Background: Phage display technology is commonly used to improve peptide and protein affinities.
Results: A system circumventing a key bottleneck in this technology is used to improve a peptide inhibitor of HIV-1 entry.
Conclusion: This system enables integration of sensitive and functional assays such as flow cytometry and viral neutralization into an iterative phage display workflow.
Significance: This approach expands the power of a critical biotechnology.
Keywords: Antimicrobial Peptides, Antiviral Agents, HIV-1, Human Immunodeficiency Virus, Phage Display, Fc Fusion Protein, Immunoadhesin
Abstract
Phage display is a key technology for the identification and maturation of high affinity peptides, antibodies, and other proteins. However, limitations of bacterial expression restrict the range and sensitivity of assays that can be used to evaluate phage-selected variants. To address this problem, selected genes are typically transferred to mammalian expression vectors, a major rate-limiting step in the iterative improvement of peptides and proteins. Here we describe a system that combines phage display and efficient mammalian expression in a single vector, pDQ1. This system permits immediate expression of phage-selected genes as IgG1-Fc fusions in mammalian cells, facilitating the rapid, sensitive characterization of a large number of library outputs for their biochemical and functional properties. We demonstrate the utility of this system by improving the ability of a CD4-mimetic peptide to bind the HIV-1 envelope glycoprotein and neutralize HIV-1 entry. We further improved the potency of the resulting peptide, CD4mim6, by limiting its ability to induce the CD4-bound conformation of the envelope glycoprotein. Thus, CD4mim6 and its variants can be used to investigate the properties of the HIV-1 envelope glycoprotein, and pDQ1 can accelerate the discovery of new peptides and proteins through phage display.
Introduction
Phage display technology is frequently used to identify protein variants that will ultimately be produced in mammalian cells or expressed in vivo (1–3). However, bacterially expressed proteins selected as fusions with a phage coat protein do not always express or retain their function in mammalian cells, and in standard phage display protocols, these defective proteins are retained throughout the selection process (2, 4). Moreover, peptides too small to be expressed by themselves are typically first evaluated on the surface of the phage by ELISA, but this approach is quantitatively imprecise and retains artifacts from the original selection (5, 6). This has led to the exploration of alternative display methods, such as yeast or mammalian cell surface display (7–9). However, the library sizes possible with these approaches, and thus the complexity of the sequence space that can be probed, are orders of magnitude lower than that routinely achieved with phage libraries. To circumvent these difficulties while retaining the power of the phage display method, library outputs can be subcloned to generate fusion proteins, a time-consuming step that limits the number of outputs that can be so evaluated (1–3, 10, 11). Expression in bacteria also precludes use of certain fusion proteins, notably those with antibody Fc domains. Fc domains facilitate the use of a broad set of commercial tools for purification, immunoprecipitation, flow cytometry, and functional studies. Ideally, one would incorporate such studies early in the validation of phage library outputs (12). Accordingly, we developed a vector that expresses library variants as phage pIII coat-protein fusions in bacterial cells and as fusions with the human IgG1 Fc domain in mammalian cells. This was achieved by inserting the machinery of bacterial expression and phage display within the introns of a mammalian expression vector. We demonstrated the utility of this system by improving the potency of a natural amino acid form of a previously described peptide inhibitor of HIV-1 entry. We further show that the resulting peptide and its variants can be used to explore conformational transitions of the HIV-1 envelope glycoprotein.
EXPERIMENTAL PROCEDURES
pDQ1 Vector Construction
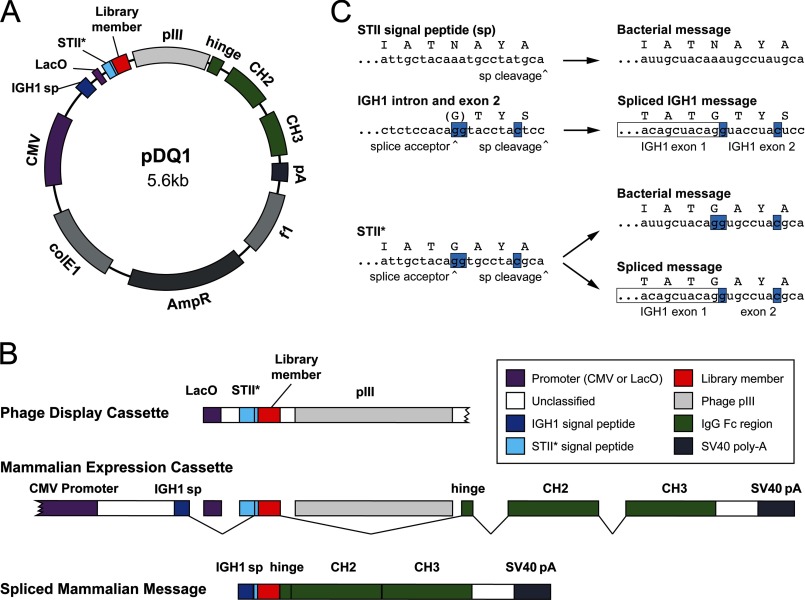
pDQ1, represented in Fig. 1A, was constructed by inserting an Fc fusion expression cassette into a pUC-derived plasmid. A segment following the CMV promoter was replaced with sequence encoding the IGH1 signal peptide. The 5′ end of the signal peptide intron was then replaced with sequence encoding the Lac operon promoter, a ribosome binding site, and the STII* signal peptide, followed by a CD4-mimetic peptide. The nucleotide and amino acid sequence of the STII* signal peptide is detailed in Fig. 1. Finally, a segment from a pComb3 vector (13) encoding the HA tag and the pIII fusion was inserted into the IgG1 hinge-CH1 intron immediately following the VH1 splice donor.
