Background: Methylation of Rho proteins by isoprenylcysteine carboxylmethyltransferase impacts their function.
Results: RhoA methylation is dynamic and impacted by carboxylesterase 1 activity.
Conclusion: Carboxylesterase 1 plays a role in controlling the methylation status and activation of RhoA and impacts cell morphology.
Significance: This study demonstrates that C-terminal methylation is under acute control and plays a major role in cell signaling.
Keywords: Carboxylesterase, Post-translational Modification, Protein Isoprenylation, Protein Methylation, Rhoa, Methylation, Carboxylesterase 1, Isoprenylcysteine Carboxylmethyltransferase, Prenylation, rhoA
Abstract
A number of proteins that play key roles in cell signaling are post-translationally modified by the prenylation pathway. The final step in this pathway is methylation of the carboxyl terminus of the prenylated protein by isoprenylcysteine carboxylmethyltransferase. Due to the impact of methylation on Rho function, we sought to determine if the process was reversible and hence could control Rho function in a dynamic fashion. Elevating isoprenylcysteine carboxylmethyltransferase activity in cells has profound effects on MDA-MB-231 cell morphology, implying the presence of a pool of unmethylated prenyl proteins in these cells under normal conditions. Using a knockdown approach, we identified a specific esterase, carboxylesterase 1, whose function had a clear impact not only on the methylation status of RhoA but also RhoA activation and cell morphology. These data provide compelling evidence that C-terminal modification of prenyl proteins, rather than being purely a constitutive process, can serve as a point of regulation of function for this important class of protein.
Introduction
Post-translational modifications of proteins play important roles in all aspects of biology. Thus, understanding the dynamics and mechanisms of post-translational processing is vital to furthering our knowledge of biology at the molecular level. Numerous proteins, including most members of the Ras superfamily of GTPases, undergo a series of three such modifications collectively termed prenylation.
Proteins are directed into the prenylation pathway by a conserved CaaX motif at their C terminus. The process is initiated by the attachment of an isoprenoid lipid to an invariant cysteine residue, the Cys of the CaaX motif ((1, 2) by protein farnesyltransferase or protein geranylgeranyltransferase-I, respectively (3). The initial prenylation step is followed by cleavage of the three C-terminal amino acids (the -aaX) by an endoplasmic reticulum-bound protease termed Rce1. The third modification occurs when the prenylated cysteine, which is now located at the C terminus, is methylated by isoprenylcysteine carboxylmethyltransferase (Icmt),2 another integral endoplasmic reticulum membrane protein (4, 5). The final result of these modifications is a protein that contains a prenylated and methylated cysteine at its C terminus. The prenylation pathway plays a significant role in cancer biology because many oncogenic proteins require modification by this pathway to be active; thus, the enzymes involved in this pathway have emerged as anti-cancer targets (6, 7). However, many fundamental questions remain unanswered concerning the biological regulation and ramifications of prenylation-dependent processing.
The methylation of prenyl proteins has been shown to facilitate protein association with cellular membranes and also influence protein-protein interactions and protein stability; much of these data come from the study of Ras family GTPases (8, 9). Recently, we demonstrated a link between Icmt activity and Rho-mediated metastatic behavior in breast cancer cells (10). When Icmt activity was suppressed by either genetic or pharmacologic methods, RhoA and Rac1 activation was reduced, resulting in impaired actin cytoskeletal organization and inhibition of migration. These findings led us to postulate that Icmt-mediated methylation of small GTPases might be more highly regulated than previously envisioned.
In the current study, we found that MDA-MB-231 breast cancer cells contain a surprisingly large pool of unmethylated RhoA and that elevating Icmt activity has a profound impact on morphology of these cells. Furthermore, we identify a specific carboxylesterase termed CES1 whose function impacts the methylation status of RhoA. CES1 silencing impacted Rho function and cytoskeletal organization in a fashion similar to Icmt overexpression, demonstrating that methylation of RhoA is a controlled and dynamic process. These findings open an exciting new avenue of research for this important post-translational processing pathway.
EXPERIMENTAL PROCEDURES
Generation of Stable Cell Lines
Retroviral plasmids containing enhanced GFP or enhanced GFP-Icmt (11) were transfected along with the helper plasmid VSVG into GP2–293 human embryonic kidney cells using FuGENE (Roche Applied Science). The following day, medium was replaced, and after 8 h, the virus-containing medium was harvested and replaced with fresh medium. The process was repeated after another 16 h, and the two preparations were pooled. The virus-containing medium was then filtered with 0.45 μm SFCA filter (Corning) and used to infect MDA-MB-231 cells for 24 h. The cells were allowed to recover for another 24 h and then selected in 2 μg/ml puromycin for ∼3 weeks.
Methylation Assays
Icmt activity was determined by a modification of the in vitro assay described previously (12, 13). Approximately 130 h after siRNA transfection, MDA-MD-231 cells were harvested in lysis buffer (100 mm Hepes, pH 7.5, 5 mm MgCl2, 1 mm DTT supplemented with the protease inhibitors) and lysed by sonication. Following lysis, NaCl was added to a final concentration of 150 mm, and the lysate was precleared by centrifugation 5,000 × g for 5 min. Membranes were isolated by ultracentrifugation 100,000 × g for 30 min and resuspended in lysis buffer containing 150 mm NaCl, and protein concentrations were determined. Icmt activity was assessed by using equal amounts of membrane protein in the presence of biotin S-farnesylcysteine as the prenylcysteine substrate and [3H]S-adenosylmethionine.
