP data (Fig. 4b, STab. S1 and S2). For this equilibrium torque-twist curve, the best fit J’ = 0.5 kcal/mol reinforces our conclusion that atomic spacers efficiently abrogate coupling. The fit values Δθ0 = −0.6 rad/bp and ΔG = 0.6 kcal/(mol bp) are consistent with expectations for strand separation, based on (1) assuming complete loss of B-form helicity (B-form twist = 0.60 rad/bp) and (2) predicting the thermodynamic stability of AAT repeats from the unified view of polymer nearest-neighbor thermodynamics for strand separation [16] or more recent force spectroscopy analysis [17]. The best fit J = 7 kcal/mol provides a direct measurement of the domain wall penalty underlying the cooperativity of DNA melting, and is slightly larger than previous estimates based on thermal melting [18] and bulk topoisomer analysis [19, 20]. The AAT120-2SP measurements provide a model system for superhelically driven destabilization in AT-rich sequences, and are also directly relevant to the behavior of AAT triplet repeats in vivo; previous bulk measurements have shown that AAT repeats undergo helix opening in supercoiled plasmids [21]. Spacer-free AAT120 constructs more closely represent AT-rich sequences in their biological context; torque-twist plots for these constructs also show clear transitions, which display the fingerprint of fixed boundary conditions as expected (SFig. S5).
FIG. 4. The effect of SOI length on transitions in AAT-repeats.

(a) Series of calculated torque-twist plots using the model with free boundary conditions and SOI lengths ranging from N=20 to N=120 bp (all other parameters fixed based on AAT120-2SP, STab. S1). (b) Torque-twist plots of three independent AAT120-2SP unwinding (dashed colored lines) and rewinding (solid colored lines) experiments are overlaid with model fits (gray bold line). (c) Torque-twist plots of three independent AAT50-2SP unwinding (dashed) and rewinding (solid) experiments together with a fit in which ΔG0, J, and J’ are fixed based on AAT120-2SP (STab. S1). (d) Torque-twist plots of three independent AAT21-2SP unwinding (dashed) and rewinding (solid) experiments. The model displayed (gray bold line) uses the parameters shown in supplementary table S1 (AAT21-2SP).
Short AAT repeats show two-state behavior
As discussed, SOIs with large Ns under free boundary conditions show two abrupt torque jumps flanking a plateau. Lowering N is expected to shorten the transition plateau until the torque jumps merge into a single rip, approximating a two-state transition (Fig. 4a, SFig. S3 and S4). Measurements on AAT-repeats show the expected behavior. We tested AAT-repeats with a total of N = 50 (AAT50-2SP) and N = 21 (AAT21-2SP) basepairs (Fig. 4b and 4c). The measured torque response of AAT50-2SP and AAT21-2SP match our predictions and are consistent with parameters measured for AAT120-2SP (Fig. 4 and STab. S1).
Short-lived dynamic duplex opening (DNA “breathing”) has been studied because of its relevance to kinetic mechanisms of biological processes [22, 23]. AAT21-2SP exhibits 2-state behavior and shows reversible transitions under our conditions (Fig. 4c, 5a and SFig. S4). We observe reversible hopping between the two states (strand-separated or “open” and B-form or “closed”) during twist ramps (Fig. 5a). Long observations at fixed twist show many reversible transitions and allow the measurement of state lifetimes for open and closed conformations. An increase in negative twist shifts the equilibrium from the closed to the open state (Fig. 5b,c and SFig. S6).
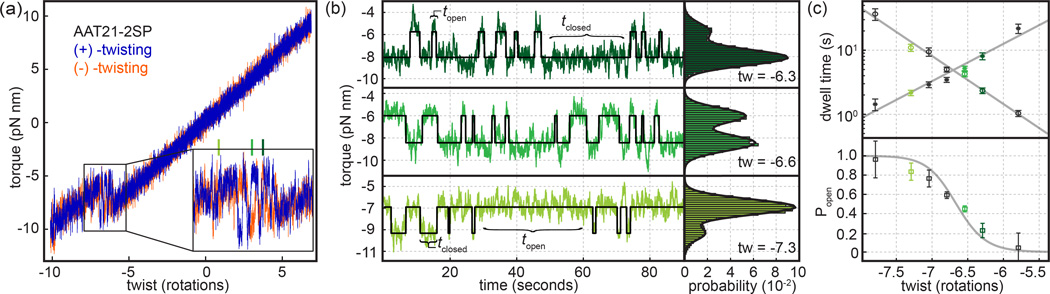
FIG. 5. “Breathing” dynamics of 21 bp AAT-repeats.
(a) Torque response of the AAT21-2SP SOI during linear twist ramps (10 deg/s) in the (−)-twisting (red) and (+)-twisting (blue) directions. The inset contains an expanded view of the transition region, showing reversible hopping. Green lines are used to indicate twist values used for fixed-twist dynamic experiments shown in subsequent panels. (b), Raw data (green traces) are overlaid with 2-state idealizations (black line) used to measure topen and tclosed as shown. Torque histograms at the right of each panel were generated from the entire 800 s of data collected at each twist condition, and are overlaid with double Gaussian fits. (c) Semi-log plot of mean dwell times <topen> and <tclosed> as a function of total imposed twist (top panel). Probabilities of being in the open state at fixed twists are fit using a two-state model (Equation S2). The fit gives a total ΔG0 of 12 kcal/mol; κ0, Δθ0 and Δct were fixed based on linear fits to the torque-twist curve for this SOI (AAT21-2SP) as described in STab. S1.
