Background: Resistance to β-lactams in Burkholderia is mediated by different β-lactamases (e.g. PenA and PenI).
Results: PenA from B. multivorans is a carbapenemase, and PenI from B. pseudomallei is an extended-spectrum enzyme.
Conclusion: Subtle changes within the active site of β-lactamases result in major phenotypic changes.
Significance: Future antibiotic design must consider the distinctive phenotypes of PenA and PenI β-lactamases.
Keywords: Antibiotic Resistance, Antibiotics, Microbiology, Molecular Modeling, Structural Biology
Abstract
Burkholderia cepacia complex and Burkholderia pseudomallei are opportunistic human pathogens. Resistance to β-lactams among Burkholderia spp. is attributable to expression of β-lactamases (e.g. PenA in B. cepacia complex and PenI in B. pseudomallei). Phylogenetic comparisons reveal that PenA and PenI are highly related. However, the analyses presented here reveal that PenA is an inhibitor-resistant carbapenemase, most similar to KPC-2 (the most clinically significant serine carbapenemase), whereas PenI is an extended spectrum β-lactamase. PenA hydrolyzes β-lactams with kcat values ranging from 0.38 ± 0.04 to 460 ± 46 s−1 and possesses high kcat/kinact values of 2000, 1500, and 75 for β-lactamase inhibitors. PenI demonstrates the highest kcat value for cefotaxime of 9.0 ± 0.9 s−1. Crystal structure determination of PenA and PenI reveals important differences that aid in understanding their contrasting phenotypes. Changes in the positioning of conserved catalytic residues (e.g. Lys-73, Ser-130, and Tyr-105) as well as altered anchoring and decreased occupancy of the deacylation water explain the lower kcat values of PenI. The crystal structure of PenA with imipenem docked into the active site suggests why this carbapenem is hydrolyzed and the important role of Arg-220, which was functionally confirmed by mutagenesis and biochemical characterization. Conversely, the conformation of Tyr-105 hindered docking of imipenem into the active site of PenI. The structural and biochemical analyses of PenA and PenI provide key insights into the hydrolytic mechanisms of β-lactamases, which can lead to the rational design of novel agents against these pathogens.
Introduction
Burkholderia cepacia complex is group of highly diverse bacteria that includes 17 different species (1, 2). B. cepacia complexes are typically found in soil, water, and the rhizosphere of crop plants (i.e. rice and wheat) and are used in both bioremediation and as biopestidicides. Unique among prokaryotes, B. cepacia complex has a large complex genome of seven megabases with three chromosomes and a plasmid (3, 4). However, these bacteria are also opportunistic-nosocomial pathogens of which Burkholderia multivorans and Burkholderia cenocepacia are most prevalent in this setting (3). Alarmingly, the bacteria can survive for months in solutions, such as disinfectants, saline, and mouthwash. Nosocomial infections can arise through patients interacting with contaminated solutions (5–17). In addition, B. cepacia complex is notorious for causing respiratory infections in cystic fibrosis patients, who received prolonged courses of broad spectrum antibiotics (1). As a result, B. cepacia complex is inherently resistant to multiple antibiotics. Adding to this, B. cepacia complex possesses many mobile genetic elements and genomic islands that were acquired through horizontal transfer (18).
Burkholderia pseudomallei is an intracellular pathogen and the causative agent of melioidosis (i.e. Whitmore disease) (19). This pathogen is also found in water and soil and is spread to humans and animals through direct contact with these sources. B. pseudomallei has the potential to be used in biological warfare and bioterrorism. Primary routes of infection by B. pseudomallei include inhalation, aspiration, and percutaneous inoculation. Moreover, the bacterium can persist for long periods (years) in low nutrient environments (20, 21). Notably, melioidosis has a 20–50% mortality rate even with treatment. Melioidosis is predominantly a disease of tropical climates, such as Southeast Asia and Northern Australia (22–24). Similar to B. cepacia complex, B. pseudomallei also has a large genome with two chromosomes; the transmission of genetic material between B. cepacia complex and B. pseudomallei is highly probable (25, 26).
Currently, β-lactam antibiotics are recommended as a treatment option for infections due to B. cepacia complex and B. pseudomallei (27, 28). However, resistance to β-lactams is present, and the mechanistic basis for resistance is correlated to the expression of β-lactamases (29–32) as well as decreased expression of penicillin-binding proteins in B. pseudomallei (33). B. multivorans and B. pseudomallei possess related β-lactamases, PenA and PenI, respectively, that share 71% similarity and 62% identity (34).
At present, β-lactamases (EC 3.5.2.6) are classified into four different classes based on structural similarities (A–D) (35). Class A, C, and D β-lactamases use a serine (e.g. Ser-70 in class A enzymes; Ambler amino acid-numbering scheme (35)), as the nucleophile, and class B β-lactamases are metalloenzymes using one or two Zn2+ ions for catalysis. PenA and PenI are class A β-lactamases. In general, class A β-lactamases hydrolyze β-lactams through the reaction mechanism represented in Fig. 1. Here, we use biochemical assays, crystallography, and molecular modeling to dissect the hydrolytic mechanisms of PenA and PenI. Understanding how β-lactamases interact with their target ligand, β-lactams, is critical for devising strategies to circumvent them and treat infections by these drug-resistant and virulent pathogens.
FIGURE 1.
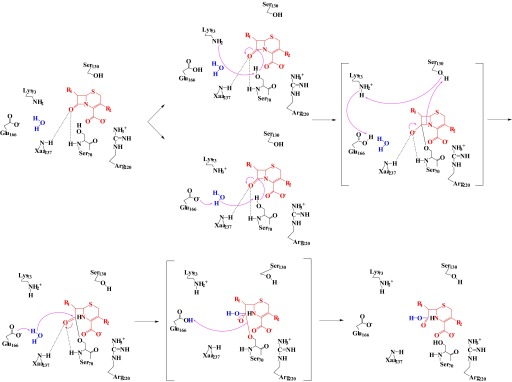
Proposed reaction mechanism for a cephalosporin (red) with a class A serine β-lactamase (black). The carbonyl of β-lactam sits in the oxyanion hole (electrophilic center) formed by the backbone nitrogens of Ser-70 and Xaa-237. The carboxylate forms a salt bridge to a positively charged amino acid, Arg-220 with PenA and PenI. Either Lys-73 directly removes a proton from Ser-70 or Glu-166 removes a proton from a water molecule (blue), which activates the nucleophilic Ser-70 to initiate acylation. Unprotonated Ser-70 attacks the β-lactam bond forming an acyl-enzyme. A proton shuttle starting with the β-lactam and Ser-130 to Lys-73 followed by Glu-166 results in an unprotonated Glu-166. Unprotonated Glu-166 removes a proton from a water molecule (i.e. DW), which attacks the acyl-enzyme complex resulting in water being added across the bond and deacylation of the inactivated β-lactam (69, 80).
EXPERIMENTAL PROCEDURES
Protein Analysis
Protein sequences were analyzed by BlastP and Bl2Seq from the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). Multiple sequence alignments were generated using Clustal 2.0.12 and BOXSHADE version 3.31C from the Mobyle@Pasteur website. Phylogenetic trees were constructed by submitting amino acid sequences and using the “one-click” mode of the phylogenetic analysis tool developed by Information Génomique et Structurale and funded by Réseau National des Génopoles (36–42).
Bacterial Strains and Plasmids
The blaPenA gene was obtained by PCR of genomic DNA isolated from B. multivorans ATCC 17616. The blaPenI gene was a kind gift from Dr. Herbert P. Schweizer (Colorado State University, Fort Collins, CO). The full-length blaPenA and blaPenI genes were cloned into the pET24a(+) plasmid using NdeI and BamHI restriction sites. Both genes were subsequently subcloned from the pET24a(+) plasmid into pBC SK(+) phagemid, including the ribosome-binding site of pET24a(+) (pBC SK(+) lacks a ribosome-binding site) by using SacII and BamHI restriction sites for blaPenA and SacI and BamHI restriction sites for blaPenI. The blaPenA gene minus the first 81 nucleotides, which encode the signal peptide for PenA, was cloned into the pGEX-6p-2 using the BamHI and XhoI restriction sites with a glutathione S-transferase gene at the 5′ end of the gene. The blaPenI gene minus the first 90 nucleotides, which encode its signal peptide, was cloned into the pET24a(+) plasmid using the NdeI and BamHI restriction sites. blaPenA and blaPenI genes cloned into pBC SK(+) phagemid were expressed in Escherichia coli DH10B cells for susceptibility testing. The blaPenA gene (Δ1–81) cloned into the pGEX-6p-2 and the blaPenI gene (Δ1–90) cloned into the pET24a(+) plasmid were expressed in E. coli Origami 2 DE3 cells for protein purification. The QuikChange XL site-directed mutagenesis kit (Stratagene) was used to perform site-directed mutagenesis of pBC SK(+) blaPenA to generate the R220G substitution in PenA using the manufacturer's protocol.