Immunofluoresence Microscopy
MDA-MB-231 cells were harvested as described above, plated 10,000 cells/well on coverslips coated with fibronectin (10 μg/ml), and allowed to adhere overnight or for 2 h as described in Figs. 1 and 5, respectively. Coverslips were washed with PBS to remove non-adherent cells, and the remaining cells fixed with paraformaldehyde. Actin was visualized with rhodamine phalloidin (Fluka). Images were obtained with a Zeiss Axio Imager fluorescence microscope and analyzed using Metamorph software.
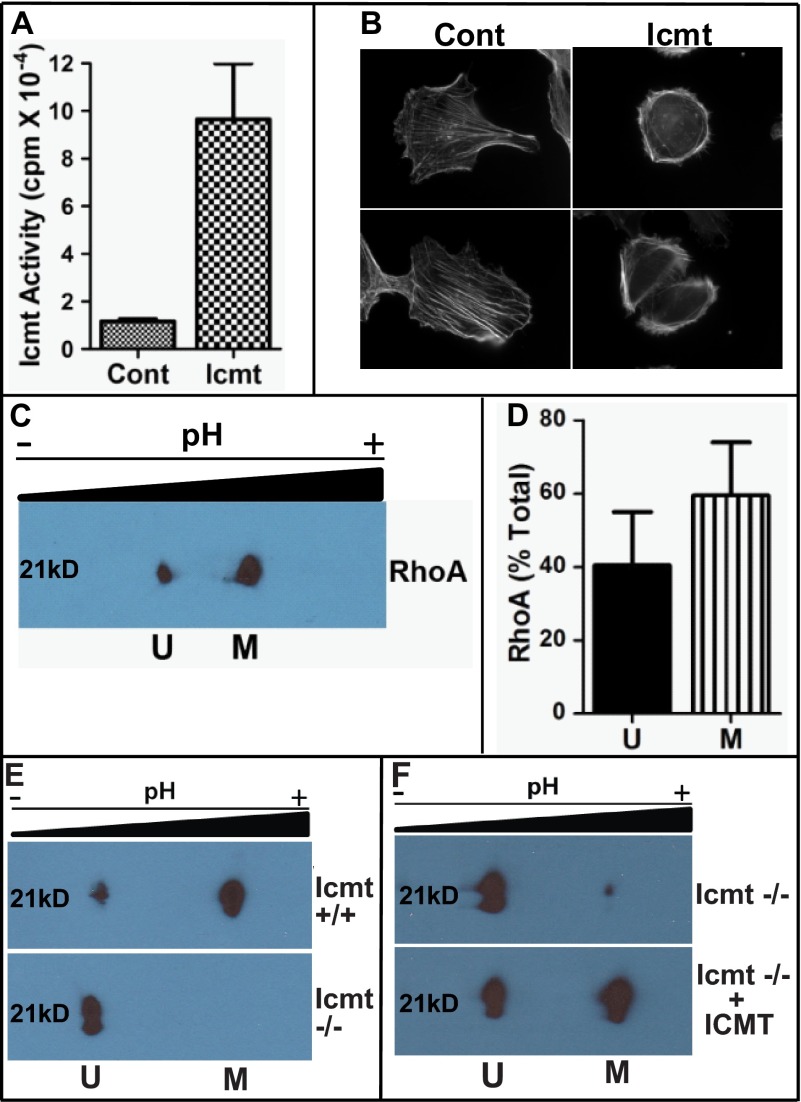
FIGURE 1.
Overexpression of Icmt impacts cell morphology. A, increased Icmt enzymatic activity upon overexpression in MDA-MB-231 cells. Cells expressing GFP (Cont (control)) of GFP-Icmt (Icmt) were harvested, membranes were isolated, and Icmt activity was determined as described under “Experimental Procedures.” Data are shown as the mean ± S.E. (n = 4 for each condition). B, visualization of actin cytoskeletal structure in MDA-MB-231 cells in the presence of increased Icmt activity. Cells were plated on coverslips coated with fibronectin and allowed to adhere overnight. Representative images are displayed showing the more rounded morphology and altered actin cytoskeletal structure in GFP-Icmt-overexpressing cells (Icmt) as compared with cells only expressing GFP (Cont (control)). C, two-dimensional gel analysis of RhoA methylation status. MDA-MB-231 cells were harvested and processed, and the presence of unmethylated Rhoa (U) in the more acidic pool or methylated RhoA (M) in the more basic pool was detected by immunoblot analysis with a RhoA-specific antibody. D, quantitative representation of unmethylated and methylated RhoA levels from C, expressed as a fraction of the total amount of RhoA. Data represent the mean ± S.E. from pooled results of two independent experiments. E, validation of two-dimensional gel analysis to identify methylated and unmethylated RhoA. Cell lysates from wild-type (Icmt +/+) and null Icmt (Icmt −/−) mouse embryonic fibroblasts were harvested and processed as described under “Experimental Procedures,” and the presence of unmethylated RhoA (U) in the more acidic pool or methylated RhoA (M) in the more basic pool was detected by immunoblot analysis with a RhoA-specific antibody. F, in vitro methylation of RhoA. To confirm methylation of RhoA, recombinant Icmt was incubated with Icmt null lysate as described under “Experimental Procedures.” Icmt −/− lysate was mock-treated (Icmt −/−) or incubated with recombinant Icmt (Icmt−/− + ICMT) prior to two-dimensional gel analysis.