FIGURE 1.
Design of pDQ1, a vector for phage display and mammalian cell expression of protein-Fc fusions. A, map of pDQ1, with mammalian (CMV) and bacterial (LacO) promoters indicated (purple); exon 1 of the heavy chain of IgG1 (IGH1), encoding most of the IgG1 signal peptide (blue); STII*, encoding a form of the bacterial signal peptide STII (heat-stable enterotoxin II) modified to include a splice acceptor (light blue); the diversified library member (red); the phage pIII protein; individual exons comprising the Fc domain of human IgG1 (green); and an SV40 polyadenylation termination signal (gray). B, bacterial and mammalian expression cassettes. Phage display uses a cassette including the LacO promoter and sequence encoding the STII* signal peptide and a library component fused to the phage PIII protein. In contrast, mammalian expression utilizes a CMV promoter and encodes a chimera of the IgG1 signal peptide and STII*, the same library component, and the Fc domain of IgG1, terminated with an SV40 polyadenylation signal. Note that in mammalian cells splicing excises the LacO promoter, most of STII*, and the PIII gene. C, modifications of the bacterial STII signal peptide necessary for expression in mammalian cells. The DNA and amino acid sequences of the C terminus of the STII signal peptide are shown at the top, with the signal peptidase cleavage site indicated. This signal peptide functions in bacteria but not in mammalian cells. Below that, the intronic sequence and the 5′ region of IGH1 exon 2 are shown. This region encodes the C terminus of the IgG1 signal peptide. The 3′ region of the splice acceptor and the site of signal peptidase cleavage are indicated. In mammalian cells, IGH exon 2 is spliced to exon 1, encoding the complete IGH1 signal peptide. Underneath, sequence encoding the C terminus of the STII* signal peptide is shown. Blue highlighting indicates nucleotides from exon 2 of IGH1 that introduce an efficient splice acceptor while retaining signal peptide function in both bacterial and mammalian cells. In bacteria, the entire STII* signal peptide is used, whereas in mammalian cells, exon 1 of IGH1 is spliced to a short 3′ region of STII*.
Library Design and Assembly
Primers were designed using four handmixes, each containing 85% of the template nucleotide and 5% of the remaining three nucleotides. Library 1 was constructed using CD4mim2 as a template. In all cases positions 6, 10, 21–24, and 26 were held fixed as indicated in Fig. 2B. The remaining positions were soft randomized with the appropriate handmix for the first two codon positions, and 50:50 G:C mix was used for the wobble position. Library 2 was constructed using CD4mim4 as a template. In this iteration, the wobble position was soft-randomized for amino-acids encoded by fewer than four codons (Lys, Asn, His, Gln, Asp, Glu, Met, Ile, Tyr, and Trp), and a 50:50 G:C mix was used in the remaining cases (see supplemental Fig. 1). The 3′ end of the primer extended outside the randomization region and was of sufficient length to have a melting temperature ≥ 72 °C. Primers were phosphorylated with T4 PNK (New England Biosciences) and used for inverse PCR in 96-well plates with Phusion Flash (Finnzymes). The PCR product was pooled, column-purified (Qiagen), and ligated with T4 DNA Ligase (New England Biolabs). The ligation product was precipitated using yeast tRNA carrier (14) and transformed into MC1061 F′ (Lucigen), titered on LB agar ampicillin, and spread onto multiple 140-mm LB agar carbenicillin/tetracycline/glucose plates.
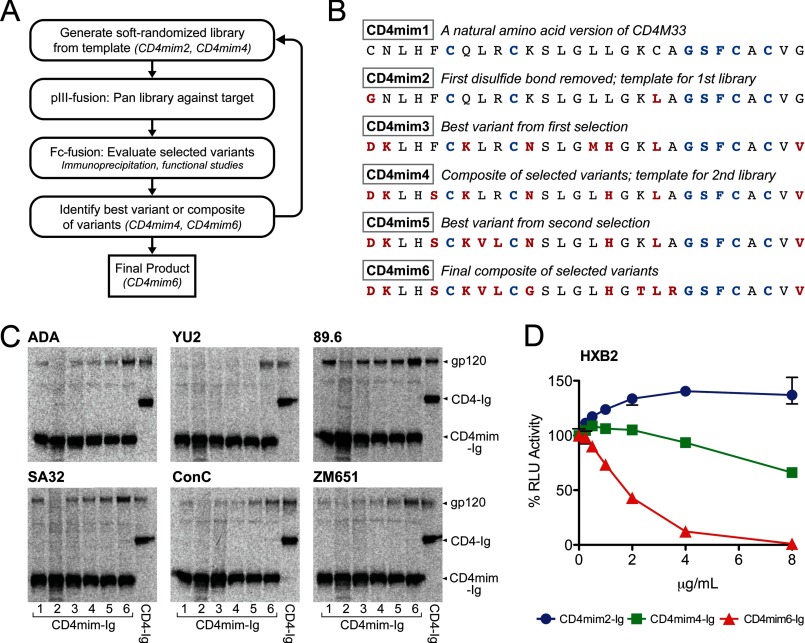
FIGURE 2.
