Background: Chitinase C from Ralstonia sp. A-471 (Ra-ChiC) is a chitinase that was first found in glycohydrolase family 23.
Results: The crystal structure of Ra-ChiC exhibited a tunnel-shaped conformation in its active site.
Conclusion: The tunnel-shaped conformation is essential for a unique arrangement of the catalytic residues and substrate specificity.
Significance: This is the first report on the tunnel-shaped binding site of an inverting chitinase.
Keywords: Chitinase, Crystal Structure, Glycoside Hydrolases, Mutagenesis Site Specific, Oligosaccharide
Abstract
Chitinase C from Ralstonia sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose-type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptidoglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu-141 acts as a catalytic acid, and that Asp-226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
Introduction
Chitin, a β-1,4 polymer of N-acetyl-d-glucosamine (NAG),3 is one of the most abundant biopolymers in nature. It is easily obtained from natural resources such as shrimp and crab shell waste. It is estimated that at least 10 gigatons (1 × 1013 kg) of chitin are synthesized each year, of which the vast majority is discarded (1). Chitin could be a significant biological resource because N-acetyl-chitooligosaccharides ((NAG)n) and chitooligosaccharides, products of chitin hydrolysis, have a variety of biological functions including antibacterial activity and antitumor activity (2). In nature, chitin is degraded by chitinases (EC 3.2.1.14) that belong to glycohydrolase (GH) families 18 and 19 (3, 4). These two families do not have sequence or structural similarities and act via different catalytic mechanisms. GH family 18 chitinases are distributed in various organisms from microbes to human. Their catalytic domain has an (α/β)8-barrel structure and utilizes a double-displacement mechanism, which results in the net retention of an anomeric configuration (retaining mechanism) (5–7). In contrast to GH18 chitinases, GH family 19 chitinases are found only in plants and some bacteria (8, 9). Their overall structures show high α-helical contents (10–12), and possess an active site topology similar to those of lysozymes from hen (GH22), goose (GH23), and phages (GH24) (13–15). GH family 19 chitinases utilize a single-displacement mechanism, which results in an inverted anomeric configuration (inverting mechanism) (16).
We previously cloned the gene encoding a chitinase from a moderate thermophilic strain Ralstonia sp. A-471 (Ra-ChiC) (17). Ra-ChiC comprises a putative signal peptide (residues 1–35), a chitin-binding domain (residues 36–80), an interdomain linker (residues 81–102), and a catalytic domain (residues 103–252). Ra-ChiC produces α-anomers by hydrolyzing β-1,4-glycosidic linkages, indicating that the enzyme is an inverter like GH family 19 chitinases. Ra-ChiC is stable up to 50 °C and retains considerably high activity when incubated with high concentrations of organic solvents such as 50% acetonitrile, 50% methanol, and 50% ethanol. This enzyme is also stable over pH range 5.0–10.0. In addition, the enzyme is also active toward various chitin derivatives, including ethylene glycol chitin, CM-chitin, and soluble chitin (50% deacetylated chitin). Because the enzymatic activity is barely affected by metal ions, except for Hg+, Ra-ChiC could be useful for industrial applications (17).
Ra-ChiC is the first chitinase classified as GH family 23 (CAZy database) (18), which has been recognized to include goose-type (G-type) lysozymes, peptidoglycan lyases, and peptidoglycan lytic transglycosylases. Ra-ChiC cleave the β-1,4-glycosidic bonds between NAG residues in chitin, whereas the other GH23 members act toward the bonds between N-acetyl-d-muramic acid (NAM) and NAG in peptidoglycan. Because of the chemical similarities of their substrates, some chitinases can hydrolyze peptidoglycans less efficiently in addition to chitin and, conversely, some lysozymes can hydrolyze chitin (19–22). However, Ra-ChiC exhibits only chitinase activity but not the activity toward peptidoglycan (17). In addition, Ra-ChiC differs from G-type lysozymes in their splitting mode of NAG oligosaccharide substrates, (NAG)n: for example, the NAG hexamer substrate, (NAG)6, is split predominantly into (NAG)3 + (NAG)3 by G-type lysozymes, whereas the same substrate is split also into (NAG)2 + (NAG)4 by Ra-ChiC. The subsite structure of Ra-ChiC appears to differ from those of G-type lysozymes.
Inverting glycoside hydrolases have been recognized to require two acidic amino acids for their catalytic reaction; one acts as a general acid and the other as a general base. The distance between the carboxyl groups of acidic residues was proposed to be ∼8–10 Å (23, 24). G-type lysozymes have a highly conserved glutamic acid residue, Glu73, as a general acid (25–27). In addition, Helland et al. (28) suggested that Asp101 in Atlantic cod G-type lysozyme (AcLYZ), which is at a distance of 8.5 Å from Glu73 acts as a general base in the catalytic reaction. Sequence alignments of Ra-ChiC and AcLYZ suggested that Glu73 and Asp101 of AcLYZ are conserved in Ra-ChiC as Glu141 and Glu162, respectively (17), which are the best candidates for catalytic residues. However, this is only a suggestion from sequence comparison, and the catalytic residues of the unusual chitinase, Ra-ChiC, remains to be elucidated by the structural and mutation analyses.
Here we report the binding specificity of Ra-ChiC as determined by NMR spectroscopy and the first crystal structures for the catalytic domain of Ra-ChiC in the presence and absence of (NAG)2, an E141Q mutant of the catalytic domain in the presence of (NAG)4, and an E162Q mutant of the catalytic domain in the presence and absence of (NAG)2. We found that Ra-ChiC has a unique substrate-binding site compared with other GH family 23 enzymes. In addition, we carried out a mutation analysis of acidic amino acid residues located at the active site, and found that Glu141 is a catalytic acid. Contrary to the suggestion from the sequence comparison, Asp226 rather than Glu162 was found to act as a catalytic base. Based on structural and mutation analyses, we also discuss a catalytic mechanism for Ra-ChiC.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Protein expression and purification of Ra-ChiC segments, which included residues 36–252 (Ra-ChiCmature) and 89–252 (Ra-ChiCcat) with an N-terminal His6 tag, were previously described (29). The mutants derived from Ra-ChiCmature and Ra-ChiCcat were produced by site-directed mutagenesis using a Stratagene QuikChange II kit (Stratagene, La Jolla, CA). The forward and reverse oligonucleotide primers are shown in supplemental Table S1. The mutants E141Q, E141A, E141D, E141N, E162Q, E162A, E162D, E162N, D226N, and D226A were created using the synthesized oligonucleotide primers. Selenomethionine-labeled Ra-ChiCcat was overexpressed using the methionine pathway inhibition in LeMaster Broth (30–32). The E141Q, E162Q, and selenomethionine Ra-ChiCcat proteins were purified using the same procedure as wild-type Ra-ChiCcat. To produce 15N-labled Ra-ChiCmature, Escherichia coli cells harboring the expression plasmid were grown in M9 minimal medium containing 15N-NH4Cl (0.5 g/liter). Production and purification of the labeled Ra-ChiCmature was done as described above.
NMR Spectroscopy
NMR samples contained 0.1 mm protein in 50 mm sodium acetate buffer, pH 5.0 (90% H2O/10% D2O). All NMR spectra were acquired at 300 K using a Bruker AV500 spectrometer controlled with TopSpin 3.0 software and equipped with a triple-resonance pulsed-field gradient cryoprobe head. All spectra were processed using NMRPipe software and analyzed using Sparky software. Two ligands, (NAG)2 and a peptide-glycan fragment, N-acetylglucosaminyl-(β-1,4)-N-acetylmuramyl-l-alanyl-d-isoglutamine (NAG-NAM-AiQ, Calbiochem Co.), were used for NMR titration experiments.
Crystallization
Purified Ra-ChiCcat was concentrated to 4.5–6.5 mg/ml for crystallization. Crystallization conditions are shown in supplemental Table S2. Crystallization was carried out by the hanging-drop vapor diffusion method at 20 °C. All drops included equal volumes of the protein and reservoir solutions. Crystals of E141Q and E162Q mutants were obtained by the micro-seeding method using wild-type crystals as a seed, and the (NAG)n-bound forms were obtained by the soaking method.