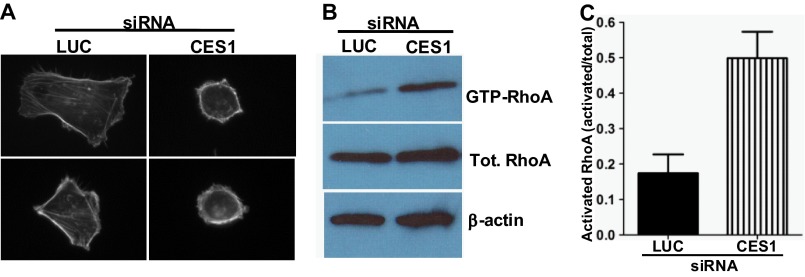
FIGURE 5.
Inhibition of CES1 impacts MDA-MB-231 cell morphology and RhoA activation status. A, inhibition of CES1 alters cell morphology of MDA-MB-231 cells. Cells were treated for 72 h with luciferase (LUC) or CES1 siRNA as described under “Experimental Procedures.” Cells were then plated on coverslips coated with fibronectin and allowed to adhere for 2 h prior to imaging. Representative images are displayed showing the more rounded morphology and altered actin cytoskeletal structure in cells treated with siRNA targeting CES1 as compared with those treated with siRNA targeting luciferase (LUC). B, knockdown of CES1 increases RhoA-GTP levels. MDA-MB-231 cells were treated with luciferase (LUC) or CES1 siRNA for 72 h as described above and then plated on dishes coated with fibronectin for 2 h. Lysates were prepared and activated RhoA (RhoA-GTP) was isolated by pulldown with GST-Rhotekin as described under “Experimental Procedures.” Levels of precipitated and total (Tot.) RhoA were determined by immunoblot analysis using a RhoA-specific antibody; β-actin was utilized as a loading control. C, quantitative representation of data in B. The level of activated RhoA is expressed as a fraction of total RhoA l. Data represent the mean ± S.E. from three independent experiments.
Isoelectric Focusing and Visualization of RhoA
Mouse embryonic fibroblast cells or MDA-MB-231 cells were lysed in MLB lysis buffer (25 mm HEPES pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 10 mm MgCl2, 1 mm EDTA, 2% glycerol, 1 mm DTT, containing protease inhibitors, and 20 μg/ml ebelactone B) for ∼15 min at 4 °C. The lysate was cleared by centrifugation for 5 min at 14,000 rpm at 4 °C, and protein concentration was determined. Equal amounts of protein, generally 1 mg, were precipitated. Protein pellets were resuspended in rehydration buffer (8 m urea, 1% Chaps, 50 mm DTT, 0.2% biolytes (Bio-Rad, Bio-Lyte 3/10 Ampholyte catalog no. 163-1112), 0.001% bromphenol blue), and samples were loaded onto Bio-Rad ReadyStrip IPG strips (11 cm, pH 5–8) for separation. Following isoelectric focusing, proteins were separated by SDS-PAGE using 14% criterion gel (Bio-Rad). RhoA was visualized by immunoblot analysis with a monoclonal Rho-specific antibody (Santa Cruz Biotechnology). Quantitation was performed using NIH ImageJ software.
siRNA Transfections
MDA-MB-231 cells were plated 100,000 cells/well in six-well dishes 24 h prior to transfection. Stock solutions of siRNA oligonucleotides (Thermo Scientific) CES1 (catalog no. L-009051-00), CES2 (catalog no. L-008938-00), luciferase (catalog no. D-001100-01) were prepared at 20 μm. siRNA oligonucleotides were mixed with Opti-Mem I medium (Invitrogen) following the manufacturer's instructions and then mixed with Lipofectamine RNAi Max (Invitrogen) prior to transfection. Cells were harvested, and assays were conducted ∼72 h post-transfection.
Quantitative PCR
Cells were transfected with siRNA as described above and harvested 72 h post-transfection; cell pellets were frozen until RNA isolation. RNA was extracted according to the manufacturer's instructions using QIAshredder homogenization and the RNAeasy Plus mini kit (Qiagen). RNA (500 ng) from each sample was reverse-transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Invitrogen). Quantitative PCR was performed using TaqManTM primers (Applied Biosystems, Invitrogen) for CES1 (catalog no. Hs00275607_m1), CES2 (catalog no. Hs01077945_m1), and β-actin (catalog no. 4333762F) expression, using the Taqman Universal Master Mix II on an ABI StepOne PCR System (Applied Biosystems, Invitrogen). All PCR reactions were performed in duplicate. The relative amounts of CES1 and CES2 mRNA expression were normalized to β-actin mRNA (ΔCT = [CT(CES) − CT(β-actin)]), where CT is the threshold cycle. The relative expression compared with the siLuciferase control was then calculated by 2 −ΔΔCT.
In Vitro Enzymatic Assays
Mouse embryonic fibrobasts or MDA-MD-231 cells were harvested in lysis buffer (100 mm Hepes, pH 7.5, 5 mm Mg Cl2, 1 mm DTT) and lysed by sonication. Following lysis, NaCl was added to a final concentration of 150 mm, and then the lysate was cleared by centrifugation at 14,000 × g for 5 min, and protein concentrations were determined. Approximately 1 mg of lysate was incubated with recombinant Icmt (10 μg of Sf9 membrane protein (14)) or with 1 μm CES1 (purified as described (15)). For methylation assays, AdoMet (10 μm) was added as substrate. The reactions were incubated at 37 °C for 30 min, whereupon Nonidet P-40 detergent was added to a final concentration of 1%, and incubation was continued at 4 °C for 30 min. The detergent extract was cleared by centrifugation for 14,000 × g for 5 min, and proteins were precipitated from the cleared lysate with the addition of trichloroacetic acid. The resulting protein pellets were then processed by two-dimensional gels for analysis of the methylation status of RhoA and CDC42 as described above.