Antibiotic Susceptibility
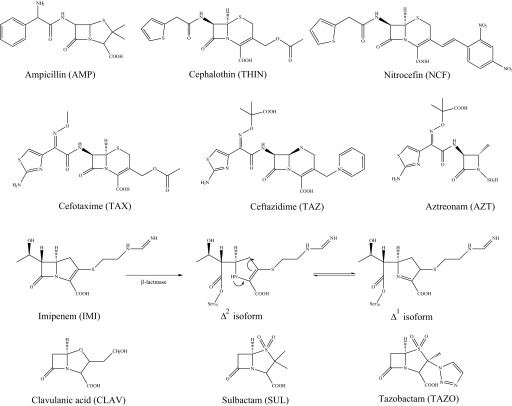
Mueller-Hinton agar dilution MICs,2 according to the Clinical and Laboratory Standards Institute (43, 44), were used to phenotypically characterize E. coli DH10B pBC SK(+) with blaPenA and blaPenI, as described previously (45). Etest MIC assays were conducted using the manufacturer's instructions. Colonies, directly resuspended into sterile water equivalent to a 0.5 McFarland standard, were used to inoculate Mueller-Hinton agar plates. The IMI Etests (Biomerieux Diagnostics) were placed on each plate; the bacteria were grown at 37 °C for 18 h, and MICs were read. Chemical structures of compounds tested are presented in Fig. 2, and the sources of these compounds were described previously (46).
FIGURE 2.
Compounds used in this study. Antibiotics from each of the four classes of β-lactams were studied, including penicillins, AMP; the cephalosporins, THIN, nitrocefin (NCF), TAX, and TAZ; the monobactam, AZT; and the carbapenem, IMI. The tautomerization of the double bond within IMI upon β-lactamase hydrolysis to form the Δ2 isoform to the Δ1 isoform is depicted in the 3rd row. β-Lactamase inhibitors utilized in this work include CLAV, SUL, and TAZO.
β-Lactamase Purification
The E. coli Origami 2 DE3 cells producing the PenA and PenI β-lactamases were grown and lysed as described previously (47). PenA with a cleavable N-terminal GST tag was purified from crude extracts using GSTrap FF column as described previously (48). Crude extracts from PenI pellets were used for preparative isoelectric focusing using a pH 3.5–10 gradient, as described previously (48, 49).
Purity of the fractions from both preparations was determined by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250. Protein concentrations were determined by measuring absorbance at λ280 nm and using the protein's extinction coefficient (Δϵ, 32,555 m−1 cm−1 for PenA and Δϵ, 21,555 m−1 cm−1 for PenI at 280 nm), which were obtained using the ExPASy ProtParam tool. To maintain full activity, PenA and PenI were stored in 10 mm PBS with 25% glycerol at −20 °C.
Kinetics
Steady-state kinetic parameters were determined using an Agilent 8453 diode array spectrophotometer (Santa Clara, CA), as described previously (45, 50). Using Enzfitter (Biosoft Corp., Ferguson, MO), a nonlinear least square fit of the data (Henri-Michaelis-Menten equation) determined the kinetic parameters, Vmax and Km as shown in Equation 1,
The β-lactamase inhibitors (CLAV, SUL, and TAZO) inactivate β-lactamases by interfering with the catalytic mechanism. For class A enzymes, CLAV, SUL, and TAZO are recognized as β-lactams. The apparent Km (Km, app) value of the inhibitors for the enzyme was determined using a direct competition assay under steady-state conditions, as described previously (45, 50).
Partition ratios (kcat/kinact) at 15 min for PenA and PenI were obtained by incubating the enzymes with increasing concentrations of inhibitor at room temperature (45, 50). The ratio of inhibitor to enzyme (I/E) necessary to inhibit the hydrolysis of nitrocefin by greater than 99% was determined.
Crystallization and Structure Refinement
PenA and PenI were crystallized by the vapor diffusion method using a 250-μl reservoir with a 4-μl hanging drop (2 μl reservoir solution + 2 μl protein solution). For PenA, the well solution contained 25% polyethylene glycol of 8 kilodaltons (PEG8K), 0.2 m sodium chloride, 0.1 m sodium phosphate/citric acid at pH 4.2 and 15 mg/ml of PenA in 2 mm HEPES, pH 7.5. Two successful conditions were obtained for PenI, one at pH 7.5 and the other at pH 9.5. One well solution contained 25% PEG8K, 0.1 m ammonium sulfate, 0.1 m HEPES, pH 7.5, and 10 mg/ml protein in 2 mm HEPES, pH 7.5, and the second contained 25% PEG8K, 0.1 m N-cyclohexyl-2-aminoethanesulfonic acid, pH 9.5 (reservoir), and 10 mg/ml protein in 2 mm HEPES, pH 7.5. Crystals appeared in 1–2 weeks reaching sizes of 0.4–0.8 mm. The crystals were cryoprotected by dipping them into a reservoir solution containing 20% glycerol, flush-cooled, and kept at 100 K with a nitrogen gas stream.
The 0.5° oscillation images were collected on a Q270 CCD detector with synchrotron radiation (λ = 1.00 Å at beamline NE3A of the Advanced Ring of the Photon Factory, Tsukuba, Japan). The HKL2000 program was used to index and scale x-ray intensities (51). For the PenI structure, the β was close to 90° (89.996° as the HKL2000 output) in the C2 space group. Other bravais-lattice candidates, which were suggested by HKL2000, were also examined. Only in the case of the C Centered Monoclinic was a reasonable Rmerge value (0.04 versus >0.4) obtained.
Molecular replacement for PenI structure obtained at pH 9.5 was completed at 2.0 Å using CTX-M-9 β-lactamase (PDB entry 2P74) as a search model with the program Phaser with Phenix Auto-MR wizard (52). For molecular replacement of the other two structures, the working model of PenI at pH 9.5 structure was used as a search model. With the highest resolution data, anisotropic B-factors with SIMU restraint 0.025 were introduced. At this step, Rfactor and Rfree values were reduced from 0.176/0.205 to 0.136/0.170, from 0.201/0.218 to 0.159/0.186, and from 0.201/0.242 to 0.141/0.189 for PenI at pH 9.5, PenI at pH 7.5, and PenA, respectively. The validity of these steps in the refinement was verified with PARVATI, a program that analyzes the modeled anisotropy (53). Riding hydrogen atoms were later added. Rfactor and Rfree values were reduced from 0.126/0.167 to 0.115/0.154, from 0.144/0.171 to 0.134/0.159, and from 0.143/0.187 to 0.132/0.174 for PenI at pH 9.5, PenI at pH 7.5, and PenA, respectively.
The occupancies of the DWs of PenI structures were refined, because the electron densities in the 2Fo − Fc map were poor compared with the PenA structure. The occupancies of the amino acid residues with poor density with the 1.0 σ 2Fo − Fc map were set as 0.6. The atoms with no electron density in the map were not included in the final model.
The results showed clear initial solutions and a reasonable molecular arrangement of each of the β-lactamases in an asymmetric unit. The model was then subjected to numerous cycles of conjugate gradient refinement and stepping to add higher resolution data with the program SHELX-97 followed by manual model building with the program COOT (54, 55). See Table 1 for data collection and refinement statistics. The coordinates were deposited in the Protein Data Bank (PenA, PDB code 3W4Q; PenI, pH 7.5, PDB code 3W4P; and PenI, pH 9.5, PDB code 3W4O).
TABLE 1.
X-ray data collection and crystallization refinement statistics
| PenI at pH 9.5 | PenI at pH 7.5 | PenA at pH 4.2 | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | C2 |
| Cell constant | 41.4, 52.8, 50.5 | 41.4, 52.7, 50.5 | 121.0, 69.9, 84.4 |
| a, b, c | 90.0, 92.6, 90.0 Å | 90.0, 92.5, 90.0 Å | 90.0, 90.0, 90.0 Å |
| Wavelength | 1.000 Å | 1.000 Å | 1.000 Å |
| Resolution | 50 to 1.18 Å (1.22 to 1.18 Å | 50 to 1.05 Å (1.07 to 1.05 Å) | 50 to 1.20 Å (1.22 to 1.20 Å) |
| Observations | 274,380 (21,669) | 375,483 (8696) | 845,028 (36,428) |
| Unique reflections | 68,595 (6191) | 98,393 (3781) | 208,965 (10,119) |
| Rmerge | 0.088 (0.286) | 0.092 (0.350) | 0.039 (0.450) |
| Completeness | 96.4% (87.5%) | 97.8% (75.7%) | 95.5% (93.1%) |
| Average I/σ(I) | 14.8 (4.3) | 13.6 (2.6) | 31.6 (2.7) |
| Data refinement | |||
| Resolution range | 15 to 1.18 | 15 to 1.05 | 15 to 1.2 |
| No. of reflections used (F > 0σ(F)) | 66,491 | 95,315 | 202,528 |
| Rwork/Rfreea | 0.1138/0.1528 | 0.1345/0.1602 | 0.1322/0.1749 |
| Rtotal | 0.1150 | 0.1354 | 0.1334 |
| Residue in Ramachandran zone | |||
| Favored/allowed | 260 (98.5%)/4 (1.5%) | 262 (99.2%)/2 (0.8%) | 759 (98.8%)/9 (1.2%) |
| Disallowed | 0 | 0 | 0 |
| r.m.s.d. values from ideality | |||
| Bond lengths | 0.013 Å | 0.014 Å | 0.013 Å |
| Bond angles | 0.031 Å | 0.029 Å | 0.031 Å |
| Zero chiral volumes | 0.080 Å3 | 0.081 Å3 | 0.074 Å3 |
| Nonzero chiral volumes | 0.088 Å3 | 0.096 Å3 | 0.080 Å3 |
| Mean B-factor (no. of atoms) | |||
| Protein | 9.35 (2130) | 12.57 (2104) | 16.39 (6039) |
| Solvent | 21.83 (249) | 26.27 (171) | 31.16 (929) |
| Others | 14.06 (19) | 35.38 (23) | None |
| Total | 10.68 (2398) | 13.82 (2298) | 18.36 (6968) |
| No. of hydrogen atoms | 2137 | 2088 | 5949 |
a Rfree values were calculated from 3% of reflections, respectively.