Data Collection, Processing, Phasing, and Structure Refinement
Diffraction data were collected at 100 K with 20% (v/v) 2-methyl-2,4-pentanediol in the mother liquid as a cryoprotectant at beamlines BL38B1 and BL41XU at SPring-8 (Harima, Japan) and at beamlines NW12A, NE3A, BL-1A, BL-5A, and BL-17A at Photon Factory (Tsukuba, Japan). The data sets for the selenomethionine crystal were collected at 0.9641 Å (remote (high)), 0.9950 Å (remote (low)), 0.9789 Å (peak), and 0.9730 Å (edge) for multiple wavelength anomalous diffraction phasing (supplemental Table S3). Data were processed and scaled using the HKL2000 suite (33). Initial phasing, phase improvement, and auto model building were performed by SOLVE, DM, and buccaneer, respectively (34–36). The crystals belonged to space group P6122 with unit cell parameters a = b = 100 Å, c = 243 Å and contained four Ra-ChiCcat molecules (MolA, MolB, MolC, and MolD) in the asymmetric unit. Refinement was carried out using REFMAC5, including refinement of atomic displacement parameters by translation, liberation, and screw method, with each monomer in the asymmetric unit treated as a single translation, liberation, and screw group (37). Manual model modification was performed by Coot (38). Data collection statistics and refinement parameters are given in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses correspond to the highest resolution shell.
| WT | WT-NAG2 | E141Q-NAG4 | E162Q-HEPES | E162Q-NAG2 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 1.0000 | 1.0000 | 1.1000 | 0.9800 | 0.9800 |
| Space group | P6122 | P6122 | P6122 | P6122 | P6122 |
| Cell dimensions (Å) | |||||
| a, b | 99.7 | 99.0 | 99.4 | 99.9 | 99.3 |
| c | 243 | 242 | 243 | 241 | 242 |
| Resolution (Å) | 1.90 (1.97-1.90) | 2.00 (2.03-2.00) | 2.15 (2.19-2.15) | 2.15 (2.19-2.15) | 2.10 (2.14-2.10) |
| Completeness (%) | 97.5 (95.5) | 97.6 (92.4) | 99.9 (100.0) | 99.9 (99.8) | 99.6 (99.7) |
| Rmerge (%)a | 7.6 (39.8) | 13.7 (29.6) | 7.8 (35.8) | 10.2 (37.1) | 10.3 (37.7) |
| 〈I/σ(I)〉 | 42.2 (3.5) | 31.5 (4.8) | 73.4 (15.6) | 36.6 (6.8) | 28.1 (3.6) |
| Refinement | |||||
| Resolution range (Å) | 32.62-1.90 | 34.62-2.00 | 42.34-2.15 | 48.92-2.15 | 49.65-2.10 |
| Rwork/Rfree (%)b | 18.5/22.9 | 18.0/22.6 | 18.2/22.1 | 18.7/21.9 | 19.8/24.2 |
| No. of atoms | |||||
| Proteins | 4834 | 4765 | 4783 | 4778 | 4740 |
| Ligands | 24 | 210 | 225 | 53 | 210 |
| Water molecules | 236 | 252 | 293 | 245 | 203 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.022 | 0.021 | 0.007 | 0.007 | 0.007 |
| Bond angles (°) | 1.81 | 1.94 | 0.98 | 0.98 | 1.01 |
a Rmerge = 100 × Σ|Ihkl − 〈Ihkl〉|/Σ Ihkl, 〈Ihkl〉 is the mean value of Ihkl.
b Rwork = 100 × Σ‖Fo| − | Fc‖/Σ|Fo|. Rfree was calculated from the test set (5% of the total data).
Catalytic Activity against Ethylene Glycol Chitin
According to the method described previously (39), the enzyme activities of wild-type and 10 mutants of Ra-ChiCmature were determined using ethylene glycol chitin as the substrate in 0.1 m sodium phosphate buffer, pH 6.0, at 37 °C. One unit of activity was defined as the amount of enzyme that liberated 1 μmol of the reducing sugar per minute.
Catalytic activity against (NAG)n
Time courses of (NAG)n (n = 3, 4, 5, and 6) hydrolysis catalyzed by wild-type Ra-ChiCmature were determined by quantifying the amounts of substrate and products by HPLC. The enzymatic reaction was performed in 50 mm acetate buffer, pH 6.0, at 40 °C for a given incubation time. Enzyme and substrate concentrations were 0.82 μm and 4.75 mm, respectively. The reaction mixture was rapidly chilled in liquid nitrogen to terminate the enzymatic reaction, and then applied to HPLC using a column of TSK-gel NH2-60 (Tosoh Co.). Elution was carried out with 70% (v/v) acetonitrile at a flow rate of 0.8 ml/min. (NAG)n fragments were monitored by absorption at 220 nm. From the peak area obtained by HPLC, (NAG)n concentrations at each reaction time were calculated using a standard curve obtained with an authentic saccharide solution, and plotted against the reaction time.
RESULTS
Interaction Analysis by NMR Spectroscopy
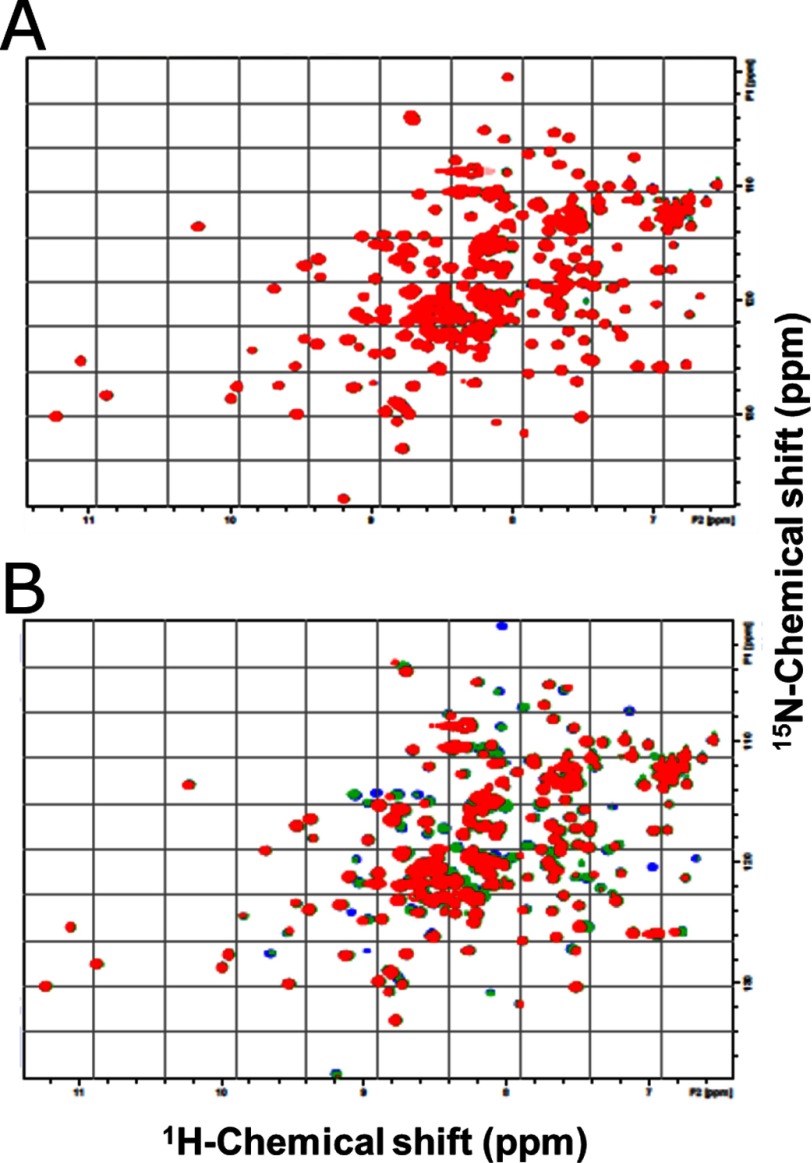
1H-15N heteronuclear single quantum coherence spectra of the 15N-labeled Ra-ChiCmature were obtained in the presence of (NAG)2 (0.6 or 1.2 mm) or NAG-NAM-AiQ (0.6 or 1.2 mm). The overlaid spectra are shown in Fig. 1. Only a trivial change was observed, when the NAG-NAM-AiQ was added to the protein solution (Fig. 1A). However, the numerous resonances were affected upon addition of (NAG)2 (Fig. 1B). This indicates that Ra-ChiC specifically interacts with the chitin oligomer, but not with the peptidoglycan fragment.
FIGURE 1.
Superimposition of the 1H-15N heteronuclear single quantum coherence spectra of Ra-ChiCmature in the absence or presence of ligands, NAG-NAM-AiQ (A) and (NAG)2 (B). The titration was conducted in 50 mm sodium acetate buffer, pH 5.0, containing 10% D2O. Solution temperature was 300 K. The molar ratios of ligand:Ra-ChiCmature were 0:1 (blue), 6:1 (green), and 12:1 (red). No appreciable changes were observed upon further addition of the ligands.
