Background: Some proteins are ubiquitinated on their N terminus, yet the enzymes that facilitate N-terminal ubiquitination are unknown.
Results: Ube2w has a novel active site and ubiquitinates the N terminus of substrates.
Conclusion: Ube2w is an N terminus-ubiquitinating E2.
Significance: Ube2w is the first identified E2 that ubiquitinates the N terminus of substrates.
Keywords: Neurodegeneration, Post-translational Modification, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination, N-terminal Ubiquitination
Abstract
Attachment of ubiquitin to substrate is typically thought to occur via formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and a lysine residue in the substrate. In vitro, Ube2w is nonreactive with free lysine yet readily ubiquitinates substrate. Ube2w also contains novel residues within its active site that are important for its ability to ubiquitinate substrate. To identify the site of modification, we analyzed ubiquitinated substrates by mass spectrometry and found the N-terminal -NH2 group as the site of conjugation. To confirm N-terminal ubiquitination, we generated lysine-less and N-terminally blocked versions of one substrate, the polyglutamine disease protein ataxin-3, and showed that Ube2w can ubiquitinate a lysine-less, but not N-terminally blocked, ataxin-3. This was confirmed with a second substrate, the neurodegenerative disease protein Tau. Finally, we directly sequenced the N terminus of unmodified and ubiquitinated ataxin-3, demonstrating that Ube2w attaches ubiquitin to the N terminus of its substrates. Together these data demonstrate that Ube2w has novel enzymatic properties that direct ubiquitination of the N terminus of substrates.
Introduction
The attachment of ubiquitin to a substrate occurs through a cascade of three enzymes: the E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase. Ubiquitination typically results in the formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and a lysine residue in the substrate protein. Less commonly than lysine, other residues including serine, threonine, and cysteine have been reported to be conjugated with ubiquitin via formation of oxyester and thioester bonds, respectively (1–8). In addition to conjugation to the side chain of amino acids, ubiquitin can also be conjugated to the N-terminal -NH2 group of some proteins (9–17).
N-terminal ubiquitination has been shown to target a number of proteins for degradation including transcription factors, cell cycle regulators, and viral proteins (18). The enzymes mediating N-terminal ubiquitination, however, remain elusive, and this gap in knowledge represents a major barrier to achieving a mechanistic understanding of N-terminal ubiquitination. Recently, we identified Ube2w as an E2 that functions with the ubiquitin ligase C terminus of Hsc70-interacting protein (CHIP)3 (19). Here we show that the E2 Ube2w is not reactive toward free lysine and contains novel residues in its active site that are important for activity. We further show that Ube2w modifies the N terminus of its substrates rather than lysine residues. Together the results identify Ube2w as the first known N terminus-modifying E2.
EXPERIMENTAL PROCEDURES
Nucleophile Reactivity Assays
Reaction mixtures containing 20 μm E2, 20 μm Ub, 0.1 μm E1, and 5 mm ATP/MgCl2 in 50 mm Tris, 50 mm KCl, pH 7.5, were incubated for 15 min at 37 °C prior to the addition of indicated l-amino acids (Sigma). Amino acids were added to a final concentration of 50 mm and incubated for 15 min at 37 °C, quenched with nonreducing sample buffer, and run on 12% SDS-PAGE without boiling. Samples were visualized by colloidal blue Coomassie stain (Invitrogen).
Sequence Alignment
Amino acid sequences of human E2s were downloaded from National Center for Biotechnology Information (NCBI), entered into the Biology Workbench, and aligned with CLUSTALW.
Thioester Assay
Reaction mixtures containing 20 μm E2, 20 μm Ub, 0.1 μm E1, and 5 mm ATP/MgCl2 in 50 mm Tris, 50 mm KCl, pH 7.5, were incubated for the indicated time at 37 °C. Reactions were quenched with sample buffer with or without βME. Reactions with βME were heated for 4 min at 95 °C, and all samples were then separated by 12% SDS-PAGE and visualized by colloidal blue Coomassie stain (Invitrogen).
Ubiquitination Assays
Ubiquitination was typically performed at 37 °C in 10-μl mixtures containing reaction buffer E1mix (5 mm ATP, 5 mm MgCl2, 100 nm Ube1, and 500 μm ubiquitin), 1 μm of the indicated E2, 1 μm CHIP, and 1 μm substrate (ataxin-3 or Tau). Reactions were stopped by the addition of SDS-Laemmli buffer and boiling followed by separation of proteins by SDS-PAGE and visualization by Western blotting with appropriate antibodies.