Molecular Modeling
The atomic coordinates of PenA and PenI, from the crystal structures reported here were used to construct Michaelis-Menten and acyl-enzyme complexes with IMI as described previously using Discovery Studio 3.1 (Accelrys, Inc. San Diego) molecular modeling software (45). IMI was constructed using Fragment Builder tools and was minimized using a Standard Dynamics Cascade protocol of Discovery Studio 3.1. IMI was automatically docked into the active site of PenA using the CDOCKER module of Discovery Studio 3.1 (56). This protocol uses a CHARMm-based molecular dynamics scheme to dock ligands into a receptor-binding site. For IMI docking into the PenI active site, LibDock algorithm was employed (57–59). LibDock is a high throughput algorithm for docking, which uses a set of pre-generated ligand conformations to dock ligands into a receptor-active site. From a maximum of 1000 pre-calculated IMI conformations, 100 hits were retained and scored (with a minimum LibDock score of 100). The best conformations were automatically aligned to polar and apolar active site hot spots, and the best scoring poses were reported. At this step, the hydrogen atoms were not maintained. To further optimize the docked poses (i.e. add hydrogens and prevent the clashes between the receptor and ligand), a CHARMm minimization step was used. For this step the Smart Minimization algorithm was employed (i.e. 1000 steps of steepest descent with a root mean square gradient tolerance of 3 Å, followed by Conjugate Gradient minimization, with a root-mean square deviation (r.m.s.d.)-minimization gradient of 0.001 Å). For the final minimization of the IMI conformations into the active site of PenI, a r.m.s.d. cutoff of 1 Å was chosen.
The resulting conformations of both PenA and PenI complexes were analyzed; the most favorable positioning of the IMI was chosen, and the complexes between the enzyme and inhibitor created, as described previously (60).
To check the stability of the complexes, a 6-ps Molecular Dynamics Simulation (MDS) was conducted for all enzyme-IMI complexes, as described previously (46, 50). During the heating/cooling, equilibration and production stages of MDS, a temperature of 300 K and a constant pressure were maintained. The long range electrostatics were treated with Particle Mesh Ewald and explicit solvation with Periodic Boundary Condition. The MDS for PenA-IMI complexes was run without any constraints. The first 4000 steps (4 ps) of MDS (heating/cooling and equilibration) of PenI with IMI were run using distance constraints to tether the carbonyl oxygen of IMI toward the oxyanion hole, which is made up of the amide backbone nitrogens of Ser-70 and Thr-237. For the production step of MDS, the system was free without any constraints.
Imipenem Hydrolysis Assay
Periplasmic extracts were prepared from E. coli DH10B pBC SK(+), E. coli DH10B pBC SK(+) blaPenA, and E. coli DH10B pBC SK(+) blaPenA carrying the R220G substitution as described previously (47). Total protein concentrations were measured for periplasmic extracts using protein assay according to the manufacturer's instructions (Bio-Rad). Imipenem hydrolysis was monitored at 297 nm for 20 min using 100 μm imipenem and 12 μg of protein from the periplasmic extracts.
RESULTS AND DISCUSSION
Divergent Phenotypes, PenA Is a Carbapenemase and PenI Is an ESBL
One of the most significant threats in clinical medicine is resistance to carbapenems, as they are the “last line” of therapy for many types of infections (61, 62). Thus, the identification of class A β-lactamases, like PenA, with carbapenemase activity is highly alarming (63–67). Similarly, our analysis of PenI reveals it possesses ESBL properties. Thus, Burkholderia spp. harbor intrinsic β-lactamases that confer multidrug resistance phenotypes.
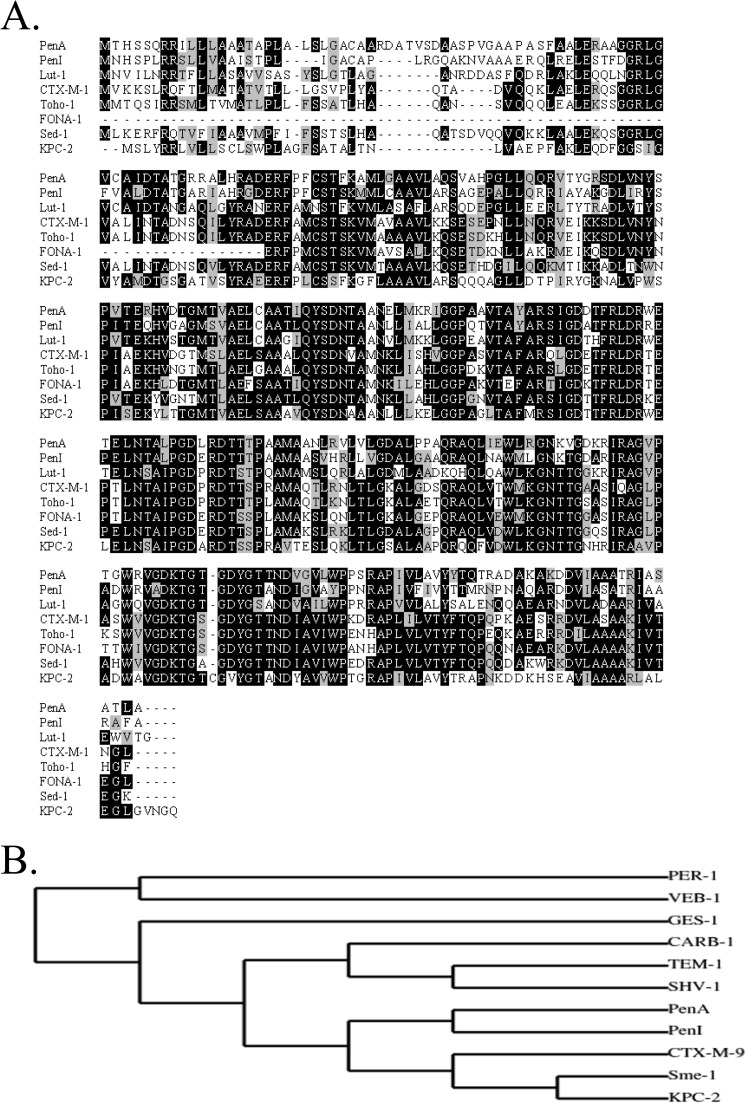
PenA and PenI possess the most amino acid sequence similarity and identity with the β-lactamases, LUT-1, Sed-1, FONA-1, CTX-M-1, and Toho-1, and the carbapenemase, KPC-2 (Fig. 3A). A phylogenetic comparison of the primary sequences of PenA and PenI to other commonly identified β-lactamases illustrates that PenA and PenI cluster together and are mostly related to the inhibitor-resistant carbapenemase, KPC-2, and the ESBL, CTX-M-9 (Fig. 3B).
FIGURE 3.
Comparisons of PenA and PenI with other β-lactamases. A, amino acid sequence alignments based on BlastP search. Amino acid sequence in black background indicates identity; gray background indicates similarity, and white background indicates differences. B, phylogenetic tree comparing PenA and PenI to prevalent β-lactamases identified by Dr. George Jacoby of the Lahey Clinic.
In vivo susceptibility testing reveals that E. coli expressing blaPenA demonstrate resistance to all four classes of β-lactam antibiotics (i.e. AMP, THIN and TAX, AZT, and IMI) as well as all currently available β-lactam-β-lactamase inhibitor combinations (i.e. AMP-CLAV, AMP-SUL, and PIP-TAZO) (Table 2 and Fig. 2). On the contrary, E. coli-producing blaPenI shows lower levels of resistance to all tested compounds compared with cells carrying blaPenA, and it is only resistant to AMP, THIN, and to a lesser extent, TAZ, TAX, and AZT (Table 2).
TABLE 2.
In vivo susceptibility testing for blaPenA and blaPenI represented in mg/liter
β-Lactam abbreviations are as follows: AMP, ampicillin; THIN, cephalothin; TAZ, ceftazidime; TAX, cefotaxime; AZT, aztreonam; IMI, imipenem; CLAV, clavulanic acid; SUL, sulbactam; PIP, piperacillin; and TAZO, tazobactam.
| AMP | THIN | TAZ | TAX | AZT | IMI | AMP/CLAVa | AMP/SULa | PIP/TAZOb | |
|---|---|---|---|---|---|---|---|---|---|
| E. coli DH10B pBC SK(+) | 4 | 2 | 0.25 | 0.06 | 0.25 | 0.5 | 50/0.06 | 50/1 | 2/0.25 |
| E. coli DH10B pBC SK(+) blaPenA | 8192 | 2048 | 8 | 16 | 64 | 4 | 50/32 | 50/256 | 128/16 |
| E. coli DH10B pBC SK(+) blaPenI | 512 | 256 | 4 | 4 | 8 | 0.25 | 50/0.125 | 50/4 | 4/0.5 |
a AMP was maintained at a constant concentration of 50 mg/liter, and CLAV and SUL concentrations were varied.
b For PIP/TAZO, both were varied at a ratio 8:1.