Crystal Structures of Wild-type Ra-ChiCcat
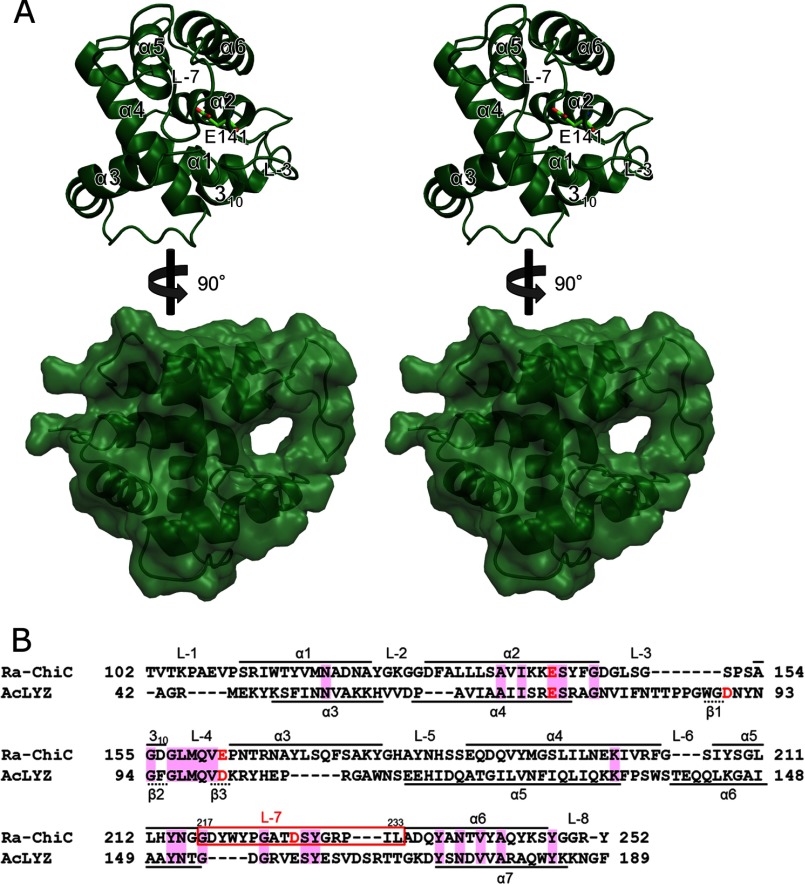
The structure of wild-type Ra-ChiCcat (WT) contains four molecules per asymmetric unit, and these molecules are superposed each other with root mean square deviation (r.m.s. deviation) values from 0.3 to 0.4 Å for the 152 corresponding Cα atoms. The N-terminal region of MolA penetrates to and interacts with an adjacent molecule, MolB, and electron densities are observed from Pro95. In the other molecules, interdomain linker regions were disordered, and electron densities were observed from Thr102. The overall structure is rich in α-helical structures, which included six α-helices, α1 (residues 111–123), α2 (residues 129–141), α3 (residues 163–177), α4 (residues 186–204), α5 (residues 207–216), and α6 (residues 234–247), and one 310 helix (residues 154–156) (Fig. 2A, upper). The catalytic residue, Glu141, is located at the end of α2 and covered with the longest loop, L-7 (residues 217–233), which connects α5 and α6 (Fig. 2A, upper). L-7 is clinched by a hydrogen bond network, and then forms a tunnel-shaped cavity (Fig. 2A, lower).
FIGURE 2.
Crystal structure of the Ra-ChiC catalytic domain. A, stereo diagrams of the front and side views of the WT structure. Front view is shown as a ribbon model, and Glu141 is shown as a stick model. Secondary structures are indicated in the figure. Side view is shown as a ribbon model with a transparent surface model. B, sequence alignment of the Ra-ChiC catalytic domain with AcLYZ calculated with ClustalW (45). Acidic residues located in the active site are shown in red. Conserved residues are shown by the magenta background. Residues that form the secondary structures of Ra-ChiC and AcLYZ are indicated above and below, respectively. L-7 is indicated by a red box.
Ra-ChiCcat has a topology similar to G-type lysozymes despite their low homology at the primary structure level (Fig. 2B). A structure similarity search using DALI indicated that AcLYZ (Z.11.6; PDB code 3GXR), the G-type lysozyme from Atlantic salmon (Z.11.4; PDB code 3MGW), the G-type lysozyme from Australian black swan egg white (Z.11.3; PDB code 1LSP), and goose egg-white lysozyme (GEWL, Z.11.3; PDB code 154L) exhibited high Z scores. Superposition of the Ra-ChiCcat structure on the AcLYZ structure gave an r.m.s. deviation value of 1.4 Å over 100 corresponding Cα atoms. Except for GH family 23 members, GH19 chitinases including those from Streptomyces griseus (Z.7.3; PDB code 2DBT), Picea abies (Z.7.0; PDB code 3HBE), and Carica papaya (Z.6.4; PDB code 3CQL) exhibited relatively high Z scores. Superposition of the Ra-ChiCcat structure with the structure of S. griseus chitinase gave an r.m.s. deviation value of 3.5 Å over 123 corresponding Cα atoms. Although Ra-ChiC does not share sequence similarities (9–12% identities) with GH family 19 chitinases, this analysis revealed that they share similar overall structures.
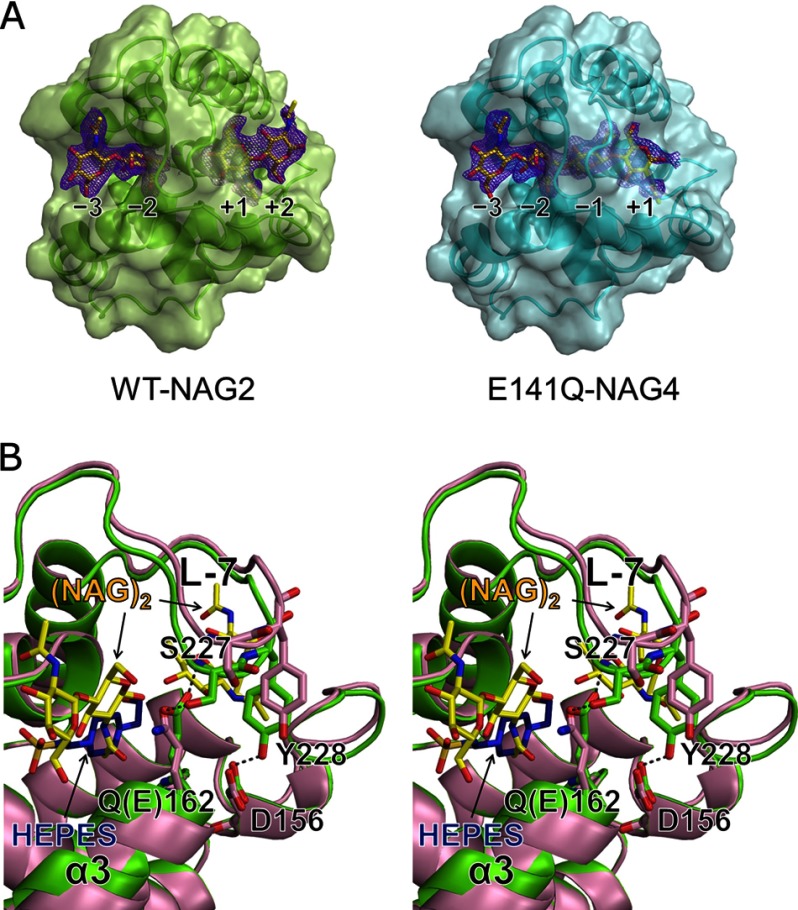
In the structure of WT complexed with (NAG)2, WT-NAG2, electron densities corresponding to (NAG)2 molecules are clearly observed in −3 to −2 and +1 to +2 subsites (Fig. 3A, left), using the nomenclature proposed by Davies et al. (40). Although the overall structure of WT was not affected upon (NAG)2 binding (r.m.s. deviation value of 0.3 Å for 153 Cα atoms), only the main chain of Gly147–Gly150 on loop 3 (L-3) had slightly moved away (≈1.1 Å) from the disaccharide bound at +1 to +2 subsites (supplemental Fig. S1).
FIGURE 3.
Comparison of Ra-ChiCcat structures. A, crystal structures of Ra-ChiCcat complexed with (NAG)n. WT-NAG2 (left) and E141Q-NAG4 (right) structures with transparent surface models are shown in light green and cyan, respectively. Bound NAG residues are shown as yellow stick models. Initial Fo − Fc maps (contoured at 2.0σ) before the sugar model were included are shown in purple. B, stereo diagram showing conformational changes observed at α3 and L-7 between WT-NAG2 (light green) and E162Q-HEPES (light magenta) structures. Bound NAG residues and HEPES molecule are shown as yellow and blue stick models, respectively. Hydrogen bonding interactions are shown as gray dashed lines.