Protein Identification by LC-MS/MS
Protein identification and ubiquitination sites on ataxin-3 were conducted based on previously described protocols (20). Briefly, protein bands corresponding to unmodified and modified AT3 were excised and destained with 30% methanol for 4 h. Upon reduction (10 mm DTT) and alkylation (65 mm 2-chloroacetamide or iodoacetamide, with similar results) of the cysteines, proteins were digested overnight with sequencing grade modified trypsin (Promega). The resulting peptides were resolved on a nano-capillary reverse phase column (PicoFrit column, New Objective) using a 1% acetic acid/acetonitrile gradient at 300 nl/min and directly introduced into a linear ion-trap mass spectrometer (LTQ Orbitrap XL, Thermo Fisher). Data-dependent MS/MS spectra on the five most intense ions from each full MS scan were collected (relative collision energy ∼35%). Proteins were identified by searching the data against the Human International Protein Index database (version 3.5) appended with decoy (reverse) sequences using the X!Tandem/Trans-Proteomic Pipeline (TPP) software suite. All peptides and proteins with a PeptideProphet and ProteinProphet probability score of >0.9 (false discovery rate <2%) were considered positive identifications and manually verified.
Carbamylation of Proteins
Carbamylation of substrates was performed as described previously (21). Purified ataxin-3 or Tau proteins were incubated in 0.2 m potassium phosphate, pH 6.0, 6 m urea, 50 mm potassium cyanate at 37 °C for 8 h. Reactions were stopped by the addition of 150 mm Gly-Gly (Sigma), and the pH was adjusted to pH 8.1 with 30% K2HPO4, pH 11.0. Samples were then incubated for 1 h at 37 °C prior to dialysis against H2O overnight with two H2O changes. The carbamylated protein was then quantified, aliquoted, and frozen at −80 °C prior to use in assays.
Edman Sequencing
Samples for Edman sequencing were separated by SDS-PAGE, transferred to PVDF (Immobilon-PSQ, Millipore), and stained with Ponceau stain. Bands were excised, and samples were submitted for Edman sequencing (Alphalyse).
RESULTS
Ube2w Is Nonreactive with Free Lysine
E2s that function with RING and U-box E3s directly transfer ubiquitin to substrate, typically to a lysine residue. In some cases, E2s transfer ubiquitin to residues other than lysine, and some E2s are incapable of transferring ubiquitin to lysine (22). To assess the reactivity profile of Ube2w, we compared the intrinsic activity of Ube2w∼Ub (∼Ub indicates a thioester bond with E2) and UbcH5c∼Ub with a panel of free amino acids including residues known to be ubiquitinated: lysine, cysteine, serine, and threonine as well as arginine, asparagine, glutamine, and histidine (Fig. 1A, top panel). Although UbcH5c∼Ub, a known lysine-reactive E2, discharged ubiquitin in the presence of lysine and cysteine, Ube2w∼Ub reacted only with cysteine (Fig. 1A). Similar results have been observed for another E2, UbcH7, which functions with HECT and RBR (RING-between-RING) E3s to transfer ubiquitin to the active site cysteine of the E3 (22). Ube2w, however, transfers ubiquitin in complex with RING/U-box type E3s that do not have active site cysteine residues to form thioester bonds with ubiquitin (23). Furthermore, substrates that are ubiquitinated by Ube2w are not sensitive to reducing agents, arguing against a role for Ube2w in transferring ubiquitin to a cysteine residue on the substrate (19).
FIGURE 1.