Steady-state kinetic analyses parallel susceptibility testing as PenA can hydrolyze AMP, THIN, TAX, AZT, and IMI with kcat/Km values of 3.6 ± 0.4 s−1, 3.1 ± 0.3 s−1, 0.48 ± 0.05 s−1, 0.16 ± 0.02 s−1, and 0.05 ± 0.01 s−1, respectively (Table 3). PenI demonstrates lower catalytic efficiencies for all β-lactams compared with PenA, but the highest kcat value (i.e. 9.0 ± 0.9 s−1) is for TAX (Table 3). Interestingly, PenA and PenI display distinct substrate spectra.
TABLE 3.
Steady-state kinetic parameters for PenA and PenI with selected β-lactams
Km and kcat values were determined under pseudo-first order conditions, and a nonlinear least square fit of the data to the Henri-Michaelis-Menten equation was determined using the kinetic parameters. Each experiment was completed in triplicate, and the error values represent the mean ± S.E.
| Km | kcat | kcat/Km | |
|---|---|---|---|
| μm | s−1 | μm−1 s−1 | |
| AMP | |||
| PenA | 88 ± 9 | 285 ± 29 | 3.2 ± 0.3 |
| PenI | 12 ± 3 | 0.69 ± 0.07 | 0.06 ± 0.01 |
| THIN | |||
| PenA | 71 ± 7 | 221 ± 21 | 3.1 ± 0.3 |
| PenI | 42 ± 4 | 4.2 ± 0.4 | 0.10 ± 0.01 |
| TAX | |||
| PenA | 295 ± 30 | 142 ± 14 | 0.48 ± 0.05 |
| PenI | 410 ± 41 | 9.0 ± 0.9 | 0.022 ± 0.002 |
| Nitrocefin | |||
| PenA | 142 ± 14 | 460 ± 46 | 3.2 ± 0.3 |
| PenI | 35 ± 4 | 4.9 ± 0.4 | 0.14 ± 0.01 |
| AZT | |||
| PenA | 378 ± 40 | 60 ± 6 | 0.16 ± 0.02 |
| PenI | 104 ± 10 | 2.1 ± 0.2 | 0.020 ± 0.002 |
| IMI | |||
| PenA | 7 ± 1 | 0.38 ± 0.04 | 0.05 ± 0.01 |
| PenI | NDa | ND | ND |
a ND means no measurable hydrolysis of imipenem was detected.
The β-lactamase inhibitors CLAV, SUL, and TAZO were developed to circumvent the production of β-lactamases. Both PenA and PenI exhibit Km, app values in the low micromolar range (i.e. 0.5 ± 0.1 to 4.1 ± 0.5 μm) to the β-lactamase inhibitors CLAV, SUL, and TAZO (Table 4). However, the partition ratios (kcat/kinact) tested at 15 min differ greatly between the two enzymes. PenA possesses much larger kcat/kinact values of 2000, 1500, and 50 for CLAV, SUL, and TAZO, respectively, whereas the values for PenI are 5, 5, and 1.
TABLE 4.
Steady-state kinetic parameters for PenA and PenI with β-lactamase inhibitors
The Km, app value was determined using a direct competition assay under steady-state conditions. kcat/kinact represents the ratio of inhibitor to enzyme (I/E) necessary to inhibit the hydrolysis of NCF by greater than 99%. Each experiment was completed in triplicate, and the error values represent the mean ± S.E.
| Km, app | kcat/kinact | |
|---|---|---|
| μm | ||
| CLAV | ||
| PenA | 3.1 ± 0.3 | 2000 |
| PenI | 2.3 ± 0.2 | 5 |
| SUL | ||
| PenA | 4.1 ± 0.4 | 1500 |
| PenI | 2.4 ± 0.2 | 5 |
| TAZO | ||
| PenA | 0.5 ± 0.1 | 50 |
| PenI | 1.9 ± 0.2 | 1 |
The ability of PenA to hydrolyze inhibitors as well as all classes of β-lactams (i.e. penicillin, cephalosporins, monobactams, and carbapenems) is worrisome; currently, β-lactamase inhibitors are not available that can inhibit this enzyme.
Crystallography Reveals Key Differences between PenA and PenI That Explain Phenotype and Catalytic Properties
The PenA crystal structure was obtained at a 1.2 Å resolution at pH 4.2, and the PenI structures were obtained at 1.05 Å resolution at pH 7.5 and at 1.18 Å resolution at pH 9.5. The PenA β-lactamase crystal was in the space group C2 with three molecules per asymmetric unit (Table 1). The PenI β-lactamases at pH 7.5 and pH 9.5 were both crystallized in space group P21 with one molecule in the asymmetric unit (Table 1).
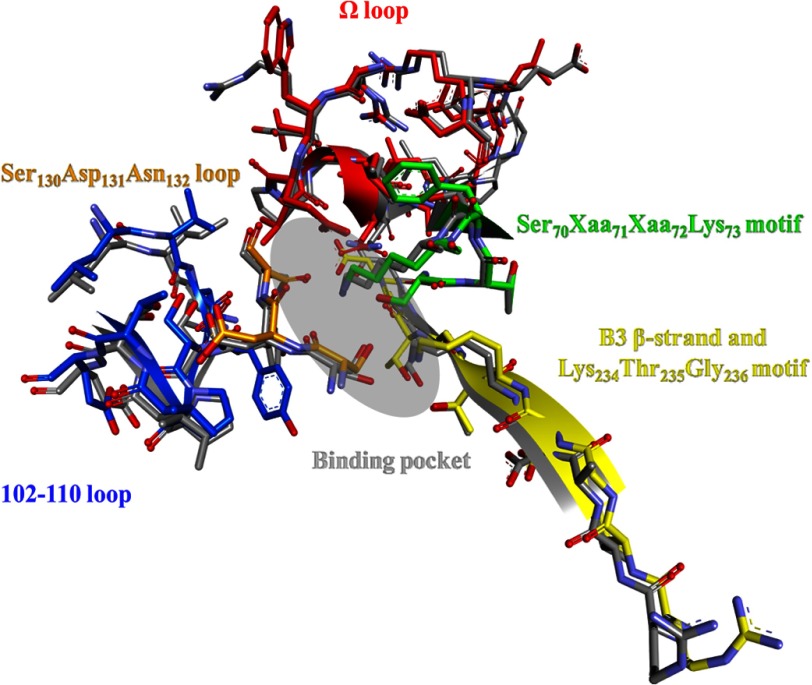
Despite differences in amino acid sequence, PenA and PenI, pH 7.5, have very similar overall three-dimensional structures (i.e. r.m.s.d. = 0.59 Å for Cα atoms between positions 32 and 289). To identify differences that may contribute to the contrasting kinetic properties of PenA and PenI, the 70SXXK73 motif, 130SDN132 loop, 234KTG236 motif, B3 β-strand, 102–110 loop, and Ω loop were compared; r.m.s.d. values comparing PenA to PenI, pH 7.5, are 0.41, 0.53, 0.23, 0.49, and 0.27 Å for these motifs, respectively (Fig. 4).
FIGURE 4.
Overlay of the PenA (colored) and PenI (gray) active sites highlighting the following major motifs: 70SXXK73 motif (green), 130SDN132 loop (orange), 234KTG236 motif (yellow), B3 β-strand (yellow), 102–110 loop (blue), and Ω loop (red), and the approximate binding pocket (gray oval) for β-lactams and β-lactamase inhibitors.
70SXXK73 Motif
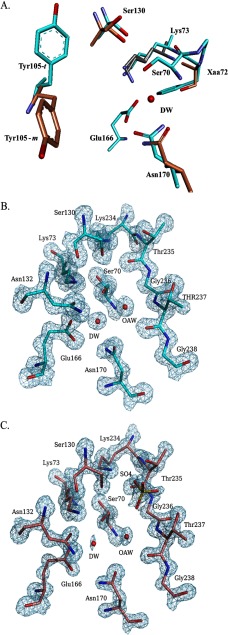
The 70SXXK73 motif contains the nucleophilic Ser-70 as well as Lys-73, which can serve as a general base to initiate β-lactam acylation and is also involved in the proton shuttle during β-lactam hydrolysis (Fig. 1). Fig. 5A shows that two conformations of Lys-73 are found in the structure of PenI obtained at pH 9.5. An overall decreased hydrolytic profile is observed in PenI compared with PenA, and the positioning of Lys-73 may be an important contributing factor. In conformation 1, which is identical to the conformation observed in PenA (Fig. 5A, cyan), the Nζ of Lys-73 can form hydrogen bonds with the Oδ1 of Asn-132 (distance 2.6 Å) and the Oγ of Ser-70 (distance 2.5 Å), and in conformation 2, the Nζ of Lys-73 is within hydrogen bonding distance of the Oϵ1 of Glu-166 (distance 2.6 Å) and the Oγ of Ser-70 (distance 2.9 Å). Not only are Lys-73 and Ser-70 important for β-lactam hydrolysis, but Glu-166 can also serve as a general base important for acylation and deacylation of β-lactams (Fig. 1). As a consequence, both the interactions observed between Lys-73, Glu-166, and Ser-70 in conformation 2, as well as the ability of Lys-73 to have two conformations, may decrease substrate hydrolysis due to inhibiting the proper function of these residues (e.g. decreased ability of Lys-73 to abstract a proton from Ser-70, altered proton shuttling, and changes in the pKa of Glu-166).