The tunnel-shaped cavity formed by L-7 represents a substrate-binding site. NAG residues that occupied the −2 and +1 subsites are almost buried within this cavity (Fig. 3A, left). In the −2 subsite, six direct and four water-mediated hydrogen bonds were observed between the NAG residue and WT (Table 2, supplemental Fig. S2). Glu162, which is a conserved acidic residue in G-type lysozymes strongly contributes to substrate recognition at the −2 subsite by forming three direct hydrogen bonds. In the +1 subsite, seven direct and one water-mediated hydrogen bonds were observed. The side chain oxygen (Oϵ2) of Glu141, a highly conserved glutamic acid, is 2.7 Å from the O4 atom of the NAG residue at the +1 subsite. In contrast, NAG residues at the −3 and +2 subsites are exposed to solvent (Fig. 3A, left), and recognition of these NAG residues are relatively loose. An NAG residue at the −3 subsite forms only one direct and three water-mediated hydrogen bonds. An NAG residue at the +2 subsite is recognized by this enzyme via two direct hydrogen bonds and one water-mediated hydrogen bond. In fact, in the WT-NAG2 structure, the average B-factor values of NAG residues at the −3 and +2 subsites are higher than those at the −2 and +1 subsites (−3 subsite: 35.1 Å2; −2 subsite: 25.4 Å2; +1 subsite: 18.8 Å2; +2 subsite: 43.2 Å2).
TABLE 2.
Hydrogen-bonding interactions between NAG residues and Ra-ChiC
| Subsite | NAG | Protein | Distance | |
|---|---|---|---|---|
| WT-NAG2 | ||||
| −3 | N2 | Thr225 | O | (Water-mediated) |
| O5 | Lys200 | Nζ | (Water-mediated) | |
| O5 | Asp218 | N | (Water-mediated) | |
| O7 | Tyr219 | N | 2.8 Å | |
| −2 | O1 | Glu162 | Oϵ1 | 3.5 Å |
| N2 | Tyr214 | O | 2.8 Å | |
| O3 | Thr165 | Oγ1 | 2.7 Å | |
| O3 | Lys200 | Nζ | (Water-mediated) | |
| O3 | Asp218 | N | (Water-mediated) | |
| O6 | Glu162 | Oϵ1 | 2.8 Å | |
| O6 | Asn164 | Nδ2 | (Water-mediated) | |
| O6 | Thr165 | Oγ1 | (Water-mediated) | |
| O7 | Glu162 | N | 2.9 Å | |
| O7 | Thr165 | Oγ1 | 3.5 Å | |
| +1 | N2 | Glu141 | O | 2.9 Å |
| O3 | Gln160 | Nϵ2 | 2.8 Å | |
| O3 | Arg230 | Nη2 | 3.3 Å | |
| O4 | Glu141 | Oϵ2 | 2.7 Å | |
| O4 | Asn215 | Oδ1 | 3.2 Å | |
| O6 | Asn215 | Nδ2 | (Water-mediated) | |
| O7 | Ser151 | Oγ | 2.5 Å | |
| O7 | Arg230 | Nη1 | 2.9 Å | |
| +2 | O5 | Lys139 | O | (Water-mediated) |
| O6 | Gly147 | O | 3.2 Å | |
| O7 | Lys140 | Nζ | 2.6 Å | |
| E141Q-NAG4 | ||||
| −3 | O6 | Thr165 | Oγ1 | 3.4 Å |
| O7 | Tyr219 | N | 2.9 Å | |
| −2 | N2 | Tyr214 | O | 2.9 Å |
| O3 | Thr165 | Oγ1 | 2.7 Å | |
| O3 | Lys200 | Nζ | (Water-mediated) | |
| O3 | Asp218 | N | (Water-mediated) | |
| O6 | Glu162 | Oϵ1 | 2.8 Å | |
| O7 | Glu162 | N | 2.9 Å | |
| O7 | Thr165 | Oγ1 | 3.3 Å | |
| −1 | O1 | Gln141 | Nϵ2 | 3.3 Å |
| N2 | Asp226 | Oδ1 | 3.0 Å | |
| O3 | Asn215 | O | 3.0 Å | |
| O6 | Gln141 | Oϵ1 | 2.9 Å | |
| O7 | Asn215 | Nδ2 | (Water-mediated) | |
| O7 | Leu233 | N | (Water-mediated) | |
| +1 | N2 | Gln141 | O | 2.8 Å |
| O3 | Gln160 | Nϵ2 | 2.7 Å | |
| O3 | Arg230 | Nη2 | 3.3 Å | |
| O6 | Asn215 | Nδ2 | (Water-mediated) | |
| O7 | Leu233 | N | (Water-mediated) | |
| O7 | Ser151 | Oγ | 2.6 Å | |
| O7 | Arg230 | Nη1 | 3.1 Å | |
Crystal Structure of the E141Q Mutant of Ra-ChiCcat
In the structure of the E141Q mutant of Ra-ChiCcat (E141Q) complexed with (NAG)4, E141Q-NAG4, electron densities corresponding to (NAG)4 are observed well at −3 to +1 subsites except for a part of an NAG residue at the +1 subsite (Fig. 3A, right). The overall structure of E141Q-NAG4 is essentially identical with that of the WT structure (r.m.s. deviation value of 0.2 Å for 153 Cα atoms), whereas a slight conformational change is observed in L-7 and L-3 (supplemental Fig. S1). The maximum shift of 0.8 and 0.5 Å were confirmed at the Cα atom position of Leu148 in L-3 and Pro231 in L-7, respectively. Because such a conformational change in L-7 is not observed in the WT-NAG2 structure, those conformational changes in L-3 and L-7 seem to be derived from the NAG residue binding at +1 and −1 subsites, respectively. In addition, the positions of NAG residues at the −3 and −2 subsites are slightly different between the WT-NAG2 and E141Q-NAG4 structures. The mean distances of the corresponding atoms of NAG residues at the −3 and −2 subsites in these structures are 0.4 and 0.6 Å, respectively, when the overall structures are superposed. These shifts of NAG residues may be influenced by the presence of an NAG residue at the −1 subsite.
From the structural differences noted above, the interactions between this enzyme and NAG residues at these subsites observed in the E141Q-NAG4 structure are also slightly different from those in the WT-NAG2 structure. In the E141Q-NAG4 structure, an additional hydrogen bond was observed between O6 of the NAG residue at the −3 subsite and Oγ1 of Thr165, whereas a direct hydrogen bond and two water-mediated hydrogen bonds observed at O1 and O6 of the NAG residue at the −2 subsite were not observed (Table 2, supplemental Fig. S3). The NAG residue at the −1 subsite is completely buried within the tunnel-shaped cavity of E141Q and adopts a chair conformation (Fig. 3A, right). Direct hydrogen bonds are observed between O1 and Nϵ2 of Gln141, N2 and Oδ1 of Asp226, O3 and the main chain carbonyl of Asn215, and O6 and Oϵ1 of Gln141. In addition, two water-mediated hydrogen bonds were observed at O7 of the NAG residue (Table 2).
Crystal Structures of the E162Q Mutant of Ra-ChiCcat
In the structure of the E162Q mutant in the absence of (NAG)n, E162Q-HEPES, electron density corresponding to the HEPES molecule derived from the mother liquid for crystallization was observed over the −3 to −1 subsite of three E162Q molecules (MolA, MolC, and MolD) in the asymmetric unit. As was observed in the WT structure, the N-terminal region of MolA penetrates to the substrate-binding site of MolB. In MolA and MolD of E162Q-HEPES, the characteristic structural change observed at α3 and L-7 results in a slight exposure of the substrate-binding site compared with other structures (supplemental Fig. S1). In contrast, this exposure is not observed in the MolC structure despite the presence of a HEPES molecule. The Cα distances of Gln162 to Ser227 and Asn156 to Tyr228 in MolD are 2.4 and 1.4 Å, respectively, longer than those in the WT-NAG2 structure and, as a result, hydrogen bonding interactions between these residues are broken in the E162Q-HEPES structure as shown in Fig. 3B.