Incorporation of mammalian cell expression and functional assays into the workflow of phage display. A, streamlined phage display workflow used in this study. Plasmids selected in panning against target were isolated and used to transfect 293T cells. Library outputs were produced as Fc fusion proteins which facilitated immunoprecipitation, flow cytometry, and neutralization studies. Elements of high performing variants were combined and reassayed. A best composite variant was used as a template for a subsequent round of phage selection and characterization of Fc fusions. B, sequences of the CD4mim variants with descriptions. Blue indicates seven residues held fixed through the process, red indicates changes from CD4mim1, a natural amino acid form of a previously described CD4-mimetic peptide. C, example immunoprecipitation study used to validate library outputs. Here, isotopically labeled CD4mim1–6 and CD4-Ig are compared for their abilities to precipitate labeled gp120 of the indicated isolate. ADA, YU2, and 89.6 are clade B isolates; SA32, ConC, and ZM651 are clade C isolates. The experiment is representative of three with similar results. D, example of a functional study used to characterize library outputs. Here, CD4mim2-Ig, CD4mim4, and CD4mim6-Ig are compared using a TZM-bl HIV-1 neutralization assay. Error bars represent a range of triplicates.
Phage Production and Panning
Libraries were resuspended from bacterial plates using Super Broth + 0.5% glucose and transduced with VCSM13 helper phage (Stratagene) as described by Rader and Barbas (15). Briefly, expression was induced overnight with SB autoinduction medium (2% yeast extract, 3% tryptone, 1% MOPS, 0.5% glycerol, 1 mm MgCl2, 0.05% glucose, 0.05% lactose) containing 100 mg/liter carbenicillin and 50 mg/liter kanamycin. The supernatant was cleared by centrifugation, passed through a 0.45-μm filter, and precipitated with PEG/NaCl + EDTA. The pellet was resuspended in cold Tris-buffered saline (TBS) and EDTA was added. gp120-Ig or Fc-only protein A or protein G magnetic beads (Invitrogen) was prepared by incubation of gp120-Ig or Fc control with beads for 1 h at 4 °C in TBS-TC (0.01% Tween 20, 1% sodium caseinate) followed by two washes. Freshly prepared phage were incubated for 10 min with beads and washed 10–20 times with TBS-T. Retained phage were eluted with freshly prepared 1% trypsin in TBS, and eluate was used to transform cultures of NEB F′Iq, and dilutions of gp120 and Fc control cultures were plated to monitor enrichment. After 1 h of recovery in Super Broth + glucose, cultures were plated as before. Colonies from the titer plates were sequenced to monitor library diversity, and the expansion and panning process was repeated for a total of three or four pans until the library size was reduced to ∼30 unique clones of 200 sequences.
Immunoprecipitation
Miniprep DNA prepared from sequenced colonies was transfected into 12-well plates of 50% confluent 293T cells at 1 μg of DNA/well by standard calcium phosphate technique. HIV-1 gp120 constructs were transfected into 6-well plates. 12 h after transfection, medium was replaced with cysteine/methionine-negative DMEM containing ExpressLabel (Cole Parmer) and 5% dialyzed calf serum (Invitrogen). Supernatants were collected 24 h later and were cleared by centrifugation 2 min at 3000 rpm followed by 1 min at 10,000 rpm, and Complete protease inhibitor mixture (Roche Applied Science) was added. Immunoprecipitations were conducted by incubating normalized quantities of Ig fusions and gp120 supernatants with protein A beads for 2 h at 4 °C with rocking. Beads were washed three times with PBS-Tween (0.01%), and protein was eluted by heating 10 min at 70 °C in NuPage sample buffer. Samples were run on NuPage bis-tris3 gels, treated with methanol/acetic acid fixative and dried on Whatman paper. Radiolabeled protein was quantified with a PhosphorImager (Fuji).
Protein Production
Maxiprep DNA was prepared from 250-ml overnight LB cultures of DH5α carrying the pDQ1-CD4mimetic plasmid using PurLink maxiprep kits (Invitrogen) according to the manufacturer's instructions. 293T cells in 140-mm plates were transfected with 25 μg/plate at 50% confluence by standard calcium phosphate transfection. At 12 h after transfection, 10% FCS-DMEM was replaced with serum-free 293 Freestyle medium (Invitrogen). Medium was collected 48 h later, and debris was cleared by centrifugation for 10 min at 1500 × g and was filtered in 0.45-μm filter flasks (Millipore). Complete protease inhibitor mixture was added to the filtered supernatants. A 500-μl bed volume of protein A-Sepharose beads (GE Healthcare) was added and was agitated 4 °C overnight. The bead/medium mixture was collected by gravity flow column (Bio-Rad) and was washed with 30 ml of PBS (Lonza) + 0.5 m NaCl (0.65 m NaCl final) followed by 10 ml of PBS. Protein was eluted with 5 ml of 2 m arginine, pH 4, into 1 ml 1 m Tris, pH 7.5. Buffer was exchanged for PBS, and protein was concentrated to 1 mg/ml by ultrafiltration (Amicon Ultra) at 3000 × g.
Surface Plasmon Resonance Studies
Surface plasmon resonance biosensor data were collected on a Biacore 3000 optical biosensor (GE Healthcare). gp120 of the clade C Du151 isolate (Immune Technology Corp.) was coupled to cell 2 or cell 4 of a CM5 chip using the Amine Coupling Kit (Biacore). Kinetic data were collected at several concentrations of mimetic-Fc fusion proteins, with two repeated concentrations. Surface regeneration was by injection of 1 m potassium thiocyanate, 1% CHAPSO. Data were analyzed using BIAevaluation software (GE Healthcare) and fit to a 1:1 Langmuir binding model.