FIGURE 5.
Crystal structures of PenA and PenI. A, overlay of active site of PenA (cyan), PenI at pH 7.5 (orange), and PenI at pH 9.5 (gray) highlighting the alternate conformations of Lys-73, Ser-130, Tyr-105, and Asn-170. B, electron density of PenA active site showing the occupancy of the DW molecule is at 100%. Also represented is the water molecule in the oxyanion hole (OAW). C, electron density of PenI, pH 7.5, active site showing the occupancy of the DW molecule is at 21.7%. Also represented is the water molecule in the oxyanion hole (OAW).
Unlike PenI, PenA possesses a Phe at position 72. The S72F substitution was implicated in promoting CLAV resistance in PenI by increasing the Ki of the enzyme for CLAV (31, 68). This hydrophobic amino acid may contribute to the CLAV resistance phenotype found in PenA by altering the overall geometry of the oxyanion hole.
130SDN132 Loop
The 130SDN132 loop contains Ser-130, which participates in the proton shuttle for β-lactam hydrolysis (Fig. 1) and is critical for inactivation of a β-lactamase by a β-lactamase inhibitor (69). Also present in the loop is Asn-132, which stabilizes the Michaelis complex. Two conformations of Ser-130 are found in PenI at pH 7.5. In conformation 1, the Oγ of Ser-130 is within hydrogen bonding distance of the Nζ of Lys-73 (distance 3.0 Å), which is similar to the orientation observed in PenA (Fig. 5A, cyan). In conformation 2, the Oγ of Ser-130 can form hydrogen bonds with the Nζ of Lys-234 (distance 2.7 Å); and an additional hydrogen bond can be observed between the Oγ of Ser-130 and a sulfate molecule (SO42010, distance 2.6 Å) trapped in the active site of PenI. Similar to Lys-73, flexibility of Ser-130 may impede hydrolysis of β-lactams by PenI, further supporting the lower -catalytic efficiencies for β-lactams observed with PenI compared with PenA (70). As with Lys-73, the decreased ability to shuttle protons by Ser-130 affects hydrolysis rates.
234KTG236 Motif and B3 β-Strand
The hydrogen bonding network between residues 236, 237, 220, and 276 was previously demonstrated to be critical for binding and hydrolysis of β-lactams and β-lactamase inhibitors by the carbapenemase, KPC-2 (46, 47). In both PenA and PenI, the Oγ1 of Thr-237 and the O of Gly-236 can form a hydrogen bonding network with the Nη2 and the Nη1 of Arg-220, respectively, in addition the Nϵ of Arg-220 is within hydrogen bonding distance of the Oδ1 Asp-276.
102–110 Loop
The importance of 105 in β-lactam binding was previously shown in KPC-2 (50), and as a result attention was directed to the 102–110 loop. The crystal structure shows that the Oη of Tyr-105 of PenA can form a hydrogen bond with the hydroxyl side chain of Tyr-129 (distance 2.8 Å) and possesses a t rotamer orientation (trans, t80°); conversely, the Tyr-105 of PenI is in the m rotamer orientation (minus, m −85°/m −30°) (Fig. 5A) (71). Typically, the m rotamer is only observed in crystal structures of β-lactamases bound by an inhibitor and is hypothesized to stabilize hydrophobic ligand side chains (71). Molecular dynamics studies revealed that Tyr-105 is a dynamic residue, and for optimal β-lactam binding the t rotamer is necessary (71). The decreased catalytic activity of PenI compared with PenA may also be the result of the orientation of Tyr-105 obstructing the active site thus preventing the β-lactam from binding.
Ω Loop and Deacylation Water
The Ω loop includes residues 164–179 and contains two critical active-site residues, Glu-166 and Asn-170; both residues anchor the DW molecule required for β-lactam hydrolysis (Fig. 1). Hydrogen bonding interactions between DW molecules and the three structures are different. In the PenA structure, the DW molecule forms hydrogen bonds with the Oϵ2 of Glu-166, the Nδ2 of Asn-170, the N of Ser-70, and Oγ of Ser-70. Compared with PenA (Fig. 5A), in PenI at pH 7.5 and pH 9.5, the side chains of Asn-170 and Glu-166 are rotated 180°, and thus the hydrogen bonding interactions with the DW are different (Fig. 5A). Specifically, in the pH 7.5 structure, the DW is not anchored appropriately to Asn-170; this suggests that the DW may be impaired in promoting deacylation of the β-lactam. In addition, the rotation of Glu-166 changes the hydrogen bonding partner from the Oϵ2 of Glu-166 to the Oϵ1 of Glu-166 with the DW. In the PenI structure at pH 9.5, the DW possesses additional hydrogen bonds to the Nδ2 and Oδ1 of Asn-170, which may decrease the rate of deacylation. In addition to the alterations in hydrogen bonding pattern of the DW in the three structures, the DW molecule is at a lower occupancy in PenI about 25% (21.7% at pH 7.5 and 27.4% at pH 9.5) compared with PenA, which is at 100% (Fig. 5, B and C). The lower DW occupancy may further explain why PenI demonstrates lower catalytic activity compared with PenA.
In class A β-lactamases, substitutions within the Ω loop can confer cephalosporin resistance (72, 73). A substitution from Pro-167 to Thr results in TAZ resistance in the class A ESBL, CTX-M-23 (74). PenA possesses Thr-167; conversely, PenI has Pro-167. PenA demonstrates a TAZ resistance level one dilution higher than PenI.
Hydrogen bonding interactions between the Ω loop and B3 β-strand have been shown to decrease the flexibility of the B3 β-strand and narrow the spectrum of the β-lactamase (75). Specifically, Arg-241, which is near the B3 β-strand in the SHV-1 class A β-lactamase, was shown to be important for stabilizing the Ω loop; however, in PenA and PenI, a tyrosine is present at position 241. This change may allow the B3 β-strand to be flexible and thus be displaced from the Ω loop, increase the distance between the two, and allow larger cephalosporins (e.g. TAX) to be hydrolyzed.
PenA Shares Significant Structural Similarities with KPC-2
PenA was compared with KPC-2 as this β-lactamase is the most prevalent and clinically important class A carbapenemase (76). r.m.s.d. for PenA to KPC-2 is 1.0 Å. KPC-2 is similar to PenA in the five major motifs and demonstrates similar differences from PenI as observed in the PenA-PenI comparison. A major difference is the lack of a disulfide bond in PenA and PenI that is present between Cys-69 and Cys-238 in KPC-2 as well as other class A carbapenemases. Notably, KPC-2 contains the 237, 220, and 276 hydrogen bonding network, which is necessary for binding and hydrolysis of β-lactams as well as CLAV resistance (46). The occupancy of DW in KPC-2 is at 100% and is anchored between Glu-166 and Asn-170. The phenotype of PenA, being so comparable to that of KPC-2, may be attributed to their similarities.
Molecular Modeling of PenA and PenI with IMI, Insights into Carbapenemase Activity
Carbapenems are the last line of therapy for many types of infections (61, 62). PenA hydrolyzes carbapenems with a kcat of 0.38 ± 0.4 s−1, although carbapenem hydrolysis by PenI was not detectable. In addition, E. coli DH10B carrying blaPenA is resistant to imipenem (MIC 4 mg/liter), whereas E. coli DH10B-producing blaPenI is susceptible. As a result, PenA and PenI were modeled with the carbapenem, IMI, to gain insight into why PenA is a carbapenemase and PenI is not. Michaelis-Menten complexes of IMI with PenA reveal that IMI is primed for acylation with PenA (Fig. 6A). The carbonyl of IMI is oriented toward the backbone amides of Ser-70 and Thr-237, which is the oxyanion hole or electrophilic center of β-lactamases. In contrast, the positioning of Tyr-105 hindered the docking of IMI into the active site of PenI. Docked complexes were eventually achieved, and the carbonyl of IMI is positioned toward the oxyanion hole (Fig. 6B).
FIGURE 6.