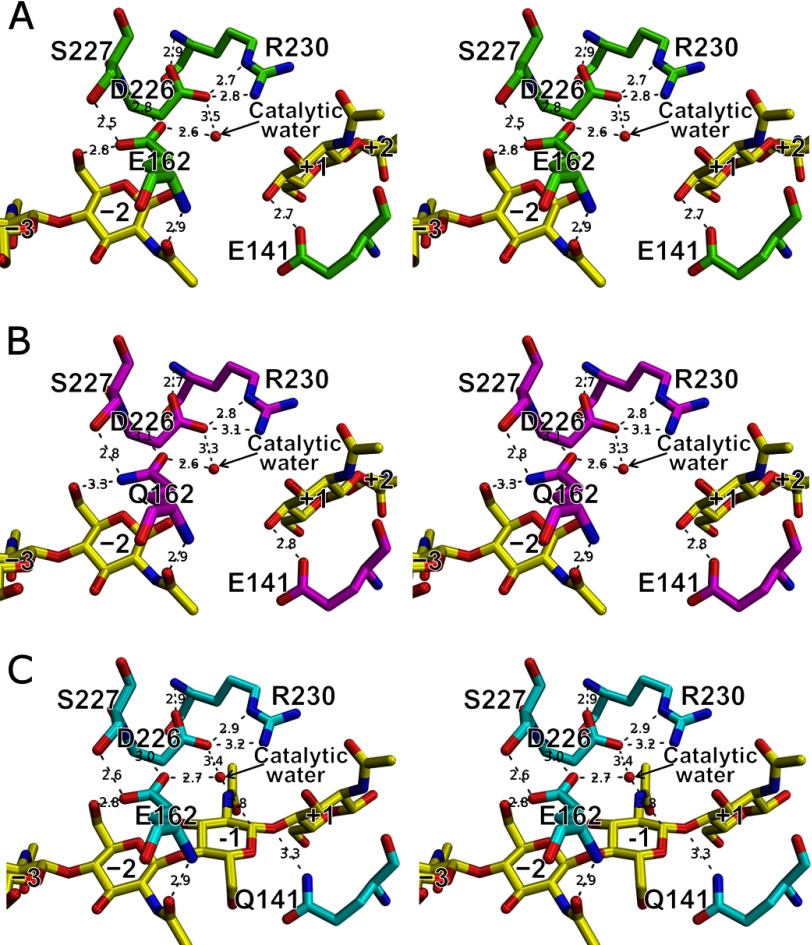
In the structure of the E162Q mutant complexed with (NAG)2, E162Q-NAG2, electron densities corresponding to (NAG)2 molecules were observed in the −3 to −2 and +1 to +2 subsites analogous to the WT-NAG2 structure. In contrast to the MolA and MolD in the E162Q-HEPES structure, the structure of E162Q-NAG2 is quite similar to the WT-NAG2 structure including L-3, L-7, α3, and NAG molecules (r.m.s. deviation value of 0.1 Å for 153 Cα atoms) (supplemental Fig. S1). The hydrogen bonding pattern between NAG residues and WT in the WT-NAG2 structure (listed in Table 2) is conserved in the E162Q-NAG2 structure even with the substitution of Glu162 to Gln (Fig. 4, A and B).
FIGURE 4.
Ra-ChiC active site. A, WT-NAG2; B, E162Q-NAG2; and C, E141Q-NAG4. All figures are shown as stereo diagrams. Hydrogen bonds are shown as gray dashed lines (bond lengths are indicated).
Mutation Analysis of Ra-ChiC
Mutation analyses of Glu141 and two acidic residues (Glu162 and Asp226) located within 10 Å distance from Glu141 were performed to determine the catalytic residues of Ra-ChiC. We prepared 10 single mutants of Ra-ChiCmature (E141A, E141Q, E141D, E141N, E162A, E162Q, E162D, E162N, D226A, and D226N) and determined their chitinolytic activities against glycol chitin (Table 3). The E141A, E141Q, and E141D mutations resulted in >250-fold decreased chitinolytic activity, and the E141N mutation decreased the activity >100-fold. In addition, two mutants relevant to Asp226 (D226A and D226N) also decreased the catalytic activities by over 100-fold. These results indicated that Glu141 and Asp226 have an important role in Ra-ChiC catalytic activity. In contrast, the E162A, E162D, and E162N mutants retained moderate activity, and the catalytic activity of the E162Q mutant was 2.5-fold higher than that of wild-type. Steady state kinetics of the enzymatic reaction toward glycol chitin did not afford accurate values of kcat and Km for the mutant enzymes, except for Glu162 mutants, because of their low activity.
TABLE 3.
Mutation analysis of Ra-ChiC
| Enzyme | Specific activity | Relative activity |
|---|---|---|
| units/mg protein | % | |
| Wild-type | 6.77 | 100.0 |
| E141A | 0.02 | 0.3 |
| E141Q | 0.02 | 0.3 |
| E141D | 0.03 | 0.4 |
| E141N | 0.05 | 0.8 |
| E162A | 1.10 | 16.2 |
| E162Q | 17.17 | 253.6 |
| E162D | 2.03 | 30.0 |
| E162N | 1.50 | 22.2 |
| D226A | 0.06 | 0.9 |
| D226N | 0.04 | 0.6 |
Catalytic Activity of Ra-ChiC against (NAG)n
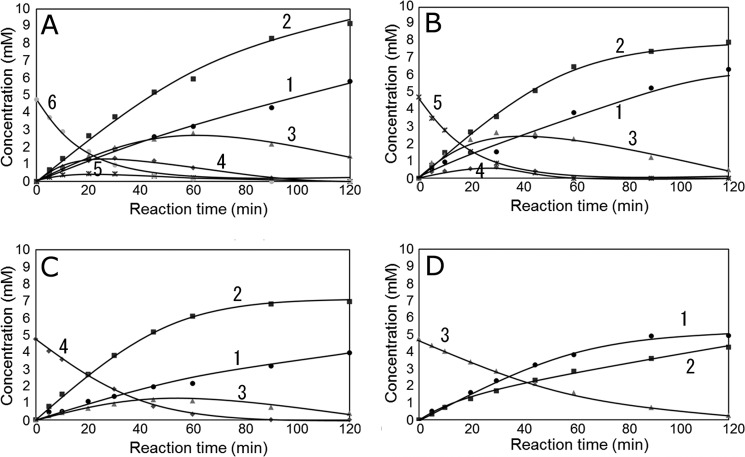
Time courses of NAG oligomer hydrolysis catalyzed by Ra-ChiCmature were determined by HPLC, and the results are shown in Fig. 5. Specific activities toward (NAG)3, (NAG)4, (NAG)5, and (NAG)6, were calculated to be 16.6, 31.6, 52.6, and 52.6 mm/min/mg, respectively, based on the initial velocity of the substrate degradation. (NAG)3 was hydrolyzed into (NAG)2 + NAG (Fig. 5D). As reported in a previous article (17), the rate of β-(NAG)3 hydrolysis was higher than that of α-(NAG)3, and the products (NAG)2 and NAG were rich in α- and β-forms, respectively. Because Ra-ChiC is an inverting enzyme, (NAG)3 is supposed to bind predominantly to −2, −1, and +1 subsites. (NAG)4 was hydrolyzed into (NAG)2 + (NAG)2 and (NAG)3 + NAG (Fig. 5C). Because the products (NAG)3 and NAG were rich in α- and β-forms, respectively (17), (NAG)4 is supposed to bind to −2, −1, +1, and +2 subsites and −3, −2, −1, and +1 subsites. Thus the affinity at the glycon-binding site (negatively numbered subsites) is likely higher than that at the aglycon-binding site (positively numbered subsites).
FIGURE 5.
Time courses of Ra-ChiCmature-catalyzed hydrolysis of (NAG)6 (A), (NAG)5 (B), (NAG)4 (C), and (NAG)3 (D) as determined by HPLC. The enzymatic reaction was performed in 50 mm acetate buffer, pH 6.0, at 40 °C for a given incubation time. Enzyme and substrate concentrations were 0.82 μm and 4.75 mm, respectively. HPLC determination was performed using a column of TSK-gel NH2-60 (Tosoh Co.), eluting with 70% (v/v) acetonitrile at a flow rate of 0.8 ml/min. Symbols: ●, NAG; ■, (NAG)2; ▴, (NAG)3; ♦, (NAG)4; ×, (NAG)5; closed circle (gray), (NAG)6. Numbers in the figure indicate the n value of (NAG)n.
Profiles of the product formations from the substrates (NAG)5 and (NAG)6 were quite complicated because of the subsequent breakdown of the products of (NAG)3, (NAG)4, and (NAG)5 (Fig. 5, A and B). In the early reaction stage, however, simple hydrolysis of the substrates was observed. (NAG)5 was hydrolyzed frequently into (NAG)2 + (NAG)3, and into (NAG)4 + NAG in a lesser extent. All five glycosidic bonds in (NAG)6 were able to be hydrolyzed by these enzymes. No evidence for transglycosylation was found from the time course data. The mutant enzymes except Glu162 mutants (E162A, E162Q, E162D, and E162N) did not hydrolyze the NAG oligomers at all.
DISCUSSION
We have already reported that Ra-ChiC belonging to the GH-23 family hydrolyzes ethylene glycol chitin, carboxyl methyl chitin, and other soluble chitin but not the cell wall of Micrococcus lysodeikticus (17). Because the specificity was judged only from the data obtained with the polysaccharide substrates, it was still uncertain if Ra-ChiC interacts only with the chitinous substrates. Thus, we tested (NAG)2 and NAG-NAM-AiQ for NMR titration experiments. As shown in Fig. 1, the backbone N-H resonances were strongly affected by the addition of (NAG)2 but not by NAG-NAM-AiQ. Ra-ChiC indeed recognizes the chitin oligomer, but not the fragment derived from the lysozyme substrate, peptidoglycan.