HIV-1 Neutralization Studies
Pseudotyped virus was produced by coexpression of envelope glycoproteins of the indicated HIV isolates, or the VSV-G protein, with NL43ΔEnv. 293T cells at 50% confluence in T175 (Falcon) flasks were transfected with 25 μg of plasmid encoding envelope glycoprotein and 45 μg of NLΔEnv by the standard calcium phosphate technique. 10% FCS-DMEM was changed at 12 h, and medium was collected at 48 h. Viral supernatants were cleared by centrifugation for 10 min at 1500 × g, passed through a 0.45-μm syringe filter (Millipore), and stored at −80 °C. Neutralization by Fc fusion constructs was performed according to the protocol described in Li et al. (16) In brief, varying concentrations of CD4 mimetic-Ig or free CD4-mimetic peptide (NeoBioLab) were incubated with virus at a volume of 100 μl/well in 96-well plates for 1 h at after which 10,000 cells/well TZM-bl cells in 100 μl of medium were added. Plates were incubated 72 h at 37 °C after which 100 μl of medium was replaced with freshly prepared Brite-Lite reagent (PerkinElmer Life Sciences), and luminescence data were collected on a Victor3V (Perkin Elmer Life Sciences). For peptide neutralizations, 10,000 cells/well were first plated in 96-well plates. 12 h later, peptide and pseudoviruses were mixed in V-bottom 96-well plates, incubated 1 h, and then added to the adhered cells. The medium was changed after 2-h incubation at 37 °C. All neutralizations were performed in triplicate.
Infection of CCR5-positive, CD4-negative Cells
CF2th-CCR5 cells were incubated with pseudoviruses generated with pNL4–3.Luc.R-.E bearing the indicated HIV-1 envelope glycoprotein and encoding firefly luciferase in the presence of increasing amounts of peptide forms of CD4mim6, CD4mim6W, or soluble CD4. Infection was measured as in neutralization studies.
Staining of Cell-expressed HIV-1 Envelope Glycoprotein
Cells expressing HIV-1 envelope glycoprotein trimers were prepared by transient transfection of plasmids expressing envelope glycoproteins truncated in the C-terminal cytoplasmic domains to facilitate surface expression (gp160-ΔCT). T75 flasks of 293T at 50% confluence were transfected with 20 μg of plasmid encoding gp160-ΔCT and 3 μg of plasmid encoding tat by the calcium phosphate technique and incubated overnight. DMEM + 10% FCS was changed at 18 h after transfection. Cells were collected 36 h after transfection by treatment with nonenzymatic cell dissociation solution (Sigma), followed by centrifugation 5 min at 1200 rpm. Cells were resuspended in flow cytometry buffer (PBS + 2% donor goat serum + 0.1% sodium azide). Cells were incubated with Ig fusion proteins or the HIV-1 neutralizing antibodies E51 or 2G12 diluted to varying concentrations in FACS buffer for 45 min on ice, followed by two FACS buffer washes. APC-conjugated anti-human Fc-γ (Jackson ImmunoResearch) diluted in FACS buffer was added, and cells were incubated on ice for 30 min. Cells were then washed two times with flow cytometry buffer and PBS then fixed for flow cytometric analysis with 1% paraformaldehyde in PBS. Flow cytometry data were analyzed using FlowJo.
RESULTS
Design of a Dual Expression Vector
The vector, shown in Fig. 1A, exploits the fact that splicing occurs only in eukaryotic cells. Splice donors and acceptors were engineered to excise domains specific to phage display and thereby encode a protein-of-interest fusion with the Fc domain of human IgG1 (Fig. 1B). A first splice links regions encoding a mammalian signal peptide to one encoding the protein under selection, excising sequence encoding a bacterial promoter and signal peptide. A second splice links the gene for the protein under selection to sequence encoding the hinge region of IgG1, excising the gene for phage pIII. Thus in bacteria the vector functions like standard phagemid vectors (e.g. pCOMB3) used for phage display (13), whereas when transfected into mammalian cells, protein-Fc fusions are expressed. Our design required that the bacterial and mammalian signal sequences share four C-terminal amino acids and encode an efficient splice acceptor. No native bacterial or mammalian sequence met these requirements. However, using splice site and signal peptide prediction programs (17, 18), we combined elements from the bacterial STII signal peptide and the mammalian IgG1 signal peptide to generate a novel signal peptide (STII*) that allowed for efficient expression in both systems (Fig. 1C). The resulting vector, pDQ1, expressed Fc fusion products in mammalian cells as efficiently as the original pCDM8-derived vector from which the Fc gene was cloned (data not shown) (19) and was used to produce all of the Fc fusion proteins described here.
Construction of a “Soft Randomized” Library
In a standard phage display workflow, bacteria culture supernatants, periplasmic extracts, or precipitated phage are used to initially validate and compare selected phagemid clones. The alternative workflow used here integrates mammalian expression of selected variants, as represented in Fig. 2A. As indicated, peptide Fc fusion proteins were produced directly from pDQ1 phagemid clones in mammalian cells, and these Fc fusion proteins were assayed with a range of techniques. Here, for example, immunoprecipitations of HIV-1 gp120, the target protein, were performed using metabolically labeled supernatants of transfected 293T cells. In parallel, promising peptide-Fc variants were compared using flow cytometry, HIV-1 neutralization studies, and surface plasmon resonance. Elements from optimal variants were then combined, and the composite peptide that best bound gp120 and neutralized HIV-1 then served as the template for a new phagemid library and a subsequent round of selection.