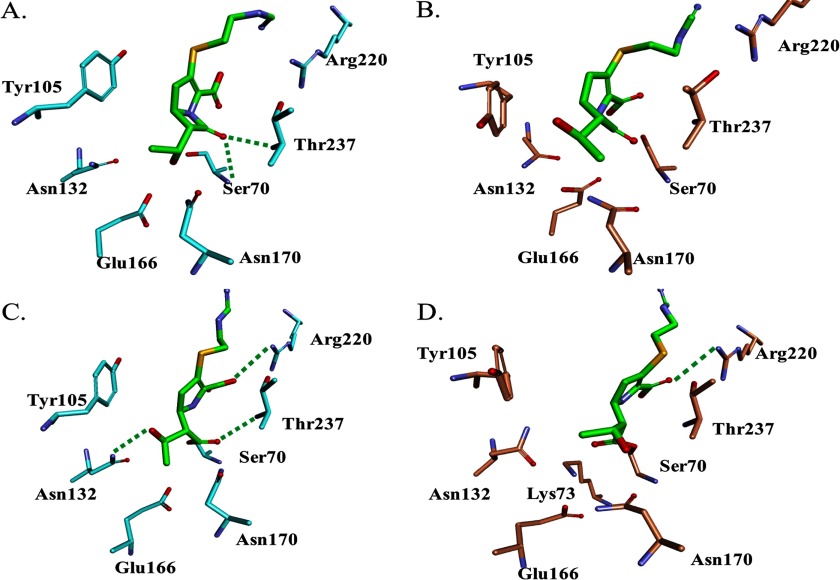
Molecular modeling of PenA and PenI with imipenem. A, Michaelis-Menten complex of PenA (cyan) with IMI (green) shows that the carbonyl of IMI is oriented toward the oxyanion hole, the backbone amides of Ser-70 and Thr-237. B, Michaelis-Menten complex of PenI (orange) with IMI (green) reveals the different conformation of Tyr-105 compared with PenA model. C, Δ2 isoform of IMI (green) with PenA (cyan) reveals the carbonyl of IMI is within the oxyanion hole and in a position favorable for deacylation. D, Δ2 isoform of IMI (green) with PenI (orange) demonstrates that the carbonyl of IMI is outside of oxyanion hole. Water molecules (not shown) are present near Arg-220; thus IMI is in a favorable orientation for tautomerization of its C2–C3 double bond for generation of the more stable Δ1 isoform.
To gain further insights into the mechanism, the acyl-enzyme complexes of PenA and PenI with IMI were constructed with the Δ2 isoform of IMI. Upon acylation, carbapenems can form two isoforms (e.g. Δ2 → Δ1) due to tautomerization of the pyrroline-double bond from C2–C3 to C3–C4 (Fig. 2). In TEM-1, a class A enzyme, deacylation of the carbapenem proceeds more rapidly with the Δ2 isoform; the Δ1 isoform deacylates slowly (77, 78). An arginine residue at 244 in the TEM-1 β-lactamase coordinates a water molecule that is the source of the proton for this tautomerization (79); the Arg-244 equivalent in PenA and PenI is Arg-220.
In the molecular representation of PenA with the Δ2 isoform of IMI, the carbonyl of IMI is docked in the oxyanion hole (2.7 Å from the N of Ser-70 and 2.9 Å from the N of Thr-237) (Fig. 6C). The O4 of the IMI carboxylate forms a hydrogen bonding interaction with the Nη1 of Arg-220 (distance 2.9 Å); no waters are found near (within 4 Å) Arg-220 to promote tautomerization. Thus, the IMI is oriented in a position for deacylation.
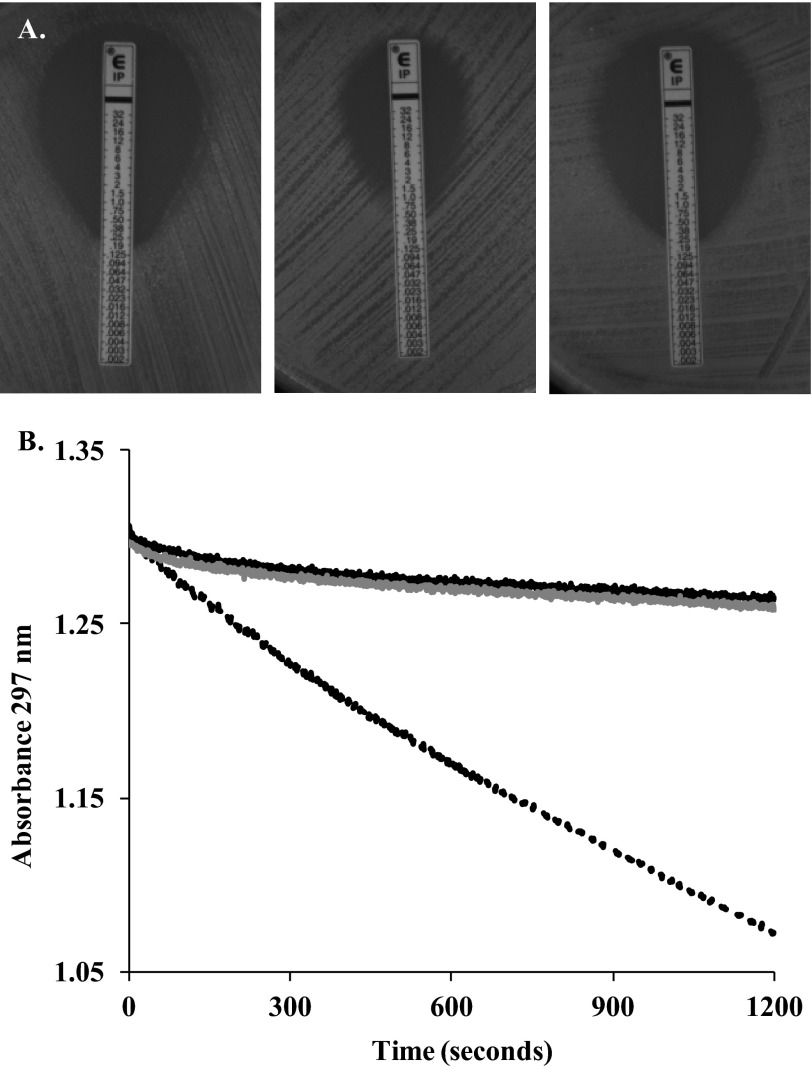
To confirm this model, we substituted Arg-220 to Gly; this resulted in a loss of IMI resistance by PenA and a significant decrease in catalytic activity when measured enzymatically (Fig. 7, A and B). Therefore, Arg-220 is critical for interacting with IMI.
FIGURE 7.
Arg at position 220 is critical for imipenem resistance and hydrolysis by PenA. A, Etests for E. coli DH10B pBC SK(+) (left), E. coli DH10B pBC SK(+) blaPenA (middle), and E. coli DH10B pBC SK(+) blaPenA carrying the R220G substitution (right) reveals MICs of 0.19, 1.5, and 0.25 mg/liter, respectively. B, imipenem hydrolysis was monitored at 297 nm using periplasmic extracts prepared from E. coli DH10B pBC SK(+) (black line), E. coli DH10B pBC SK(+) blaPenA (black dotted line), and E. coli DH10B pBC SK(+) blaPenA carrying the R220G substitution (gray striped line).
In contrast, in the model of PenI with the Δ2 isoform of IMI, the carbonyl is not in an oxyanion hole (Fig. 6D). The Arg-220 can form a hydrogen bond with the O4 of the IMI carboxylate, and water molecules are within hydrogen bonding distance of Arg-220, which may influence the tautomerization of IMI in PenI to the unhydrolyzable and stable Δ1 isoform.
The carbapenemase phenotype of the PenA enzymes compared with PenI may also be attributed to the positioning of Tyr-105, thus allowing IMI to enter the active site. In addition, the Δ2 isoform of IMI forms interactions that are favorable for deacylation; the carbonyl of IMI remains in the oxyanion hole during the 6-ps molecular dynamics simulation, unlike with PenI. Finally, the water molecules necessary for tautomerization of the IMI to the more stable Δ1 isoform are not near Arg-220 in PenA but are present in PenI.
Conclusions
B. multivorans is an opportunistic nosocomial pathogen primarily infecting patients with cystic fibrosis, and B. pseudomallei is the causative agent of melioidosis and possesses the potential to be used as a bioweapon. Both organisms are resistant to β-lactam antibiotics, and this phenotype is linked to the expression of a class A β-lactamase, PenA in B. multivorans, and PenI in B. pseudomallei.
Here, the crystal structures of two phylogenetically similar yet phenotypically distinct β-lactamases from Burkholderia were analyzed to dissect their different mechanisms. Kinetic and crystallographic analysis of both enzymes revealed key differences that may directly lead to their unique phenotypes. Alterations in the positioning of important catalytic residues (e.g. Lys-73, Ser-130, and Tyr-105) may directly contribute to PenI's lower catalytic efficiencies and kcat/kinact values toward β-lactams and β-lactamase inhibitors, respectively, compared with PenA. In addition, altered anchoring of and decreased occupancy of the DW molecule is most likely another consideration of importance. On a different note, resistance to β-lactamase inhibitors by PenA may be the result of the presence of Phe at 72, as well as the topology of the active site; PenA demonstrates strong similarity to KPC-2, an inhibitor-resistant carbapenemase.
Distressingly, these studies highlight the tremendous hurdles that appear before the medicinal chemist and clinician facing the challenge of designing drugs to treat multidrug resistance Gram-negative pathogens. For the former, designing novel β-lactams that can be effective against a pathogen harboring two different β-lactamases that possess contrasting properties is a complex endeavor. To the latter, the correct combination of drugs that will affect a durable cure must be chosen.
Acknowledgments
We thank Dr. Herbert P. Schweizer from the Colorado State University (Fort Collins, CO) for sending us the blaPenI gene. We thank also Dr. Nobutada Tanaka (Showa University) for technical advice for crystallography.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI100560 and R01 AI063517 from NIAID (to R. A. B.). This work was also supported by the Veterans Affairs Career Development Program (to K. M. P.-W.), by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program, Geriatric Research Education and Clinical Center Grant VISN 10 (to R. A. B.), and Japan Society for the Promotion of Science Grant-in-aid for Scientific Research (KAKENHI) Grant 22590401 (to M. N.). This work has been performed under the approval of the Photon Factory Program Advisory Committee (Proposal No. 2010G584).