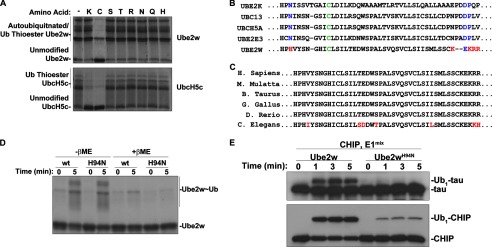
Ube2w has novel enzymatic properties. A, amino acid reactivity of Ube2w∼Ub and UbcH5c∼Ub. Ube2w or UbcH5c was charged with ubiquitin for 5 min prior to the addition of the indicated free amino acids for 25 min. Samples were analyzed by Coomassie Blue stain. B, sequence alignment of key active site residues for select E2s. Green indicates the active cysteine residue, blue indicates conserved key active site residues, and red indicates differences in Ube2w. H. Sapiens, Homo sapiens; M. Mulatta, Macaca mulatta; B. Taurus, Bos taurus; G. Gallus, Gallus gallus; D. Rerio, Danio rerio; C. Elegans, Caenorhabditis elegans. C, alignment of Ube2w from different species. Red indicates residues that are nonidentical residues across species. D, thioester formation by Ube2w and Ube2wH94N. Ube2w or Ube2wH94N was incubated with E1, ubiquitin, and ATP/MgCl2 for either 0 or 5 min prior to the addition of SDS sample buffer with or without βME (as indicated). Samples were analyzed by Coomassie Blue stain. E, Ube2wH94N has reduced capacity to ubiquitinate substrate. Tau ubiquitination reactions were performed with Ube2w or Ube2wH94N for the indicated lengths of time. Samples were analyzed by Western blot for either Tau or CHIP, as indicated.
Ube2w Has a Novel Active Site
To gain insight into differences in active site residues of Ube2w, we performed CLUSTALW alignment of E2s (Fig. 1B). Among residues known to be involved in E2 catalysis, Ube2w has a histidine (His-94) in place of the highly conserved asparagine observed in other E2s, such as Asn-77 in UbcH5a (Fig. 1B). Additionally, Asp-117 of UbcH5a and related E2s is thought to be important for suppressing the pKa of substrate lysine (24, 25), and although this negatively charged residue is conserved in Ube2w (Glu-132), it is uniquely surrounded by a cluster of basic residues, making it unlikely that it could serve a similar function in deprotonating substrate lysine (Fig. 1B). These features of the active site of Ube2w are likely important for its activity because they are highly conserved in Ube2w orthologs across evolution (Fig. 1C). To begin assessing the importance of these residues in the active site of Ube2w, we mutated His-94 of Ube2w to the more commonly found asparagine in related E2s. Although Ube2wH94N formed a thioester bond with ubiquitin as efficiently as wild-type Ube2w (Fig. 1D), the H94N mutation markedly impaired substrate ubiquitination of both Tau and CHIP (Fig. 1E). Separately, we mutated the basic cluster of amino acids in Ube2w to resemble UbcH5a, but this caused the mutant protein to become insoluble, preventing its use in ubiquitination assays (data not shown). Together these results suggest that Ube2w has a novel active site that is necessary for it to ubiquitinate substrate.
Ube2w Ubiquitinates the N Terminus of Substrates
In addition to internal lysine, serine, threonine, and cysteine residues, ubiquitin can also be attached to the N-terminal -NH2 group of select proteins (18). One HECT type E3 has been demonstrated to N-terminally ubiquitinate substrate (26), but to date no N terminus-ubiquitinating E2s have been identified, thus far excluding RING/U-box type E3s from facilitating N-terminal modification. To determine where Ube2w ubiquitinates its substrates, we monoubiquitinated several substrates with Ube2w and subjected them to mass spec analysis. This analysis indicated that the Ube2w preferentially ubiquitinates the N terminus of tested substrates including ataxin-3 (Fig. 2A), Tau, HSP70, HSP90, and Ube2w (data not shown).
FIGURE 2.