We began with a previously described CD4-mimetic peptide that included three unnatural amino acids (20–22). These amino acid were reverted to their closest natural form, introducing a cysteine at position 1, a phenylalanine at position 23, and a valine at position 27 to generate CD4mim1 (Fig. 2B). To facilitate N-terminal fusions of this peptide with CCR5-mimetic peptides (23, 24) and to expand the conformational space available to peptide variants, the first of three disulfide bonds was eliminated (CD4mim2). Note that CD4mim2 bound gp120 with lower affinity than CD4mim1, presumably due to the loss of this disulfide bond. CD4mim2 was used as a template to generate a first library of CD4mim variants. CD4mim4, the final composite product of our first selection cycle, was used as a template for a second library and another round of selection. Both libraries were soft randomized from their respective templates.
Soft randomization is a diversification technique that permits the introduction of a small number of amino acid changes (e.g. 5) distributed throughout a larger peptide (e.g. the 28 amino acids of CD4mim2 and CD4mim4). This approach allows modest changes along the length of the peptide without introducing a large number of mutations likely to be deleterious (1, 5, 25). Supplemental Fig. 1 details our procedure for library construction using pDQ1 and soft randomized oligonucleotides. Seven amino acids were held fixed: the four remaining cysteines and a GSF region identical to the key gp120-binding residues of CD4 (residues 41–43 (21)), whereas the remaining 21 residues were encoded by doped oligonucleotides favoring template amino acids at an 85:5:5:5 ratio. This library was panned against HIV-1 gp120 (those of the clade B isolate ADA and the clade C isolate ZM651; see “Experimental Procedures”). Selected CD4mim variants were expressed as peptide-Fc fusions in 293T cells and used to compare output variants for functional activity in immunoprecipitation and HIV-1 neutralization assays (Fig. 2, C and D, for example). Elements of selected variants were combined, and resulting proteins were again compared. In this way, an optimal peptide was selected as a template for a second soft-randomized library. The procedure was repeated, and an improved peptide, CD4mim6, was selected. Supplemental Fig. 2 details the process by which CD4mim6 was developed as a composite of several library outputs.
Enhanced Neutralization of HIV-1 by CD4mim6
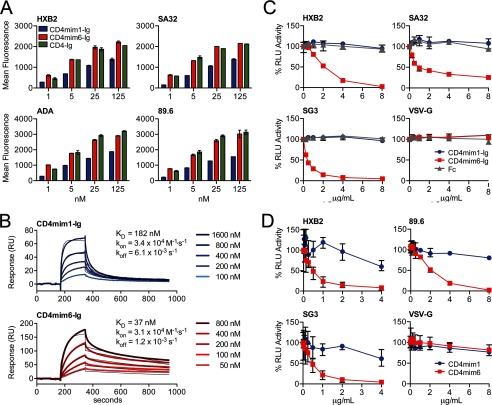
Because CD4mim1, with three disulfide bonds, outperformed in immunoprecipitation and neutralization assays our starting point, CD4mim2, with two disulfide bonds (Fig. 2D and data not shown), we compared the properties of our final product, CD4mim6, with CD4mim1, using Fc fusions of both variants. Both peptides bound measurably to 293T cell surface-expressed HIV-1 envelope glycoprotein trimers (Fig. 3A). However, at all concentrations, and for all clade B and C envelopes assayed, CD4mim6-Ig bound more efficiently than CD4mim1-Ig and similarly to CD4-Ig. Surface plasmon resonance studies with immobilized gp120 (Du151, clade C) made clear that the basis for the efficient binding was due to a slower off-rate, five times slower for CD4mim6-Ig. In fact, CD4mim6-Ig had a modestly slower on-rate, presumably due to its greater flexibility (Fig. 3B). More total CD4mim6-Ig bound to immobilized gp120, perhaps indicating that the more flexible CD4mim6-Ig accessed more gp120 conformations than the fixed CD4mim1-Ig.
FIGURE 3.
Comparisons of CD4mim1-Ig and CD4mim6-Ig. A, binding of CD4mim-Ig variants and CD4-Ig to 293T cells transfected to express the trimeric envelope glycoproteins of the indicated clade B (HXB2, ADA, 89.6) and clade C (SA32) isolates. Binding was determined by flow cytometry using a goat anti-human secondary antibody. Background binding, determined using Fc domain alone, was <15 mean fluorescence intensity in each case. Experiment is representative of two with similar results. B, surface plasmon resonance studies of CD4mim1-Ig and CD4mim6-Ig using immobilized gp120 of the clade C isolate Du151. Results are shown with thick lines. Thin black lines represent global fitting of the data to a 1:1 Langmuir binding model. C, CD4mim6-Ig neutralizes clade B (HXB2) and clade C (SG3, SA32) HIV-1 isolates with IC50 values of 1–2 μg/ml (20–40 nm), whereas no neutralization was observed for CD4mim1-Ig at 8 μg/ml (160 nm), and HIV-1 pseudotyped with VSV-G protein was unaffected by any Fc fusion. Experiment is representative of three with similar results. D, synthetic monomers of CD4mim1 and CD4mim6 were compared in the same assay. CD4mim6 neutralized the indicated isolates with IC50 values in the 0.5–2 μg/ml range, whereas IC50 values for CD4mim1 were >4–8 μg/ml. Experiment is representative of two with similar results. Error bars in all cases represent a range of duplicates (A) or triplicates (C and D).