The atomic coordinates and structure factors (codes 3W4Q, 3W4P, and 3W4O) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AMP
- ampicillin
- THIN
- cephalothin
- TAZ
- ceftazidime
- TAX
- cefotaxime
- AZT
- aztreonam
- IMI
- imipenem
- CLAV
- clavulanic acid
- SUL
- sulbactam
- TAZO
- tazobactam
- PEG8K
- polyethylene glycol at 8 kDa
- r.m.s.d.
- root mean squared deviation
- MDS
- Molecular Dynamics Simulation
- DW
- deacylation water
- PDB
- Protein Data Bank
- ESBL
- extended spectrum β-lactamase
- MIC
- minimal inhibitory concentration.
REFERENCES
- 1. Mahenthiralingam E., Vandamme P. (2005) Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chron. Respir. Dis. 2, 209–217 [DOI] [PubMed] [Google Scholar]
- 2. Vandamme P., Dawyndt P. (2011) Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 34, 87–95 [DOI] [PubMed] [Google Scholar]
- 3. Mahenthiralingam E., Baldwin A., Dowson C. G. (2008) Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551 [DOI] [PubMed] [Google Scholar]
- 4. Suárez-Moreno Z. R., Caballero-Mellado J., Coutinho B. G., Mendonça-Previato L., James E. K., Venturi V. (2012) Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266 [DOI] [PubMed] [Google Scholar]
- 5. Douce R. W., Zurita J., Sanchez O., Cardenas Aldaz P. (2008) Investigation of an outbreak of central venous catheter-associated bloodstream infection due to contaminated water. Infect. Control Hosp. Epidemiol. 29, 364–366 [DOI] [PubMed] [Google Scholar]
- 6. Heo S. T., Kim S. J., Jeong Y. G., Bae I. G., Jin J. S., Lee J. C. (2008) Hospital outbreak of Burkholderia stabilis bacteraemia related to contaminated chlorhexidine in haematological malignancy patients with indwelling catheters. J. Hosp. Infect. 70, 241–245 [DOI] [PubMed] [Google Scholar]
- 7. Lee C. S., Lee H. B., Cho Y. G., Park J. H., Lee H. S. (2008) Hospital-acquired Burkholderia cepacia infection related to contaminated benzalkonium chloride. J. Hosp. Infect. 68, 280–282 [DOI] [PubMed] [Google Scholar]
- 8. Memish Z. A., Stephens G., Balkhy H. H., Cunningham G., Francis C., Poff G. (2009) Outbreak of Burkholderia cepacia bacteremia in immunocompetent children caused by contaminated nebulized sulbutamol in Saudi Arabia. Am. J. Infect. Control 37, 431–432 [DOI] [PubMed] [Google Scholar]
- 9. Molina-Cabrillana J., Bolaños-Rivero M., Alvarez-León E. E., Martín Sánchez A. M., Sánchez-Palacios M., Alvarez D., Sáez-Nieto J. A. (2006) Intrinsically contaminated alcohol-free mouthwash implicated in a nosocomial outbreak of Burkholderia cepacia colonization and infection. Infect. Control Hosp. Epidemiol. 27, 1281–1282 [DOI] [PubMed] [Google Scholar]
- 10. Moreira B. M., Leobons M. B., Pellegrino F. L., Santos M., Teixeira L. M., de Andrade Marques E., Sampaio J. L., Pessoa-Silva C. L. (2005) Ralstonia pickettii and Burkholderia cepacia complex bloodstream infections related to infusion of contaminated water for injection. J. Hosp. Infect. 60, 51–55 [DOI] [PubMed] [Google Scholar]
- 11. Sunenshine R., Schultz M., Lawrence M. G., Shin S., Jensen B., Zubairi S., Labriola A. M., Shams A., Noble-Wang J., Arduino M. J., Gordin F., Srinivasan A. (2009) An outbreak of postoperative Gram-negative bacterial endophthalmitis associated with contaminated trypan blue ophthalmic solution. Clin. Infect. Dis. 48, 1580–1583 [DOI] [PubMed] [Google Scholar]
- 12. Yang C. J., Chen T. C., Liao L. F., Ma L., Wang C. S., Lu P. L., Chen Y. H., Hwan J. J., Siu L. K., Huang M. S. (2008) Nosocomial outbreak of two strains of Burkholderia cepacia caused by contaminated heparin. J. Hosp. Infect. 69, 398–400 [DOI] [PubMed] [Google Scholar]
- 13. Dolan S. A., Dowell E., LiPuma J. J., Valdez S., Chan K., James J. F. (2011) An outbreak of Burkholderia cepacia complex associated with intrinsically contaminated nasal spray. Infect. Control Hosp. Epidemiol. 32, 804–810 [DOI] [PubMed] [Google Scholar]
- 14. Martin M., Christiansen B., Caspari G., Hogardt M., von Thomsen A. J., Ott E., Mattner F. (2011) Hospital-wide outbreak of Burkholderia contaminans caused by prefabricated moist washcloths. J. Hosp. Infect. 77, 267–270 [DOI] [PubMed] [Google Scholar]
- 15. Martin M., Winterfeld I., Kramme E., Ewert I., Sedemund-Adib B., Mattner F. (2012) Outbreak of Burkholderia cepacia complex caused by contaminated alcohol-free mouthwash. Anaesthesist 61, 25–29 [DOI] [PubMed] [Google Scholar]
- 16. Martins I. S., Pellegrino F. L., Freitas Ad, Santos Mda S., Ferraiuoli G. I., Vasques M. R., Amorim E. L., Oliveira S., Nouér S. A., Cardoso F. L., Mascarenhas L. A., Magalhães A. C., Cleinman I. B., Figueiredo A. M., Moreira B. M. (2010) Case-crossover study of Burkholderia cepacia complex bloodstream infection associated with contaminated intravenous bromopride. Infect. Control Hosp. Epidemiol. 31, 516–521 [DOI] [PubMed] [Google Scholar]
- 17. Rosengarten D., Block C., Hidalgo-Grass C., Temper V., Gross I., Budin-Mizrahi A., Berkman N., Benenson S. (2010) Cluster of pseudoinfections with Burkholderia cepacia associated with a contaminated washer-disinfector in a bronchoscopy unit. Infect. Control Hosp. Epidemiol. 31, 769–771 [DOI] [PubMed] [Google Scholar]
- 18. Holden M. T., Seth-Smith H. M., Crossman L. C., Sebaihia M., Bentley S. D., Cerdeño-Tárraga A. M., Thomson N. R., Bason N., Quail M. A., Sharp S., Cherevach I., Churcher C., Goodhead I., Hauser H., Holroyd N., Mungall K., Scott P., Walker D., White B., Rose H., Iversen P., Mil-Homens D., Rocha E. P., Fialho A. M., Baldwin A., Dowson C., Barrell B. G., Govan J. R., Vandamme P., Hart C. A., Mahenthiralingam E., Parkhill J. (2009) The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Larsen J. C., Johnson N. H. (2009) Pathogenesis of Burkholderia pseudomallei and Burkholderia mallei. Mil. Med. 174, 647–651 [PubMed] [Google Scholar]
- 20. Lazar Adler N. R., Govan B., Cullinane M., Harper M., Adler B., Boyce J. D. (2009) The molecular and cellular basis of pathogenesis in melioidosis: How does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 33, 1079–1099 [DOI] [PubMed] [Google Scholar]
- 21. Aldhous P. (2005) Tropical medicine: Melioidosis? Never heard of it. Nature 434, 692–693 [DOI] [PubMed] [Google Scholar]
- 22. Wooten M. D., Panwalker A. P. (2001) Septic arthritis caused by Burkholderia pseudomallei: Case report and review of the literature. J. Clin. Rheumatol. 7, 242–247 [DOI] [PubMed] [Google Scholar]
- 23. Koponen M. A., Zlock D., Palmer D. L., Merlin T. L. (1991) Melioidosis. Forgotten, but not gone! Arch. Intern. Med. 151, 605–608 [DOI] [PubMed] [Google Scholar]
- 24. Falade O. O., Antonarakis E. S., Kaul D. R., Saint S., Murphy P. A. (2008) Clinical problem-solving. Beware of first impressions. N. Engl. J. Med. 359, 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holmes A., Govan J., Goldstein R. (1998) Agricultural use of Burkholderia (Pseudomonas) cepacia: A threat to human health? Emerg. Infect. Dis. 4, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pearson T., Giffard P., Beckstrom-Sternberg S., Auerbach R., Hornstra H., Tuanyok A., Price E. P., Glass M. B., Leadem B., Beckstrom-Sternberg J. S., Allan G. J., Foster J. T., Wagner D. M., Okinaka R. T., Sim S. H., Pearson O., Wu Z., Chang J., Kaul R., Hoffmaster A. R., Brettin T. S., Robison R. A., Mayo M., Gee J. E., Tan P., Currie B. J., Keim P. (2009) Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Avgeri S. G., Matthaiou D. K., Dimopoulos G., Grammatikos A. P., Falagas M. E. (2009) Therapeutic options for Burkholderia cepacia infections beyond co-trimoxazole: a systematic review of the clinical evidence. Int. J. Antimicrob. Agents 33, 394–404 [DOI] [PubMed] [Google Scholar]
- 28. Wuthiekanun V., Peacock S. J. (2006) Management of melioidosis. Expert Rev. Anti Infect. Ther. 4, 445–455 [DOI] [PubMed] [Google Scholar]
- 29. Cheung T. K., Ho P. L., Woo P. C., Yuen K. Y., Chau P. Y. (2002) Cloning and expression of class A β-lactamase gene blaA(BPS) Burkholderia pseudomallei. Antimicrob. Agents Chemother. 46, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Godfrey A. J., Wong S., Dance D. A., Chaowagul W., Bryan L. E. (1991) Pseudomonas pseudomallei resistance to β-lactam antibiotics due to alterations in the chromosomally encoded β-lactamase. Antimicrob. Agents Chemother. 35, 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tribuddharat C., Moore R. A., Baker P., Woods D. E. (2003) Burkholderia pseudomallei class A β-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob. Agents Chemother. 47, 2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trépanier S., Prince A., Huletsky A. (1997) Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother 41, 2399–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chantratita N., Rholl D. A., Sim B., Wuthiekanun V., Limmathurotsakul D., Amornchai P., Thanwisai A., Chua H. H., Ooi W. F., Holden M. T., Day N. P., Tan P., Schweizer H. P., Peacock S. J. (2011) Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 108, 17165–17170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poirel L., Rodriguez-Martinez J. M., Plésiat P., Nordmann P. (2009) Naturally occurring class A β-lactamases from the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 53, 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bush K., Jacoby G. A. (2010) Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dereeper A., Audic S., Claverie J. M., Blanc G. (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar R. C. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 40. Guindon S., Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 41. Anisimova M., Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 [DOI] [PubMed] [Google Scholar]