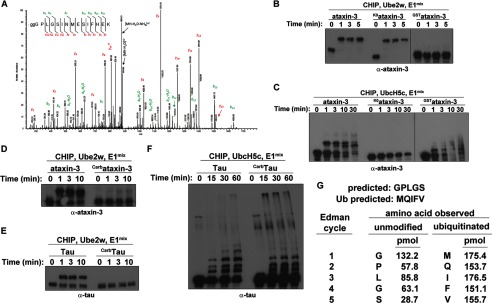
Ube2w modifies the N terminus of substrates. A, Ube2w ubiquitinates the N terminus of ataxin-3. Ataxin-3 was ubiquitinated in vitro by Ube2w, run on SDS-PAGE, and stained with Coomassie Blue. After in-gel digestion with trypsin, peptides were subjected to LC-MS/MS analysis using an orbitrap mass spectrometer. The resulting MS/MS spectra were searched against the Swiss-Prot human protein database appended with synthetic ataxin-3 using the x!Tandem/TPP software suite, considering N-terminal ubiquitination as both a fixed and a variable modification. All peptide-to-spectral matches (PSMs) with a PeptideProphet probability of >0.9 were considered correct assignments. PSMs of modified peptides were manually verified, and a representative spectrum is shown. Observed b- and y-ion series are indicated. The majority (46/50) of PSMs assigned to N-terminal peptide were ubiquitinated, indicating modification of the ataxin-3 N terminus. B, Ube2w modifies lysine-less ataxin-3 but not N-terminal GST-tagged ataxin-3. In vitro ubiquitination reactions were performed for the indicated lengths of time with ataxin-3, K0ataxin-3, or GSTataxin-3. Samples were analyzed by Western blot with anti-ataxin-3 antibody. C, as in B except employing UbcH5c as the E2 instead of Ube2w. D, chemical modification of the N terminus of ataxin-3 prevents Ube2w-mediated ubiquitination. Ubiquitination reactions were performed for the indicated lengths of time with ataxin-3 or carbamylated ataxin-3 as substrate. E, as in D but employing Tau or carbamylated Tau as substrate. F, as in E except employing UbcH5c as the E2 instead of Ube2w. G, results of Edman sequencing are consistent with N-terminal modification of substrates by Ube2w. Unmodified ataxin-3 or ataxin-3 ubiquitinated by Ube2w were transferred to PVDF and Ponceau-stained, and bands corresponding to unmodified or ubiquitinated ataxin-3 were subjected to Edman sequencing. The predicted sequences for unmodified or ubiquitinated ataixn-3 are shown. Blank and standard cycles were run prior to each run, and pmols of amino acid observed from each run were quantified.
In addition to N-terminal ubiquitination of ataxin-3, we also identified a small number of peptides in which Lys-200 of ataxin-3 had been ubiquitinated (data not shown). To determine whether Lys-200 is an important site of conjugation, we mutated Lys-200 to arginine and observed that eliminating Lys-200 did not alter ataxin-3 modification by Ube2w (data not shown). Moreover, mutating all lysines in ataxin-3 to arginine (ataxin-3K0) did not prevent ataxin-3 ubiquitination by Ube2w (Fig. 2B). By contrast, altering the N terminus of ataxin-3 either by adding a GST fusion (GSTataxin-3; Fig. 2B) or by carbamylation (Carbataxin-3; Fig. 2D), which selectively modifies the N terminus of proteins (11), inhibited ubiquitination of ataxin-3 by Ube2w (Fig. 2, B and D) but not by UbcH5c (Fig. 2C, data not shown). To confirm that the observed effects were specific to Ube2w, we confirmed that UbcH5c, a lysine-reactive E2, was unable to ubiquitinate ataxin-3K0 yet could ubiquitinate GSTataxin-3 and Carbataxin-3 (Fig. 2, C and D). Carbamylation also inhibited ubiquitination of a second substrate, Tau, by Ube2w (Fig. 2E) but not by UbcH5c (Fig. 2F).
Finally, to confirm that the N terminus is the site of conjugation by Ube2w, we purified unmodified and ubiquitinated ataxin-3 and subjected these samples to Edman sequencing. Unmodified ataxin-3 gave the expected residues for the N terminus of ataxin-3, whereas ataxin-3 ubiquitinated by Ube2w only gave chromatographs consistent with ubiquitin and lacked any signal corresponding to the N terminus of ataxin-3, consistent with N-terminal ubiquitination of ataxin-3 (Fig. 2G and data not shown). In addition, although Edman sequencing of Tau revealed multiple N-terminal sequences consistent with partial cleavage of Tau during the purification process (27), upon ubiquitination by Ube2w, only N-terminal residues consistent with ubiquitin were observed (data not shown). These results demonstrate that Ube2w preferentially ubiquitinates the N terminus of substrates.