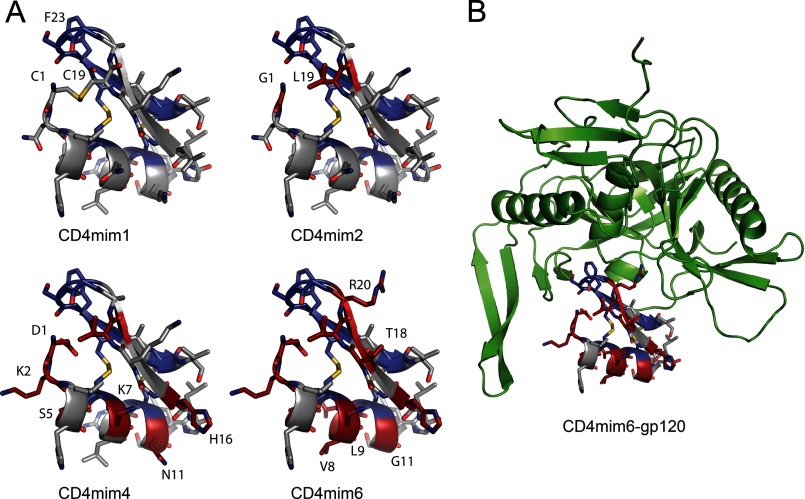
CD4mim6-Ig also efficiently neutralized clade B and clade C HIV-1 isolates at concentrations (1–2 μg/ml; 20–40 nm) where no neutralization was observed for CD4mim1-Ig (Fig. 3C). When CD4mim1 and CD4mim6 were synthesized as free peptides without an Fc domain, CD4mim6 efficiently neutralized HIV-1, again at concentrations where no neutralization was observed from CD4mim1 (Fig. 3D). Fig. 4, A and B, shows a model of CD4mim6 bound to HIV-1 gp120, with changes from CD4mim1 highlighted.
FIGURE 4.
Model of CD4mim variants with alterations indicated. A, CD4mim1 is a natural amino acid form of CD4M33, a CD4-mimetic peptide built from a scorpion toxin scaffold, with three disulfide bonds. Residues held constant during the library construction are indicated in blue. For CD4mim2, cysteines 1 and 19 were altered to glycine and leucine, respectively, shown in red. CD4mim4, the product of the first cycle of phage selection and the template the second phage library has eight total differences from CD4mim1, again indicated in red (see Fig. 2B for changes). CD4mim6, the final product of the second cycle, has 12 such differences. B, CD4mim6 modeled into the CD4 binding site of HIV-1 gp120. All models are based on the Protein Data Bank structure 1YYM by Huang et al. (20) and generated with the PyMOL mutagenesis tool.
CD4mim6 as a Tool for Understanding Neutralization of HIV-1
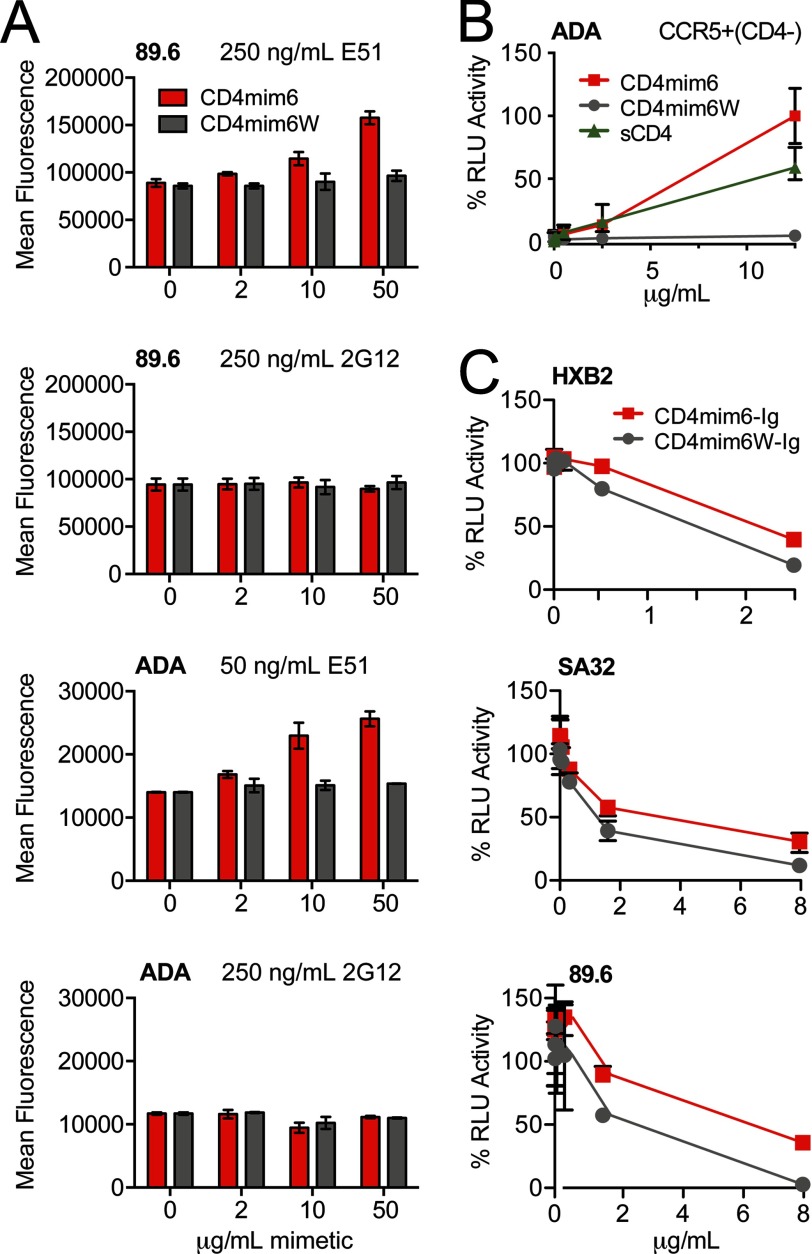
To determine the similarity between CD4mim6 and sCD4, we incubated CD4mim6 with cells expressing the HIV-1 envelope glycoprotein ADA and measured its ability to promote association with the CD4-inducible antibody E51 (Fig. 5A). CD4mim6 increased association of E51 with the envelope glycoprotein, but had no effect on an HIV-1 neutralizing antibody, 2G12, that is unaffected by CD4 association. In parallel, we assayed a CD4mim6 variant, CD4mim6W, in which phenylalanine 23 was replaced with a tryptophan. Phenylalanine 23 binds a critical pocket of HIV-1 gp120 also occupied by CD4 phenylalanine 43. Occupation of this same gp120 pocket with a tryptophan has been shown to increase the potency of the neutralizing antibody NIH-45–46 (26). Strikingly, CD4mim6W did not enhance binding of E51 to HIV-1 envelope glycoprotein. We speculated that it would therefore be unable to promote infection when cellular CD4 was limiting, in contrast to both sCD4 and CD4mim6. Indeed with CD4-negative, CCR5-high cells, infection increased with increasing concentration of sCD4 or CD4mim6 (Fig. 5B). However, little or no infection was observed at any concentration with CD4mim6W. Consistent with these observations, CD4mim6W more potently neutralized HIV-1 than CD4mim6 (Fig. 5C). Thus, the potency of CD4mim6 and perhaps CD4 binding site antibodies may be attenuated by their tendency to promote infection when cellular CD4 is limiting.