RhoA Activation Assays
Following siRNA treatment for 72 h, cells were harvested with CellstripperTM (Cellgro), and 2 million cells were plated onto 10 cm2 dishes coated with 10 μg/ml fibronectin. Following 2 h of incubation, cells were washed with PBS, harvested in MLB buffer (25 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 10 mm MgCl2, 1 mm EDTA, 2% glycerol) or then incubated for 10 min to ensure complete lysis. The lysates were then cleared by centrifugation at 15000 × g for 5 min. Rho activation assays were performed as described previously (16). Briefly, Rho-GTP in cell lysates was precipitated using purified GST-Rhotekin-RBD (10) and detected following separation on SDS-PAGE by immunoblot analysis with a monoclonal Rho-specific antibody (Santa Cruz Biotechnology) or monoclonal CDC42-specific antibody (BD Transduction Laboratories). Concurrently, an aliquot of the total cell lysate was separated by SDS-PAGE, and RhoA and β-actin levels were visualized by immunoblot analysis.
RESULTS
Elevation of Icmt Activity Impacts Morphology of MDA-MB-231 Cells
To examine the impact of elevating Icmt activity on the biology of MDA-MB-231 cells, we created stable cell lines overexpressing a GFP fusion of Icmt (11) or GFP alone as a control. Following selection, the activity of Icmt in membrane fractions derived from the cells was determined. As shown in Fig. 1A, cells stably expressing Icmt showed an 8-fold increase in enzyme activity as compared with control cells. To determine the impact of Icmt overexpression on cell morphology and actin cytoskeletal arrangement, the cells were plated on fibronectin and visualized. The elevation of Icmt activity dramatically impacted cell morphology and actin cytoskeletal structure (Fig. 1B); the Icmt-expressing cells appeared more rounded, with most of the actin bundling in the periphery as compared with control cells in which the actin was spread throughout the cell, and stress fibers were much more evident. In addition, the polarity of the control cells was much more distinct, with a clear leading edge defined by membrane ruffling and a clear trailing edge. In contrast, the cells overexpressing Icmt exhibited a much more rounded phenotype without a clear polarity.
The finding that simple elevation of Icmt activity in cells elicited a phenotype associated with enhanced RhoA signaling implied the existence of a pool of unmethylated Icmt substrates under normal conditions. To test this hypothesis, we utilized two-dimensional gel electrophoresis and Western blotting with antibodies specific to RhoA to separately identify unmethylated and methylated RhoA. As seen in Fig. 1C, under normal growth conditions, two pools of RhoA could indeed be detected. The amount of unmethylated RhoA present in MDA-MB-231 cells was surprisingly large, with roughly 40% of the RhoA being in the unmethylated state (Fig. 1D). The identity of the more acidic spot as unmethylated RhoA was confirmed via the use of Icmt-null mouse embryonic fibroblast cells (Fig. 1, E and F).
RhoA Methylation Is Dynamic: Identification of a RhoA Carboxylmethylesterase
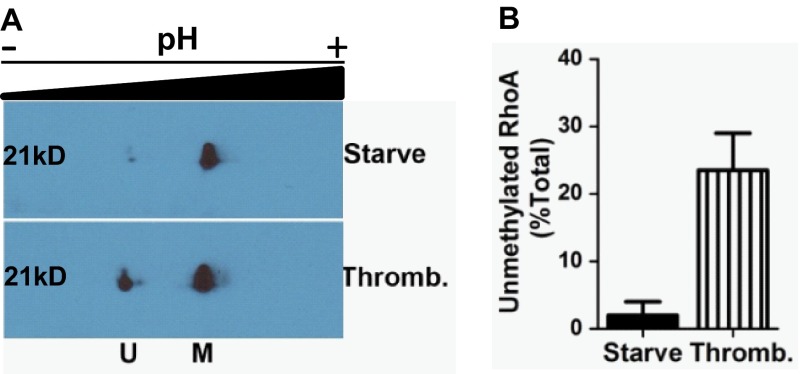
Although prenylation has long been viewed as a constitutive process, it has long been appreciated that the methylation step was in principle reversible, and thus, this could represent a mechanism for dynamic control of protein function. Because the two-dimensional gel system represented a robust technique to examine the methylation status of RhoA, we sought to determine whether this status changed as a function of cell growth conditions. This indeed turned out to be the case, as eliciting quiescence via serum starvation in the MD-MBA-231 cells led to a dramatic shift to almost complete methylation of RhoA (Fig. 2A). Furthermore, treatment of the serum-starved cells for only 30 min with thrombin, which leads to activation of Rho proteins among other actions, triggered an apparent demethylation of the RhoA. In fact, almost a quarter of the RhoA in the cells was demethylated by this brief stimulation with thrombin (Fig. 2B).
FIGURE 2.