DISCUSSION
Here we have shown that Ube2w is a non-lysine-reactive E2 with a novel active site. Mutations of the Ube2w active site to residues found in traditional, lysine-reactive E2s inhibited Ube2w activity, suggesting that Ube2w has novel enzymatic properties. We also provide multiple lines of evidence that Ube2w is an N terminus-modifying E2: 1) mass spectrometry directly identified N-terminal modification of five substrates; 2) Ube2w effectively ubiquitinated a lysine-less version of ataxin-3; 3) modification of the N terminus of two substrates, ataxin-3 and Tau, inhibited their ubiquitination by Ube2w; and 4) after Ube2w-mediated ubiquitination of these substrates, only the N-terminal sequence of ubiquitin was obtained upon Edman sequencing. Together these findings argue that Ube2w contains a novel active site that mediates N-terminal ubiquitination of substrates. Ube2w thus represents the first identified E2 to transfer ubiquitin to the N termini of substrates.
It has been known for some time that certain proteins can be targeted for degradation via N-terminal ubiquitination (18). The enzymes responsible for N-terminal modification have remained elusive. One E3, HUWE1 (HECT, UBA, and WWE domain-containing 1), has been shown to ubiquitinate lysine-less MyoD on its N terminus, but this activity may not be physiologically relevant as HUWE1 preferentially modifies lysines and fails to modify the N terminus of wild-type MyoD (26). By contrast, eliminating all lysines in ataxin-3 does not alter the rate or extent of ubiquitination of ataxin-3 by Ube2w, strongly suggesting that the physiological target of Ube2w is the N terminus of substrates.
In addition to modification of the N terminus of substrates, we also observed limited modification of internal lysine residues by Ube2w. We observed modification of Lys-200 of ataxin-3 (data not shown) and previously observed Lys-2 modification of CHIP (19). In both cases, however, mutating these lysines to arginine did not inhibit the ability of Ube2w to ubiquitinate the substrate, arguing that lysine modification by Ube2w is not likely to be its primary physiological role.
Ube2w strictly monoubiquitinates substrates. We have yet to observe appreciable amounts of multi-monoubiquitinated substrate in Ube2w reactions, supporting a restricted role for Ube2w in N-terminal modification. Intriguingly, Ube2w does not elongate linear ubiquitin chains as has been observed for the linear ubiquitin chain assembly complex (LUBAC) (28). Unlike Ube2w, which attaches ubiquitin to the N terminus of substrates, LUBAC specifically extends linear polyubiquitin chains that are attached to an internal lysine residue of one of its substrates, NF-κB essential modulator (NEMO) (29). Although LUBAC recognizes and extends polyubiquitin chains via the N terminus of ubiquitin, Ube2w does not extend polyubiquitin chains. The failure of Ube2w to extend chains likely reflects the lack of a noncovalent interaction between Ube2w and ubiquitin (30) as noncovalent interactions with ubiquitin appear to be a property common to ubiquitin chain-elongating E2s (30–35). Moreover, Ube2w does not ubiquitinate an N-terminally GST-tagged version of ataxin-3, suggesting that undefined structural features of the substrate participate in recognition of substrates by Ube2w. How Ube2w recognizes its substrates and why it does not recognize the N terminus of ubiquitin nor GST as a substrate are important unanswered questions.
Given the novel function of Ube2w as an N terminus-conjugating E2, it now will be important to define its physiological role in cells. N-terminal ubiquitination has been demonstrated to target some proteins for degradation via the proteasome (18). An alternative possible function is suggested by that fact that linear ubiquitin chain formation by LUBAC participates in signaling pathways (28). It will be important to identify bona fide in vivo targets of Ube2w to begin defining the in vivo role of Ube2w. Further work elucidating the outcome of N-terminal ubiquitination by Ube2w on its substrates will be critical to begin understanding the function of Ube2w in vivo.
During the preparation of this manuscript, Tatham et al. (36) also reported that Ube2w N-terminally modified two substrates, small ubiquitin modifier-2 (SUMO-2) and CHIP.
Acknowledgments
We thank members of the Paulson laboratory for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants K99 NS073936 (to K. M. S.), R00 NS064097 (to S. V. T.), and R01 AG034228 and R01 NS038712 (to H. L. P.).
- CHIP
- C terminus of Hsc70-interacting protein
- Ub
- ubiquitin
- Ubc
- ubiquitin-conjugating enzyme
- βME
- β-mercaptoethanol
- HECT
- homologous to the E6-AP C terminus
- LUBAC
- linear ubiquitin assembly complex
- NEMO
- NF-κB essential modulator
- PSM
- peptide-to-spectral match.