FIGURE 5.
CD4mim6, but not CD4mim6W, induces the CD4-bound conformation of the HIV-1 envelope glycoprotein. A, 239T cells transfected to express the envelope glycoprotein of the clade B isolate ADA were incubated with the indicated concentrations of CD4mim6 or CD4mim6W, a variant in which phenylalanine 23 was replaced with a tryptophan. Cells were then incubated with an anti-gp120 antibody, either the CD4-inducible antibody E51 (250 ng/ml for 89.6; 50 ng/ml for ADA) or the anti-glycan antibody 2G12 (250 ng/ml). Binding was determined by flow cytometry using a goat anti-human secondary antibody. Background binding, determined using Fc domain alone was <15 mean fluorescence intensity in each case. Experiment is representative of two with similar results. B, CCR5-high, CD4-negative Cf2Th cells were incubated with an ADA pseudovirus encoding firefly luciferase in the presence of the indicated concentrations of CD4mim6, CD4mim6W, or soluble CD4 (sCD4). Infection, measured as luciferase activity (RLU) was determined as in Fig. 3C. C, neutralization studies of CD4mim6-Ig and CD4mim6W-Ig as described in Fig. 3C are shown.
DISCUSSION
Phage display remains a powerful means of improving bioactive peptides and proteins that in many cases will ultimately be produced in mammalian cells. Current approaches for validating library outputs limit the number of outputs that can be reasonably evaluated or the precision and range of assays that can be applied. The pDQ1 vector described here permits early selection for efficient mammalian expression and expands the range of available validation techniques. It thus facilitates immediate, sensitive characterization of a large number of library outputs for their biochemical and functional properties. One potentially useful adaptation of this system will be the introduction of a Fab library into the vector, facilitating direct production and characterization of full-length antibodies in mammalian cells. With minor modifications, this system might also be adapted for mammalian cell surface display, allowing for a preliminary selection in phage followed by subsequent rounds of selection by cell surface display on subsets of manageable size. This system can also be used without phage display, for example when determining a production technology for a particular biologic or when the role of a eukaryotic post-translational modification is investigated. However, it is especially useful for phage display applications. Because both phage display and mammalian expression from pDQ1 remain as efficient as with single-function vectors, we anticipate that mammalian expression will become a standard capability of vectors used for phage-based improvement of peptides and proteins.
The CD4-mimetic peptide generated in these studies is unlikely by itself to serve as a therapeutic. However, a small peptide that mimics CD4 can be especially useful as a tool for understanding neutralization of HIV-1. Its size ensures that it will not sterically interfere with binding of antibodies to the HIV-1 envelope glycoprotein. It therefore can be used can be used with various antibodies to compare the CD4-bound and unliganded states of the envelope glycoprotein. Further study of CD4mim6W may also deepen our understanding of the different conformational states available to gp120 and the envelope glycoprotein trimer.
Although the therapeutic potential of CD4mim6 alone is limited, it may be more potent as a fusion with other inhibitors. CD4mim1 has been fused at its C terminus with tyrosine-sulfated CCR5-mimetic peptides to make a double-mimetic peptide with considerably higher potency than either peptide alone (23). CD4mim6 possess two advantages over CD4mim1 in this context. First, it binds gp120 with higher affinity and neutralizes HIV-1 more efficiently than CD4mim1. Second, some steric interference between the N terminus of CD4mim1 and gp120 has been observed (22), precluding N-terminal fusions with this mimetic. By eliminating the first disulfide bond of CD4mim1, the N terminus of CD4mim6 can now be readily fused to a CCR5-mimetic peptide, positioning the latter mimetic much closer to its binding pocket. These kinds of constructs induce the CD4-bound conformation of the trimer and therefore can potentially synergize with gp41-derived peptides such as T20/enfuvirtide. Further improvement of both double- and single-mimetic peptides, and of neutralizing antibodies that target HIV-1 and other pathogens, can be significantly accelerated using the pDQ1 system described here.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI080324 and P01 AI100263 through the NIAID (to M. F.).

This article contains supplemental Figs. 1 and 2.