- 42. Chevenet F., Brun C., Bañuls A. L., Jacq B., Christen R. (2006) TreeDyn: Toward dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clinical and Laboratory Standards Institute (2006) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard, 7th Ed., CLSI document M7-A7 (ISBN 1-56238-587-9) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 44. Clinical and Laboratory Standards Institute (2008) Performance Standards for Antimicrobial Susceptibility Testing; Eighteenth Informational Supplement. CLSI Document M100-S18 (ISBN 1-56238-653-0) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 45. Papp-Wallace K. M., Bethel C. R., Distler A. M., Kasuboski C., Taracila M., Bonomo R. A. (2010) Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob. Agents Chemother. 54, 890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Papp-Wallace K. M., Taracila M. A., Smith K. M., Xu Y., Bonomo R. A. (2012) Understanding the molecular determinants of substrate and inhibitor specificities in the carbapenemase KPC-2: exploring the roles of Arg220 and Glu276. Antimicrob. Agents Chemother. 56, 4428–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papp-Wallace K. M., Taracila M., Hornick J. M., Hujer A. M., Hujer K. M., Distler A. M., Endimiani A., Bonomo R. A. (2010) Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 β-lactamase. Antimicrob. Agents Chemother. 54, 2867–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winkler M. L., Rodkey E. A., Taracila M. A., Drawz S. M., Bethel C. R., Papp-Wallace K. M., Smith K. M., Xu Y., Dwulit-Smith J. R., Romagnoli C., Caselli E., Prati F., van den Akker F., Bonomo R. A. (2013) Resistance to clavulanate, a class A β-lactamase inhibitor: A natural variant of the SHV β-lactamase reveals the importance of Lys-234. J. Med. Chem. 56, 1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin S., Thomas M., Shlaes D. M., Rudin S. D., Knox J. R., Anderson V., Bonomo R. A. (1998) Kinetic analysis of an inhibitor-resistant variant of the OHIO-1 β-lactamase, an SHV-family class A enzyme. Biochem. J. 333, 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papp-Wallace K. M., Taracila M., Wallace C. J., Hujer K. M., Bethel C. R., Hornick J. M., Bonomo R. A. (2010) Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci. 19, 1714–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 52. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Merritt E. A. (1999) Expanding the model: Anisotropic displacement parameters in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 55, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 54. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 55. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu G., Robertson D. H., Brooks C. L., 3rd, Vieth M. (2003) Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. J. Comput. Chem. 24, 1549–1562 [DOI] [PubMed] [Google Scholar]
- 57. Diller D. J., Li R. (2003) Kinases, homology models, and high throughput docking. J. Med. Chem. 46, 4638–4647 [DOI] [PubMed] [Google Scholar]
- 58. Rao S. N., Head M. S., Kulkarni A., LaLonde J. M. (2007) Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 47, 2159–2171 [DOI] [PubMed] [Google Scholar]
- 59. Diller D. J., Merz K. M., Jr. (2001) High throughput docking for library design and library prioritization. Proteins 43, 113–124 [DOI] [PubMed] [Google Scholar]
- 60. Drawz S. M., Taracila M., Caselli E., Prati F., Bonomo R. A. (2011) Exploring sequence requirements for C3/C4 carboxylate recognition in the Pseudomonas aeruginosa cephalosporinase: Insights into plasticity of the AmpC β-lactamase. Protein Sci. 20, 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kattan J. N., Villegas M. V., Quinn J. P. (2008) New developments in carbapenems. Clin. Microbiol. Infect. 14, 1102–1111 [DOI] [PubMed] [Google Scholar]
- 62. Ke W., Bethel C. R., Thomson J. M., Bonomo R. A., van den Akker F. (2007) Crystal structure of KPC-2: Insights into carbapenemase activity in class A β-lactamases. Biochemistry 46, 5732–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Henriques I., Moura A., Alves A., Saavedra M. J., Correia A. (2004) Molecular characterization of a carbapenem-hydrolyzing class A β-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob. Agents Chemother. 48, 2321–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Girlich D., Poirel L., Nordmann P. (2010) Novel ambler class A carbapenem-hydrolyzing β-lactamase, from a Pseudomonas fluorescens in the Seine River, Paris, France. Antimicrob. Agents Chemother. 54, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Queenan A. M., Bush K. (2007) Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 20, 440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Frase H., Shi Q., Testero S. A., Mobashery S., Vakulenko S. B. (2009) Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of β-lactamases. J. Biol. Chem. 284, 29509–29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Poirel L., Weldhagen G. F., Naas T., De Champs C., Dove M. G., Nordmann P. (2001) GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45, 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rholl D. A., Papp-Wallace K. M., Tomaras A. P., Vasil M. L., Bonomo R. A., Schweizer H. P. (2011) Molecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomallei. Front. Microbiol. 2, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drawz S. M., Bonomo R. A. (2010) Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nukaga M., Bethel C. R., Thomson J. M., Hujer A. M., Distler A., Anderson V. E., Knox J. R., Bonomo R. A. (2008) Inhibition of class A β-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J. Am. Chem. Soc. 130, 12656–12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doucet N., Pelletier J. N. (2007) Simulated annealing exploration of an active-site tyrosine in TEM-1 β-lactamase suggests the existence of alternate conformations. Proteins 69, 340–348 [DOI] [PubMed] [Google Scholar]
- 72. Palzkill T., Le Q. Q., Venkatachalam K. V., LaRocco M., Ocera H. (1994) Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol. Microbiol. 12, 217–229 [DOI] [PubMed] [Google Scholar]
- 73. Petrosino J. F., Palzkill T. (1996) Systematic mutagenesis of the active site Ω loop of TEM-1 β-lactamase. J. Bacteriol. 178, 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stürenburg E., Kühn A., Mack D., Laufs R. (2004) A novel extended-spectrum β-lactamase CTX-M-23 with a P167T substitution in the active-site Ω loop associated with ceftazidime resistance. J. Antimicrob. Chemother. 54, 406–409 [DOI] [PubMed] [Google Scholar]
- 75. Nukaga M., Mayama K., Hujer A. M., Bonomo R. A., Knox J. R. (2003) Ultrahigh resolution structure of a class A β-lactamase: On the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J. Mol. Biol. 328, 289–301 [DOI] [PubMed] [Google Scholar]
- 76. da Silva R. M., Traebert J., Galato D. (2012) Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae: A review of epidemiological and clinical aspects. Expert Opin. Biol. Ther. 12, 663–671 [DOI] [PubMed] [Google Scholar]
- 77. Charnas R. L., Knowles J. R. (1981) Inhibition of the RTEM β-lactamase from Escherichia coli. Interaction of enzyme with derivatives of olivanic acid. Biochemistry 20, 2732–2737 [DOI] [PubMed] [Google Scholar]
- 78. Easton C. J., Knowles J. R. (1982) Inhibition of the RTEM β-lactamase from Escherichia coli. Interaction of the enzyme with derivatives of olivanic acid. Biochemistry 21, 2857–2862 [DOI] [PubMed] [Google Scholar]
- 79. Zafaralla G., Manavathu E. K., Lerner S. A., Mobashery S. (1992) Elucidation of the role of arginine-244 in the turnover processes of class A β-lactamases. Biochemistry 31, 3847–3852 [DOI] [PubMed] [Google Scholar]
- 80. Meroueh S. O., Fisher J. F., Schlegel H. B., Mobashery S. (2005) Ab initio QM/MM study of class A β-lactamase acylation: Dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 127, 15397–15407 [DOI] [PubMed] [Google Scholar]