RhoA methylation is dynamic. A, impact of thrombin stimulation on RhoA methylation. MDA-MB-231 cells were starved of serum overnight and then treated with vehicle (Starve) or incubated with thrombin (Thromb.) for 30 min prior to harvest and processing for two-dimensional gel analysis as described under “Experimental Procedures.” B, quantitative representation of unmethylated RhoA levels from A, expressed as a fraction of total RhoA. Data represent the mean ± S.E. from pooled results of two independent experiments. U, unmethylated RhoA; M, methylated RhoA.
The observation of dramatic changes in the methylation status of RhoA after just 30 min of thrombin treatment strongly suggested that an esterase was involved. The existence of a esterase that could demethylate CaaX proteins and thereby allow dynamic modulation of localization/function of the proteins has been proposed by several groups, but most studies have only shown that a particular esterase was capable of acting on a prenylcysteine methyl ester (17), and data that the process actually occurs in cells have been lacking. The best evidence for a biologically relevant esterase acting on prenyl proteins comes from studies in plants, where an Arabidopsis gene acting counter to the Icmt of the organism has been identified (18, 19). Our approach to identifying a specific esterase responsible for the demethylation of RhoA observed in the MDA-MB-231 cells involved database analysis for proteins containing the esterase motif and examining the transcript expression profile of potential candidates, particularly targeting those expressed in mammary tissue and breast cancer cells. These criteria, along with homology to the recently identified porcine carboxylesterase capable of hydrolyzing prenylated cysteine methyl esters (20), led us to identify the CES family of esterases, and in particular the CES1 isoform. In parallel with assessment of CES1, we also examined the next best studied member of this family, CES2, which is more highly expressed in most tissues than CES1.
To assess whether either of the CES esterases might be responsible for the demethylation of RhoA observed in the breast cancer cells, we utilized siRNA specific to each isoform to silence its expression. MDA-MB-231 cells treated with a siRNA specific to CES1, CES2, or luciferase (as a control) for 72 h were harvested, and real-time PCR was used to determine the impact of the treatment on the CES isoforms. The siRNA treatments quite selectively silenced expression of their target genes; the siRNA targeting CES1 resulted in >95% knockdown of CES1 with negligible effect on CES2, whereas the siRNA targeting CES2 resulted in nearly 95% knockdown of CES2 with no effect on CES1 expression (data not shown).
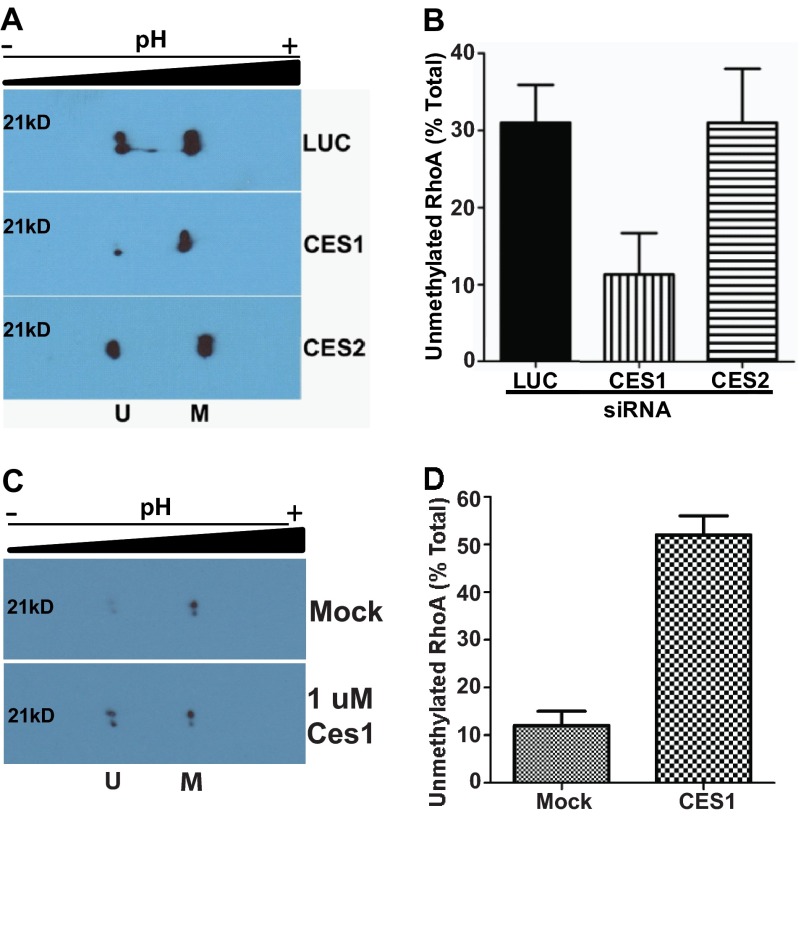
Having confirmed efficient and specific knockdown of the two CES isoforms, we assessed the impact of this knockdown on the methylation status of RhoA in the cells (Fig. 3A). As seen in Fig. 3B, knockdown of CES1 dramatically decreased the amount of unmethylated RhoA in the cells; control cells treated with siRNA targeting luciferase contained 31% unmethylated RhoA, whereas in the cells treated with CES1 siRNA, this was <12%. This effect was specific for the CES1 isoform, as cells in which CES2 expression was silenced had similar levels of unmethylated RhoA as control cells. In addition, the combination of CES1 and CES2 siRNA did not further reduce the pool of unmethylated RhoA as compared with CES1 siRNA alone, further indicating that CES2 does not play a role in RhoA methylation status (data not shown). To demonstrate that CES1 directly impacts methylation of RhoA, we incubated purified CES1 with MDA-MB-231 lysate and then utilized two-dimensional gel analysis to examine the methylation status of RhoA (Fig. 3C). The addition of recombinant CES1 shifted the pool of unmethylated RhoA to ∼50% as compared with the 10% seen in mock-treated lysate (Fig. 3D). These results provide compelling evidence that CES1 is an isoprenylcysteine methylesterase acting on RhoA.