Then we determined several structures for the catalytic domain of Ra-ChiC in the presence or absence of NAG oligomers. We also performed mutation analyses based on this structural information. The structure-function relationships provided important information for understanding the detailed catalytic mechanism and the unique substrate recognition mechanism of Ra-ChiC.
The overall structure of Ra-ChiCcat is similar to that of AcLYZ with a r.m.s. deviation value of 1.4 Å (Fig. 6A), and Glu141 in Ra-ChiC corresponds to the catalytic acid Glu73 in G-type lysozymes with respect to both the primary and tertiary structures (Figs. 2B and 6B). Indeed, the side chain atom of Gln141 in the E141Q-NAG4 structure is located close enough to the O1 glycosidic oxygen for donating proton (Fig. 4C). Furthermore, mutation analyses showed that all Glu141 mutations caused significant decreases in catalytic activities toward ethylene glycol chitin (Table 3). We tried to determine the steady state kcat and Km values. However, the very low activity of the Glu141 mutants did not allow the accurate determination of the values for the glycol chitin substrate. For the NAG-oligomer substrates, no hydrolysis was observed for the Glu141 mutants. On the other hand, binding experiments have been conducted by means of isothermal titration calorimetry, and we confirmed that the free energy changes for NAG oligomer binding were not affected by the mutation of Glu141.4 Thus, we believe that the low activities of the Glu141 mutants are exclusively derived from a decrease in kcat. These results indicated that Glu141 of Ra-ChiC could act as a general acid. The isothermal titration calorimetry data will be reported elsewhere.
FIGURE 6.
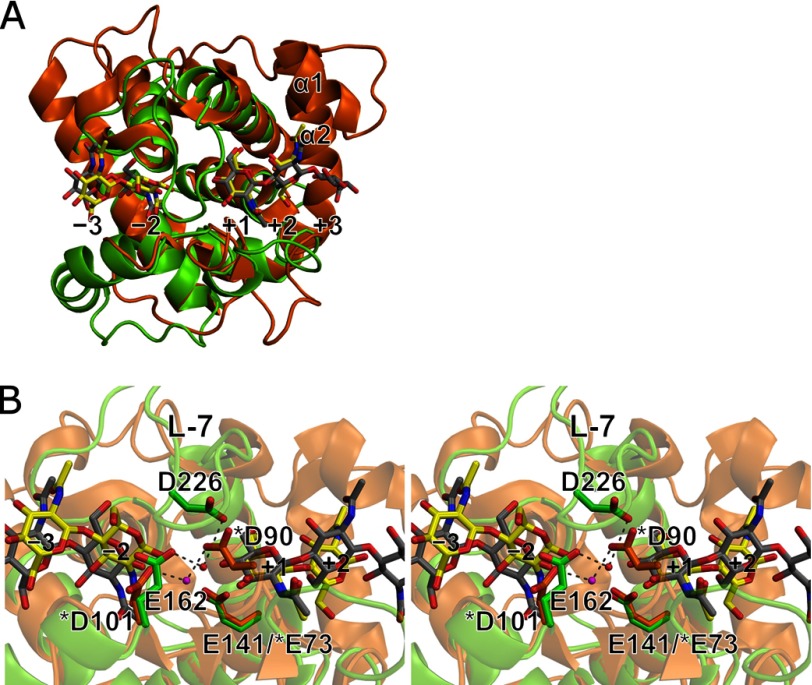
Structural comparison between Ra-ChiCcat and AcLYZ. A, superposition of the overall structures of WT-NAG2 (light green) and AcLYZ-NAG (orange). Bound NAG residues in WT-NAG2 and AcLYZ-NAG are shown as yellow and gray stick models, respectively. B, stereo diagram of active site superposition of the WT-NAG2 (light green) and AcLYZ-NAG (orange) structures. Acidic residues at each active site are shown as stick models. The catalytic water molecules observed in the WT-NAG2 and AcLYZ-NAG structures are shown as red and magenta spheres, respectively. AcLYZ residue numbers are indicated by asterisks.
The carboxyl oxygen atoms of Glu162 and Asp226 are ∼8.7 and 8.3 Å, respectively, from that of Glu141. A water molecule is observed between Glu162 and Asp226 as is the case for a catalytic water observed between Asp101 and Asp90 in the AcLYZ-NAG structure (PDB code 3GXR) in which (NAG)2 and (NAG)3 are bound at the −3 to −2 and +1 to +3 subsites (Fig. 6B). This water molecule is 3.8 Å from the C1 atom of the NAG residue at the −1 subsite in the E141Q-NAG4 structure, and consequently defined as the catalytic water (Fig. 4C). The catalytic water may be situated close to the C1 atom to be suitable for nucleophilic attack by the conformational change through the transition state. Asp226 is located at L-7, and the apparent counterpart of Asp226 is likely Glu159 in AcLYZ based on the primary structure (Fig. 2B). However, based on the crystal structure (Fig. 6B), Asp226 does not have any counterpart, and the carboxyl oxygen atom of Asp226 is located only close to Asp90, which is involved in positioning the catalytic water in AcLYZ. By comparison, Glu162 is located at a structurally analogous position to that of Asp101 in AcLYZ, a putative general base. Both Glu162 of Ra-ChiC and Asp101 of AcLYZ are next to the highly conserved sequence found among GH family 23 members (Gly157–Gln160 in Ra-ChiC numbering) (41), and sequence alignments of these enzymes indicated that Glu162 of Ra-ChiC are equivalent to Asp101 of AcLYZ (Fig. 2B). Nevertheless, mutation analysis showed that Glu162 is not essential for the catalytic activity of Ra-ChiC (Table 3). Because Glu162 forms direct hydrogen bonds with the NAG residue at the −2 subsite through its main and side chains in addition to a hydrogen bond with the catalytic water (Fig. 4), Glu162 would be involved in both NAG recognition and positioning of this water. These roles of Glu162 in Ra-ChiC correspond to those of Asp90 in AcLYZ.
Mutation analysis also indicated that Asp226 is critical for catalysis. Substitution of Asp226 not only with Ala but also with Asn caused significant decreases in catalytic activity, suggesting that the side chain carboxyl group of Asp226 is important for catalytic reactions. In the wild-type structure, the hydrogen bonding distance between Asp226 and the catalytic water is somewhat long (3.5 Å), whereas the distance between Glu162 and the water is 2.6 Å (Fig. 4A). However, Asp226 could play the role of a general base, as the pKa value of Asp226, which is calculated as −2.7 to ∼2.0, would be decreased by electrostatic interactions with the side chain of neighbor Arg230 (Fig. 4). Interestingly, similar electrostatic interactions are also constructed between the putative general base Asp101 and the neighboring Arg103 in the AcLYZ structure. These interactions are observed for the catalytic residues in some inverting enzymes, including GH family 6 cellulase (PDB code 2BVW), GH family 8 cellulase (PDB code 1KWF), and GH family 46 chitosanase (PDB code 1CHK). Therefore, we propose a catalytic mechanism for Ra-ChiC based on structural analyses, enzymatic assay, and a theoretical model (supplemental Fig. S4). Glu141 donates a hydrogen atom to the scissile glycosidic bond. A water molecule that interacts with Asp226 and Glu162 via hydrogen bonds is activated by the general base of Asp226. Then, the nucleophilc water attacks the C1 atom of the NAG residue at the −1 subsite. As a result, an NAG oligomer with an α-anomeric configuration at the reducing end is produced. It should be noted that the general base Asp226 is acting from a roof of the tunnel-shaped cavity formed by the loop, L-7. This is a unique arrangement of the catalytic residues for inverting enzymes.
As stated above, the NAG residue at the −1 subsite is completely buried within the tunnel-shaped cavity of E141Q and adopts a chair conformation. Although the general catalytic mechanism of inverting glycosidases is known, the transition state and the conformations adopted by the pyranosyl ring of the −1 sugar are still indefinite. Some reports on the inverting GH8 enzymes suggested that the −1 sugar ring displays an unfavorable boat 2,5B conformation (42, 43). We have been trying to get insight into the structural changes of the −1 sugar along the reaction pathway based on the crystal structure of the enzyme-NAG oligomer complexes. Unfortunately, however, the −1 NAG residue in the E141Q-NAG4 structure exhibits a favorable 4C1 chair conformation. It appears that the chair conformation of the −1 NAG residue reflects a nonproductive state, which does not undergo the catalytic reaction. We could not get any insight into the transition state from the crystal structures obtained in this study. Further crystallographic analysis is in progress using various mutant Ra-ChiCs.