Backbending features in isotherms are a general property of cooperative transitions in small systems under external constraints
These features have been analyzed theoretically in the IMFM [14], which has been applied to diverse phenomena [24] including transitions of adatom clusters on a surface [25]. Torque overshoots in the fixed linking number ensemble have also been studied in the case of plectonemic buckling: abrupt torque jumps at this transition have been inferred to accompany abrupt extension jumps [26, 27]; they have been predicted from theory and simulation [27, 28], measured with the help of lock-in detection [29], and directly observed using dynamic RBT and electromagnetic torque tweezers [8, 30]. At the buckling transition, the free energy associated with forming a plectonemic end loop [28] plays a role analogous to the domain wall penalty J, adding an energetic cost to the initial formation of a plectonemic domain.
Local strand separation in superhelical DNA is critical for biological functions [22, 23, 31]. In this study, we have observed duplex opening dynamics and have also measured structural, mechanical, and thermodynamic parameters for strand separation alongside measurements of the competing [32] B-Z transition (STab. S2 and accompanying note). After parameterization with simple SOIs, our theoretical treatment of cooperative structural transitions can be extended to predict the torque responses of complex biological sequences.
Supplementary Material
Acknowledgements
We would like to thank the members of the Bryant group for useful discussions and comments on the manuscript. This work was supported by a Pew Scholars Award and NIH grant OD004690 to ZB; by a Morgridge Family Stanford Graduate Fellowship to LEF; by a Stanford Interdisciplinary Graduate Fellowship and a Natural Sciences and Engineering Research Council of Canada (NSERC PGS-D) award to PL; and by an EMBO long-term fellowship, a Stanford University Dean’s Fellowship, and a Swiss National Science Foundation fellowship for advanced researchers to FCO.
REFERENCES
- 1.Mossa A, Huguet JM, Ritort F. Physica E-Low-Dimensional Systems & Nanostructures. 2010;42 [Google Scholar]
- 2.Liphardt J, et al. Science. 2002;296 doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 3.Collin D, et al. Nature. 2005;437 doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemany A, et al. Nat Phys. 2012;8 [Google Scholar]
- 5.Keller D, Swigon D, Bustamante C. Biophys J. 2003;84 doi: 10.1016/S0006-3495(03)74892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha S, Samuel J. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71 doi: 10.1103/PhysRevE.71.021104. [DOI] [PubMed] [Google Scholar]
- 7.Suzen M, Sega M, Holm C. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79 doi: 10.1103/PhysRevE.79.051118. [DOI] [PubMed] [Google Scholar]
- 8.Oberstrass FC, Fernandes LE, Bryant Z. Proc Natl Acad Sci U S A. 2012;109 doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant Z, et al. Nature. 2003;424 [Google Scholar]
- 10.Sheinin MY, et al. Phys Rev Lett. 2011;107 doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayan V, Zuzow R, O'Shea EK. Proc Natl Acad Sci USA. 2009;106 doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalski D, Natale DA, Eddy MJ. Proc Natl Acad Sci USA. 1988;85 doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Freiesleben U, Rasmussen KV. Res Microbiol. 1992;143 doi: 10.1016/0923-2508(92)90060-2. [DOI] [PubMed] [Google Scholar]
- 14.Gulminelli F, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68 doi: 10.1103/PhysRevE.68.026119. [DOI] [PubMed] [Google Scholar]
- 15.Bryant Z, Oberstrass FC, Basu A. Curr Opin Struct Biol. 2012;22 doi: 10.1016/j.sbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SantaLucia J., Jr Proc Natl Acad Sci U S A. 1998;95 doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huguet JM, et al. Proc Natl Acad Sci USA. 2010;107 [Google Scholar]
- 18.Amirikyan BR, Vologodskii AV, Lyubchenko Yu L. Nucleic Acids Res. 1981;9 doi: 10.1093/nar/9.20.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benham CJ. J Mol Biol. 1992;225 doi: 10.1016/0022-2836(92)90404-8. [DOI] [PubMed] [Google Scholar]
- 20.Bauer WR, Benham CJ. J Mol Biol. 1993;234 doi: 10.1006/jmbi.1993.1669. [DOI] [PubMed] [Google Scholar]
- 21.Ohshima K, et al. J Biol Chem. 1996;271 doi: 10.1074/jbc.271.4.1853. [DOI] [PubMed] [Google Scholar]
- 22.Krueger A, Protozanova E, Frank-Kamenetskii MD. Biophys J. 2006;90 doi: 10.1529/biophysj.105.078774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambjornsson T, et al. Biophys J. 2007;92 doi: 10.1529/biophysj.106.095935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pleimling M, Huller A. Journal of Statistical Physics. 2001;104 [Google Scholar]
- 25.Selke W, Müller T. Eur Phys J B. 1999;10 [Google Scholar]
- 26.Forth S, et al. Phys. Rev. Lett. 2008;100 doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brutzer H, et al. Biophys J. 2010;98 doi: 10.1016/j.bpj.2009.12.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marko JF, Neukirch S. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85 doi: 10.1103/PhysRevE.85.011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniels BC, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80 doi: 10.1103/PhysRevE.80.040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen XJ, et al. Nano Lett. 2012 doi: 10.1021/nl301330h. [DOI] [PubMed] [Google Scholar]
- 31.Alexandrov BS, et al. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkp1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditlevson JV, et al. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.