FIGURE 3.
CES1 impacts methylation status of RhoA. A, inhibition of CES1 specifically affects methylation status of RhoA. MDA-MB-231 cells were treated with siRNA or mock as described under “Experimental Procedures,” harvested, and processed for two-dimensional gel analysis of RhoA. A representative immunoblot is depicted showing unmethylated (U) and methylated (M) RhoA. B, quantitative representation of unmethylated RhoA from A expressed as a fraction of total RhoA. Data represent the mean ± S.E. of three independent experiments for luciferase (LUC) and CES1 and two independent experiments for CES2. C, CES1 demethylates RhoA in vitro. Lysates from MDA-MB-231 cells were prepared and incubated with purified CES1 enzyme as described under “Experimental Procedures.” A representative immunoblot is depicted showing unmethylated (U) and methylated (M) RhoA. D, quantitative representation of unmethylated RhoA from A expressed as a fraction of total RhoA. Data represent the mean ± S.E. of two independent experiments.
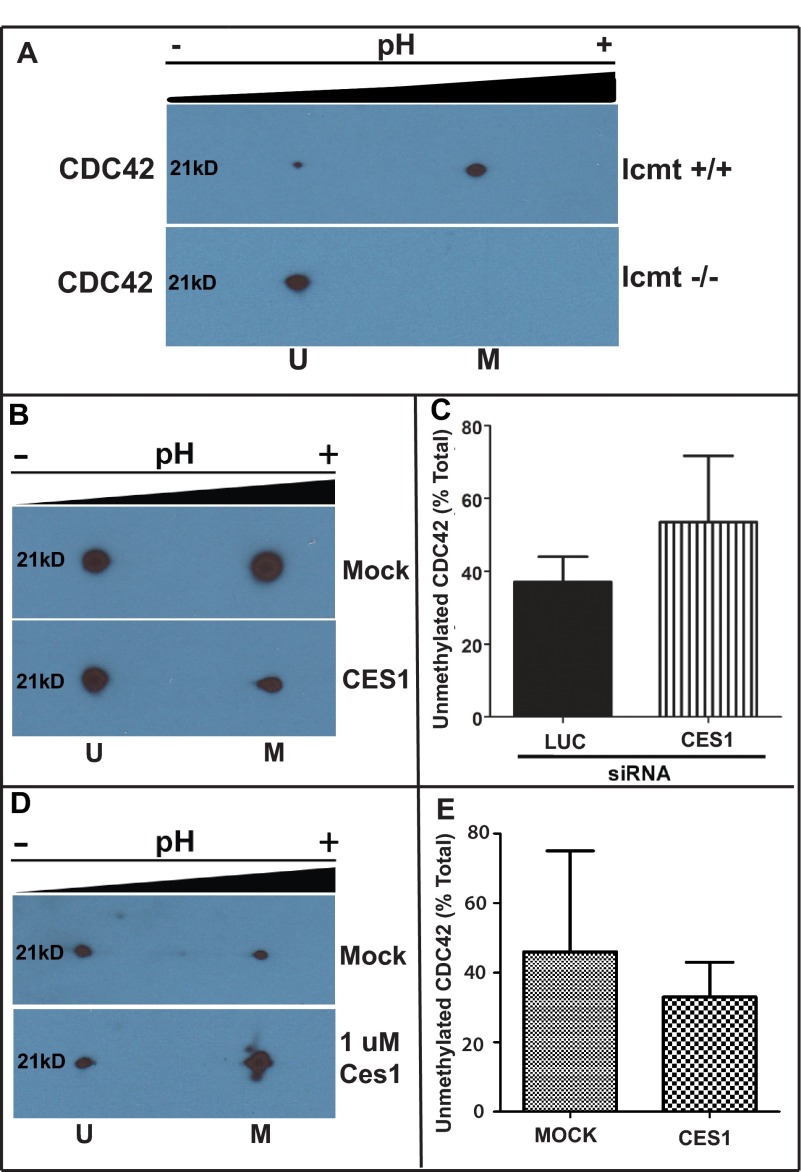
To determine whether CES1 impacted other Rho family members methylated by Icmt, we examined the impact of CES1 on the methylation status of CDC42. We again utilized wild-type and Icmt-null fibroblasts to identify a suitable antibody to visualize unmethylated and methylated CDC42 (Fig. 4A). However, CDC42 methylation was not significantly affected by CES1 gene knockdown (Fig. 4, B and C) or incubation with purified CES1 (Fig. 4, D and E). These results suggest that not all CaaX proteins are substrates for, or perhaps not accessible to, CES1.
FIGURE 4.