We defined five substrate-binding subsites (−3 to +2) in Ra-ChiCcat based on crystal structures, whereas it is well known that G-type lysozymes have six subsites. A comparison of the crystal structures for WT-NAG2 and AcLYZ-NAG illustrated that they have similar overall folds; in particular, five helices were superposed well (Fig. 6A). However, the substrate-binding site of Ra-ChiC appears to be shorter than that of AcLYZ because of a lack of two helices in its N-terminal region (α1 and α2 of AcLYZ), resulting in a lack of a sixth subsite (+3 subsite) in Ra-ChiC. Enzymatic assays for Ra-ChiC using (NAG)n showed that the activity of Ra-ChiC against (NAG)5 was equivalent to that against (NAG)6. Thus, we concluded that the Ra-ChiC catalytic domain have five substrate-binding subsites (−3 to +2) unlike G-type lysozymes. As noted above, the recognition of the NAG residues occupying the −3 and +2 subsites is looser than those of the other NAG residues. Therefore, although Ra-ChiC has five subsites at maximum, the number of core subsites might be only three (−2, −1, and +1). This proposition is consistent with the data obtained by HPLC-based time courses of NAG oligomer degradation; that is, (NAG)3 binds predominantly to subsites −2, −1, and +1, (NAG)4 binds to subsites −3, −2, −1, and +1 and subsites −2, −1, +1, and +2. When (NAG)6 is hydrolyzed by Ra-ChiC, this enzyme produces (NAG)4, (NAG)3, and (NAG)2, and minimal amounts of (NAG)5 and NAG (Fig. 5A). This phenomenon would be caused by multiple binding modes of (NAG)6 due to the weak interactions with the sugar residues at both ends of the substrate-binding site.
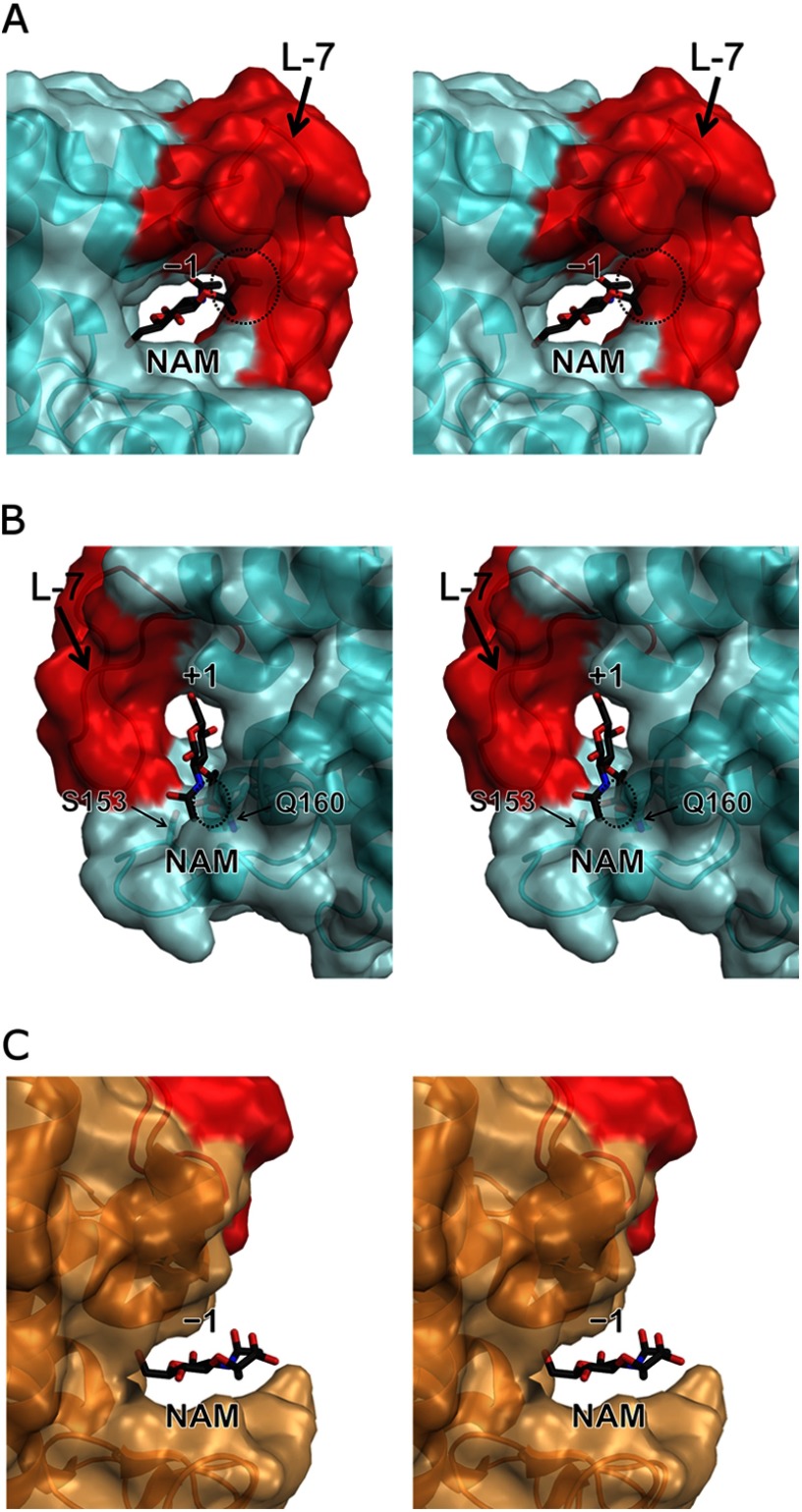
L-7 in Ra-ChiC has the characteristic conformation compared with G-type lysozymes. The tunnel-shaped conformation of Ra-ChiC makes the −1 subsite quite narrow (Fig. 3A). Ra-ChiC does not have lysozyme activity that cleaves the glycosidic bond between NAM and NAG residues in peptidoglycans, components of bacterial cell walls. Because NAM has an additional large substituent group (lactic acid group) on C3 of its pyranose ring compared with NAG, NAM binding to such a narrow space in Ra-ChiC might be impossible. In fact, steric hindrance took place between the lactic acid group of the NAM molecule and L-7 when the NAG residue was replaced at the −1 subsite in the E141Q-NAG4 structure (Fig. 7A). For binding of the NAM molecule to the −1 subsite, a large conformational change of L-7 would be needed, but it may cause rearrangement of Asp226 and result in inactivation of the enzyme. Although some lysozymes cleave the NAM-NAG bond, steric hindrance also took place between the NAM molecule and Ser153 and Gln160 of Ra-ChiC when the NAG residue at the +1 subsite in the E141Q-NAG4 structure was replaced by the NAM residue (Fig. 7B). In nature, the natural substrates of lysozyme, peptidoglycans, form more bulky mesh-like layers consisting of glycan chains cross-linked with oligopeptides. Therefore, it appears that peptidoglycan binding to Ra-ChiC would be more difficult. By comparison, G-type lysozymes have open conformations at their active sites and could readily accommodate NAM in their −1 subsite. In fact, a hydroxyl group attached to C3 of the NAG residue occupying the −1 subsite orients toward solvent in the crystal structure of GEWL in complex with (NAG)3 (PDB code 154L). Thus, we could replace the NAG residue with a NAM residue without steric hindrance (Fig. 7C). Hence, the characteristic L-7 conformation appears to generate the substrate specificity of Ra-ChiC.
FIGURE 7.
NAM-binding model at the −1 subsite (A) and the +1 subsite (B) for Ra-ChiC, and the −1 subsite (C) for GEWL. The model structure was constructed by replacing the NAG residue at the −1 and +1 subsites in the E141Q-NAG4 structure of Ra-ChiC and the −1 subsite in the GEWL structure in complex with (NAG)3 (PDB code 154L) to the NAM residue (black stick model), respectively. L-7 region (in A and B) and the loop region corresponding to L-7 (in C) are shown in red. Steric hindrance between NAM and L-7 is indicated by an ellipse in A and B. Figures are shown as stereo diagrams.
In contrast, the tunnel-shaped conformation needs to be opened to accommodate (NAG)n in an endo-spliting manner. In fact, a semi-opened conformation resulting from structural shifts of L-7 and α3 in the E162Q-HEPES structure suggested that the tunnel-shaped conformation can be opened (Fig. 3B). This mechanism has also been proposed for the catalytic reaction of the processive endo-cellulase Cel48F, which has a tunnel-shaped active site (44).
CONCLUSION
Ra-ChiC is a chitinase that was first found in GH family 23 (goose-type lysozyme). The crystal structure of Ra-ChiC exhibited a tunnel-shaped conformation in its active site, which makes a clear contrast to the other GH23 members. The catalytic base is located at the roof of the tunnel, forming a unique arrangement of the catalytic residues for inverting chitin hydrolysis and determining the substrate specificity. This is the first report on the tunnel-shaped binding site of an inverting chitinase.
Supplementary Material
Acknowledgments
We thank the staff of SPring-8 and Photon Factory, Japan, for their assistance with x-ray data collection. We are also indebted to Drs. R. Kuroki and M. Adachi for the valuable discussions.