REFERENCES
- 1. Cadwell K., Coscoy L. (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 [DOI] [PubMed] [Google Scholar]
- 2. Cadwell K., Coscoy L. (2008) The specificities of Kaposi's sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J. Virol. 82, 4184–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., Azevedo J. E. (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 [DOI] [PubMed] [Google Scholar]
- 4. Herr R. A., Harris J., Fang S., Wang X., Hansen T. H. (2009) Role of the RING-CH domain of viral ligase mK3 in ubiquitination of non-lysine and lysine MHC I residues. Traffic 10, 1301–1317 [DOI] [PubMed] [Google Scholar]
- 5. Tait S. W., de Vries E., Maas C., Keller A. M., D'Santos C. S., Borst J. (2007) Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J. Cell Biol. 179, 1453–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Herr R. A., Rabelink M., Hoeben R. C., Wiertz E. J., Hansen T. H. (2009) Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 187, 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams C., van den Berg M., Sprenger R. R., Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 9. Aviel S., Winberg G., Massucci M., Ciechanover A. (2000) Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway: targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275, 23491–23499 [DOI] [PubMed] [Google Scholar]
- 10. Ben-Saadon R., Fajerman I., Ziv T., Hellman U., Schwartz A. L., Ciechanover A. (2004) The tumor suppressor protein p16INK4a and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system: direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 279, 41414–41421 [DOI] [PubMed] [Google Scholar]
- 11. Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fajerman I., Schwartz A. L., Ciechanover A. (2004) Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem. Biophys. Res. Commun. 314, 505–512 [DOI] [PubMed] [Google Scholar]
- 13. Ikeda M., Ikeda A., Longnecker R. (2002) Lysine-independent ubiquitination of Epstein-Barr virus LMP2A. Virology 300, 153–159 [DOI] [PubMed] [Google Scholar]
- 14. Kuo M. L., den Besten W., Bertwistle D., Roussel M. F., Sherr C. J. (2004) N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18, 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reinstein E., Scheffner M., Oren M., Ciechanover A., Schwartz A. (2000) Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19, 5944–5950 [DOI] [PubMed] [Google Scholar]
- 16. Trausch-Azar J., Leone T. C., Kelly D. P., Schwartz A. L. (2010) Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1α via the N-terminal pathway. J. Biol. Chem. 285, 40192–40200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vosper J. M., McDowell G. S., Hindley C. J., Fiore-Heriche C. S., Kucerova R., Horan I., Philpott A. (2009) Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284, 15458–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ciechanover A., Ben-Saadon R. (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 19. Scaglione K. M., Zavodszky E., Todi S. V., Patury S., Xu P., Rodríguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., Gestwicki J. E., Paulson H. L. (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todi S. V., Scaglione K. M., Blount J. R., Basrur V., Conlon K. P., Pastore A., Elenitoba-Johnson K., Paulson H. L. (2010) Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J. Biol. Chem. 285, 39303–39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bloom J., Pagano M. (2005) Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol. 399, 249–266 [DOI] [PubMed] [Google Scholar]
- 22. Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 24. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yunus A. A., Lima C. D. (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 [DOI] [PubMed] [Google Scholar]
- 26. Noy T., Suad O., Taglicht D., Ciechanover A. (2012) HUWE1 ubiquitinates MyoD and targets it for proteasomal degradation. Biochem. Biophys. Res. Commun. 418, 408–413 [DOI] [PubMed] [Google Scholar]
- 27. Barghorn S., Biernat J., Mandelkow E. (2005) Purification of recombinant Tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol. Biol. 299, 35–51 [DOI] [PubMed] [Google Scholar]
- 28. Tokunaga F., Iwai K. (2012) Linear ubiquitination: a novel NF-κB regulatory mechanism for inflammatory and immune responses by the LUBAC ubiquitin ligase complex. Endocr. J. 59, 641–652 [DOI] [PubMed] [Google Scholar]
- 29. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 30. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 31. Choi Y. S., Wu K., Jeong K., Lee D., Jeon Y. H., Choi B. S., Pan Z. Q., Ryu K. S., Cheong C. (2010) The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 285, 17754–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 33. Rodrigo-Brenni M. C., Foster S. A., Morgan D. O. (2010) Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tatham M. H., Plechanovová A., Jaffray E. G., Salmen H., Hay R. T. (2013) Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 10.1042/BJ20130244 [DOI] [PMC free article] [PubMed] [Google Scholar]