CES1 has no significant impact on methylation status of CDC42. A, validation of two-dimensional gel analysis to identify methylated and unmethylated CDC42. Cell lysates from wild-type (Icmt +/+) and null Icmt (Icmt −/−) mouse embryonic fibroblasts were harvested and processed, as described under “Experimental Procedures,” and the presence of unmethylated CDC42 (U) in the more acidic pool or methylated CDC42 (M) in the more basic pool was detected by immunoblot analysis with a RhoA-specific antibody. B, inhibition of CES1 does not affect methylation status of CDC42. MDA-MB-231 cells were treated with siRNA or mock as described above, harvested, and processed for two-dimensional gel analysis of RhoA. A representative immunoblot is depicted showing unmethylated (U) and methylated (M) CDC42. C, quantitative representation of unmethylated CDC42 from (B) expressed as a fraction of total RhoA. Data represent the mean ± S.E. of two independent experiments for luciferase and CES. D, CES1 has no affect on CDC42 in vitro. Lysates from MDA-MB-231 cells were prepared and incubated with purified CES1 enzyme as described under “Experimental Procedures.” A representative immunoblot is depicted showing unmethylated (U) and methylated (M) CDC42. E, quantitative representation of unmethylated and methylated CDC42 levels from D, expressed as a fraction of the total amount of CDC42. Data represent the mean ± S.E. from pooled results of two independent experiments.
Biological Consequences of Silencing CES1 Expression
Given the dramatic changes in MDA-MB-231 cell morphology observed upon increasing Icmt activity (see Fig. 1), we hypothesized that increasing methylation status of RhoA through inhibition of CES1 should have a similar impact on the cells. To determine the biological consequences of inhibiting CES1, we examined the impact of knockdown of CES1 on cell morphology and actin cytoskeleton organization. Upon binding to fibronectin, cells treated with CES1 siRNA showed a much more rounded phenotype as compared with control cells, and also, the majority of the actin was found in bundles around the periphery of the cells (Fig. 5A). This rounded morphology was indeed similar to what was observed in cells overexpressing Icmt (Fig. 1). We also examined the impact of CES1 knockdown on RhoA activation status through the use of GST-Rhotekin pulldown to determine RhoA-GTP levels (Fig. 5B). Importantly, CES1 knockdown resulted in an ∼3-fold increase in RhoA activation levels as compared with control cells (Fig. 5C), which is completely consistent with the observed impact of CES1 knockdown on Rho-mediated morphology and actin organization. These results demonstrate a role for CES1 in controlling the activation of RhoA and hence identify a new player in RhoA-mediated biology.
DISCUSSION
The prenylation pathway has been a promising target for cancer therapy for several years, and recently, inhibition of Icmt-mediated methylation has emerged as a potential therapeutic strategy. Icmt-mediated methylation of CaaX proteins plays a role in oncogenesis, autophagy, and in the metastatic behavior of cells (10, 11, 21, 22). In addition, Icmt-mediated methylation is necessary for development and involved in many other areas of biology, including insulin secretion and endothelial monolayer permeability (23–26). Although the dynamics of CaaX protein methylation has remained largely unexamined, altering Icmt activity by pharmacological inhibition or overexpression has been reported to have converse effects on the methylation status of RhoA and endothelial monolayer permeability (24). Interestingly, in this study a significant pool of unmethylated RhoA was observed in the pulmonary artery endothelia cells. However, a previous study examining the methylation status in a macrophage cell line concluded that the majority of RhoA and CDC42 was methylated upon synthesis and that the modification was stable (27). These studies, along with our current findings, indicate that the ratio of unmethylated versus methylated Rho proteins may be cell-dependent, and altering the methylation status of Rho proteins has profound effects on cell biology.
The dramatic differences in RhoA methylation observed under serum-depleted conditions and in the presence of agonist, along with the changes in methylation after manipulating CES1, revealed that this final step of the prenylation pathway is likely regulated. Regulation of the prenylation pathway has been largely unexamined, although the recent finding that entry into, and passage through, the prenylation pathway of some small GTPases can be controlled by specific SmgGDS splice variants has highlighted the potential for control of CaaX protein function via this process (28). These recent developments in the prenylation field, coupled with the genetic evidence in plants for demethylation of prenyl proteins (18, 19) and our finding of dynamic control of RhoA methylation by CES1, opens new avenues of investigation into the dynamics of prenyl protein processing.
The carboxylesterase family of proteins is quite complex and contains many homologous subfamilies, each which have several isoforms (29). In this study, we identified CES1 as an esterase involved in RhoA methylation, whereas CES2 appears to not play a role in this regard. We also determined that CES1 does not impact CDC42 methylation, but more thorough investigations need be conducted to determine whether other CaaX proteins are affected by CES1 and whether other CES family members control methylation of specific prenyl proteins. In addition, the mechanism through which CES1 engages RhoA will be important to determine; CES1 contains an endoplasmic reticulum localization sequence, but there is evidence that the enzyme can be found in the cytoplasm as well (30).
The demonstration that RhoA methylation is dynamic and reversible in the context of regulatory inputs raises many exciting questions, including how this post-translational modification is regulated and what are the biological ramifications of altering the methylation status of CaaX proteins. In this study, we observed a shift to more methylated RhoA under starvation conditions and in the absence of CES1 activity. Interestingly, in the absence of CES1, there was more active Rho (Fig. 5). These results suggest that the methylation status of RhoA does not directly relate to its activation state. However, the proper regulation of methylation may be important in controlling its state of activation. Methylation has been shown to play an important role in interactions of RhoA with other proteins. It has been demonstrated by us and others (10, 31–34) that the methylation status of RhoA directly impacts the RhoA-RhoGDI interaction and that RhoGDI plays an important role in the regulation of Rho activity. We hypothesize that, by controlling the methylation of RhoA, CES1 may be involved in the down-regulation of RhoA activity. Perhaps more RhoA is methylated under starvation conditions so that it can be more readily accessible for activation upon stimulation. In this scenario, by having another regulatory enzyme into the pathway, the cell can more tightly control Rho activity. The present study clearly reveals intriguing questions about the regulation of prenyl protein methylation. The localization and activity of Rho GTPases are regulated by a complex system of post-translational modifications and protein interactions that are still being sorted out (34). The exciting findings of dynamic RhoA methylation and a carboxylesterase capable of demethylating RhoA provide evidence that prenylation-dependent methylation may be more analogous to other highly regulated post-translational events such as phosphorylation or sumoylation than envisioned previously.