This article contains supplemental Figs. S1–S4 and Tables S1–S3.
The atomic coordinates and structure factors (codes 3W6B, 3W6C, 3W6D, 3W6E, and 3W6F) have been deposited in the Protein Data Bank (http://wwpdb.org/).
N. Kawamoto, S. Nishimura, H. Fukada, T. Fukamizo, and M. Ueda, unpublished data.
- NAG
- N-acetyl-d-glucosamine
- NAG-NAM-AiQ
- N-acetylglucosaminyl-(β-1,4)-N-acetylmuramyl-l-alanyl-d-isoglutamine
- GH family
- glycohydrolase family
- Ra-ChiC
- chitinase C from Ralstonia sp. A-471
- AcLYZ
- Atlantic cod G-type lysozyme
- r.m.s. deviation
- root mean square deviation
- GEWL
- goose egg-white lysozyme.
REFERENCES
- 1. Muzzarelli R. A. (1999) Clinical and biochemical evaluation of chitosan for hypercholesterolemia and overweight control. EXS 87, 293–304 [DOI] [PubMed] [Google Scholar]
- 2. Kim S.K., Rajapakse N. (2005) Enzymatic production and biological activities of chitosan oligosaccharides (COS). A review. Carbohydr. Polym. 62, 357–368 [Google Scholar]
- 3. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henrissat B., Bairoch A. (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 6. Tews I., Terwisscha van Scheltinga A. C., Perrakis A., Wilson K. S., Dijkstra B. W. (1997) Substrate-assisted catalysis unifies two families of chitinolytic enzymes. J. Am. Chem. Soc. 119, 7954–7959 [Google Scholar]
- 7. Synstad B., Gåseidnes S., Van Aalten D. M., Vriend G., Nielsen J. E., Eijsink V. G. (2004) Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur. J. Biochem. 271, 253–262 [DOI] [PubMed] [Google Scholar]
- 8. Ueda M., Kojima M., Yoshikawa T., Mitsuda N., Araki K., Kawaguchi T., Miyatake K., Arai M., Fukamizo T. (2003) A novel type of family 19 chitinase from Aeromonas sp. No. 10S-24. Cloning, sequence, expression, and the enzymatic properties. Eur. J. Biochem. 270, 2513–2520 [DOI] [PubMed] [Google Scholar]
- 9. Watanabe T., Kanai R., Kawase T., Tanabe T., Mitsutomi M., Sakuda S., Miyashita K. (1999) Family 19 chitinases of Streptomyces species. Characterization and distribution. Microbiology 145, 3353–3363 [DOI] [PubMed] [Google Scholar]
- 10. Song H. K., Suh S. W. (1996) Refined structure of the chitinase from barley seeds at 2.0-Å resolution. Acta Crystallogr. D 52, 289–298 [DOI] [PubMed] [Google Scholar]
- 11. Kezuka Y., Ohishi M., Itoh Y., Watanabe J., Mitsutomi M., Watanabe T., Nonaka T. (2006) Structural studies of a two-domain chitinase from Streptomyces griseus HUT6037. J. Mol. Biol. 358, 472–484 [DOI] [PubMed] [Google Scholar]
- 12. Huet J., Rucktooa P., Clantin B., Azarkan M., Looze Y., Villeret V., Wintjens R. (2008) X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases. Biochemistry 47, 8283–8291 [DOI] [PubMed] [Google Scholar]
- 13. Holm L., Sander C. (1994) Structural similarity of plant chitinase and lysozymes from animals and phage. An evolutionary connection. FEBS Lett. 340, 129–132 [DOI] [PubMed] [Google Scholar]
- 14. Hart P. J., Pfluger H. D., Monzingo A. F., Hollis T., Robertus J. D. (1995) The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8-Å resolution. J. Mol. Biol. 248, 402–413 [PubMed] [Google Scholar]
- 15. Monzingo A. F., Marcotte E. M., Hart P. J., Robertus J. D. (1996) Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat. Struct. Biol. 3, 133–140 [DOI] [PubMed] [Google Scholar]
- 16. Iseli B., Armand S., Boller T., Neuhaus J. M., Henrissat B. (1996) Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 382, 186–188 [DOI] [PubMed] [Google Scholar]
- 17. Ueda M., Ohata K., Konishi T., Sutrisno A., Okada H., Nakazawa M., Miyatake K. (2009) A novel goose-type lysozyme gene with chitinolytic activity from the moderately thermophilic bacterium Ralstonia sp. A-471. Cloning, sequencing, and expression. Appl. Microbiol. Biotechnol. 81, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 18. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boller T., Gehri A., Mauch F., Vögeli U. (1983) Chitinase in bean leaves. Induction by ethylene, purification, properties, and possible function. Planta 157, 22–31 [DOI] [PubMed] [Google Scholar]
- 20. Jekel P. A., Hartmann B. H., Beintema J. J. (1991) The primary structure of hevamine, an enzyme with lysozyme/chitinase activity from Hevea brasiliensis latex. Eur. J. Biochem. 200, 123–130 [DOI] [PubMed] [Google Scholar]
- 21. Terwisscha van Scheltinga A. C., Kalk K. H., Beintema J. J., Dijkstra B. W. (1994) Crystal structures of hevamine, a plant defense protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure 2, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 22. Bokma E., van Koningsveld G. A., Jeronimus-Stratingh M., Beintema J. J. (1997) Hevamine, a chitinase from the rubber tree Hevea brasiliensis, cleaves peptidoglycan between the C-1 of N-acetylglucosamine and C-4 of N-acetylmuramic acid and therefore is not a lysozyme. FEBS Lett. 411, 161–163 [DOI] [PubMed] [Google Scholar]
- 23. McCarter J. D., Withers S. G. (1994) Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4, 885–892 [DOI] [PubMed] [Google Scholar]
- 24. Kuroki R., Weaver L. H., Matthews B. W. (1999) Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site. Proc. Natl. Acad. Sci. U.S.A. 96, 8949–8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weaver L. H., Grütter M. G., Matthews B. W. (1995) The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the “goose-type” lysozymes lack a catalytic aspartate residue. J. Mol. Biol. 245, 54–68 [DOI] [PubMed] [Google Scholar]
- 26. Karlsen S., Hough E., Rao Z. H., Isaacs N. W. (1996) Structure of a bulgecin-inhibited G-type lysozyme from the egg white of the Australian black swan. A comparison of the binding of bulgecin to three muramidases. Acta Crystallogr. D 52, 105–114 [DOI] [PubMed] [Google Scholar]
- 27. Kawamura S., Ohno K., Ohkuma M., Chijiiwa Y., Torikata T. (2006) Experimental verification of the crucial roles of Glu-73 in the catalytic activity and structural stability of goose type lysozyme. J. Biochem. 140, 75–85 [DOI] [PubMed] [Google Scholar]
- 28. Helland R., Larsen R. L., Finstad S., Kyomuhendo P., Larsen A. N. (2009) Crystal structures of G-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism. Cell. Mol. Life Sci. 66, 2585–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okazaki N., Arimori T., Nakazawa M., Miyatake K., Ueda M., Tamada T. (2011) Crystallization and preliminary x-ray diffraction studies of the catalytic domain of a novel chitinase, a member of GH family 23, from the moderately thermophilic bacterium Ralstonia sp. A-471. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LeMaster D. M., Richards F. M. (1985) 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 [DOI] [PubMed] [Google Scholar]
- 31. Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 32. Tamada T., Feese M. D., Ferri S. R., Kato Y., Yajima R., Toguri T., Kuroki R. (2004) Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea. Acta Crystallogr. D 60, 13–21 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cowtan K. (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D 55, 1555–1567 [DOI] [PubMed] [Google Scholar]
- 36. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 37. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 39. Imoto T., Yagishita K. (1971) A simple activity measurement of lysozyme. Agric. Biol. Chem. 35, 1154–1156 [Google Scholar]
- 40. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wohlkönig A., Huet J., Looze Y., Wintjens R. (2010) Structural relationships in the lysozyme superfamily. Significant evidence for glycoside hydrolase signature motifs. PLoS One 5, e15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guérin D. M., Lascombe M. B., Costabel M., Souchon H., Lamzin V., Béguin P., Alzari P. M. (2002) Atomic (0.94-Å) resolution structure of an inverting glycosidase in complex with substrate. J. Mol. Biol. 316, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 43. Petersen L., Ardèvol A., Rovira C., Reilly P. J. (2009) Mechanism of cellulose hydrolysis by inverting GH8 endoglucanases. A QM/MM metadynamics study. J. Phys. Chem. B 113, 7331–7339 [DOI] [PubMed] [Google Scholar]
- 44. Parsiegla G., Juy M., Reverbel-Leroy C., Tardif C., Belaïch J. P., Driguez H., Haser R. (1998) The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0-Å resolution. EMBO J. 17, 5551–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.