- bis-tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid.
REFERENCES
- 1. Li B., Xi H., Diehl L., Lee W. P., Sturgeon L., Chinn J., Deforge L., Kelley R. F., Wiesmann C., van Lookeren Campagne M., Sidhu S. S. (2009) Improving therapeutic efficacy of a complement receptor by structure-based affinity maturation. J. Biol. Chem. 284, 35605–35611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sui J., Li W., Murakami A., Tamin A., Matthews L. J., Wong S. K., Moore M. J., Tallarico A. S., Olurinde M., Choe H., Anderson L. J., Bellini W. J., Farzan M., Marasco W. A. (2004) Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 101, 2536–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Valadon P., Garnett J. D., Testa J. E., Bauerle M., Oh P., Schnitzer J. E. (2006) Screening phage display libraries for organ-specific vascular immunotargeting in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mirzabekov T., Kontos H., Farzan M., Marasco W., Sodroski J. (2000) Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat. Biotechnol. 18, 649–654 [DOI] [PubMed] [Google Scholar]
- 5. Sidhu S. S., Lowman H. B., Cunningham B. C., Wells J. A. (2000) Phage display for selection of novel binding peptides. Methods Enzymol. 328, 333–363 [DOI] [PubMed] [Google Scholar]
- 6. Smith G. P., Scott J. K. (1993) Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217, 228–257 [DOI] [PubMed] [Google Scholar]
- 7. Boder E. T., Midelfort K. S., Wittrup K. D. (2000) Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 97, 10701–10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boder E. T., Wittrup K. D. (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 [DOI] [PubMed] [Google Scholar]
- 9. Beerli R. R., Bauer M., Buser R. B., Gwerder M., Muntwiler S., Maurer P., Saudan P., Bachmann M. F. (2008) Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. U.S.A. 105, 14336–14341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ray K., Embleton M. J., Jailkhani B. L., Bhan M. K., Kumar R. (2001) Selection of single chain variable fragments (scFv) against the glycoprotein antigen of the rabies virus from a human synthetic scFv phage display library and their fusion with the Fc region of human IgG1. Clin. Exp. Immunol. 125, 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zwick M. B., Bonnycastle L. L., Noren K. A., Venturini S., Leong E., Barbas C. F., 3rd, Noren C. J., Scott J. K. (1998) The maltose-binding protein as a scaffold for monovalent display of peptides derived from phage libraries. Anal. Biochem. 264, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colwill K., Renewable Protein Binder Working Group, and Gräslund S. (2011) A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 8, 551–558 [DOI] [PubMed] [Google Scholar]
- 13. Barbas C. F., 3rd, Kang A. S., Lerner R. A., Benkovic S. J. (1991) Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. U.S.A. 88, 7978–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu H., Dean R. A. (1999) A novel method for increasing the transformation efficiency of Escherichia coli: application for bacterial artificial chromosome library construction. Nucleic Acids Res. 27, 910–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rader C., Barbas C. F. (1997) Phage display of combinatorial antibody libraries. Curr. Opin. Biotechnol. 8, 503–508 [DOI] [PubMed] [Google Scholar]
- 16. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brunak S., Engelbrecht J., Knudsen S. (1991) Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220, 49–65 [DOI] [PubMed] [Google Scholar]
- 18. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 19. Zettlmeissl G., Gregersen J. P., Duport J. M., Mehdi S., Reiner G., Seed B. (1990) Expression and characterization of human CD4: immunoglobulin fusion proteins. DNA Cell Biol. 9, 347–353 [DOI] [PubMed] [Google Scholar]
- 20. Huang C. C., Stricher F., Martin L., Decker J. M., Majeed S., Barthe P., Hendrickson W. A., Robinson J., Roumestand C., Sodroski J., Wyatt R., Shaw G. M., Vita C., Kwong P. D. (2005) Scorpion toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure 13, 755–768 [DOI] [PubMed] [Google Scholar]
- 21. Martin L., Stricher F., Missé D., Sironi F., Pugnière M., Barthe P., Prado-Gotor R., Freulon I., Magne X., Roumestand C., Ménez A., Lusso P., Veas F., Vita C. (2003) Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 21, 71–76 [DOI] [PubMed] [Google Scholar]
- 22. Vita C., Vizzavona J., Drakopoulou E., Zinn-Justin S., Gilquin B., Ménez A. (1998) Novel miniproteins engineered by the transfer of active sites to small natural scaffolds. Biopolymers 47, 93–100 [DOI] [PubMed] [Google Scholar]
- 23. Kwong J. A., Dorfman T., Quinlan B. D., Chiang J. J., Ahmed A. A., Choe H., Farzan M. (2011) A tyrosine-sulfated CCR5-mimetic peptide promotes conformational transitions in the HIV-1 envelope glycoprotein. J. Virol. 85, 7563–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dorfman T., Moore M. J., Guth A. C., Choe H., Farzan M. (2006) A tyrosine-sulfated peptide derived from the heavy chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 281, 28529–28535 [DOI] [PubMed] [Google Scholar]
- 25. Hötzel I., Chiang V., Diao J., Pantua H., Maun H. R., Kapadia S. B. (2011) Efficient production of antibodies against a mammalian integral membrane protein by phage display. Protein Eng. Des. Sel. 24, 679–689 [DOI] [PubMed] [Google Scholar]
- 26. Diskin R., Scheid J. F., Marcovecchio P. M., West A. P., Jr., Klein F., Gao H., Gnanapragasam P. N., Abadir A., Seaman M. S., Nussenzweig M. C., Bjorkman P. J. (2011) Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.