This work was supported by the by the American Lebanese and Syrian Associated Charities and the St. Jude Children's Research Hospital.
- Icmt
- isoprenylcysteine carboxylmethyltransferase
- CES1
- carboxylesterase 1.
REFERENCES
- 1. Glomset J. A., Farnsworth C. C. (1994) Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu. Rev. Cell Biol. 10, 181–205 [DOI] [PubMed] [Google Scholar]
- 2. Zhang F. L., Casey P. J. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 [DOI] [PubMed] [Google Scholar]
- 3. Casey P. J., Seabra M. C. (1996) Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292 [DOI] [PubMed] [Google Scholar]
- 4. Ashby M. N. (1998) CaaX converting enzymes. Curr. Opin. Lipidol. 9, 99–102 [DOI] [PubMed] [Google Scholar]
- 5. Young S. G., Ambroziak P., Kim E., Clarke S. (2001) Postisoprenylation protein processing: CXXX (CaaX) endoproteases and isoprenylcysteine carboxyl methyltransferase. The Enzymes (Tamanoi F., Sigman D. S., eds) 3rd Ed., vol. 21, pp. 156–213, Academic Press, San Diego, CA [Google Scholar]
- 6. Doll R. J., Kirschmeier P., Bishop W. R. (2004) Farnesyltransferase inhibitors as anticancer agents: critical crossroads. Curr. Opin. Drug Discov. Devel. 7, 478–486 [PubMed] [Google Scholar]
- 7. Gelb M. H., Brunsveld L., Hrycyna C. A., Michaelis S., Tamanoi F., Van Voorhis W. C., Waldmann H. (2006) Therapeutic intervention based on protein prenylation and associated modifications. Nat. Chem. Biol. 2, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winter-Vann A. M., Casey P. J. (2005) Post-prenylation processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5, 405–412 [DOI] [PubMed] [Google Scholar]
- 9. Wright L. P., Philips M. R. (2006) Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47, 883–891 [DOI] [PubMed] [Google Scholar]
- 10. Cushman I., Casey P. J. (2009) Role of isoprenylcysteine carboxylmethyltransferase-catalyzed methylation in Rho function and migration. J. Biol. Chem. 284, 27964–27973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winter-Vann A. M., Baron R. A., Wong W., dela Cruz J., York J. D., Gooden D. M., Bergo M. O., Young S. G., Toone E. J., Casey P. J. (2005) A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 102, 4336–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baron R. A., Casey P. J. (2004) Analysis of the kinetic mechanism of recombinant human isoprenylcysteine carboxylmethyltransferase (Icmt). BMC Biochem. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baron R. A., Peterson Y. K., Otto J. C., Rudolph J., Casey P. J. (2007) Time-dependent inhibition of isoprenylcysteine carboxyl methyltransferase by indole-based small molecules. Biochemistry 46, 554–560 [DOI] [PubMed] [Google Scholar]
- 14. Otto J. C., Kim E., Young S. G., Casey P. J. (1999) Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 274, 8379–8382 [DOI] [PubMed] [Google Scholar]
- 15. Morton C. L., Potter P. M. (2000) Comparison of Escherichia coli, Saccharomyces cerevisiae, Pichia pastoris, Spodoptera frugiperda, and COS7 cells for recombinant gene expression. Application to a rabbit liver carboxylesterase. Mol. Biotechnol. 16, 193–202 [DOI] [PubMed] [Google Scholar]
- 16. Meigs T. E., Juneja J., Demarco C. T., Stemmle L. N., Kaplan D. D., Casey P. J. (2005) Selective uncoupling of Gα 12 from Rho-mediated signaling. J. Biol. Chem. [DOI] [PubMed] [Google Scholar]
- 17. Brüsehaber E., Böttcher D., Bornscheuer U. T. (2009) Insights into the physiological role of pig liver esterase: isoenzymes show differences in the demethylation of prenylated proteins. Bioorg. Med. Chem. 17, 7878–7883 [DOI] [PubMed] [Google Scholar]
- 18. Deem A. K., Bultema R. L., Crowell D. N. (2006) Prenylcysteine methylesterase in Arabidopsis thaliana. Gene 380, 159–166 [DOI] [PubMed] [Google Scholar]
- 19. Huizinga D. H., Omosegbon O., Omery B., Crowell D. N. (2008) Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis. Plant Cell 20, 2714–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oboh O. T., Lamango N. S. (2008) Liver prenylated methylated protein methyl esterase is the same enzyme as Sus scrofa carboxylesterase. J. Biochem. Mol. Toxicol. 22, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang M., Tan W., Zhou J., Leow J., Go M., Lee H. S., Casey P. J. (2008) A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J. Biol. Chem. 283, 18678–18684 [DOI] [PubMed] [Google Scholar]
- 22. Wahlstrom A. M., Cutts B. A., Liu M., Lindskog A., Karlsson C., Sjogren A. K., Andersson K. M., Young S. G., Bergo M. O. (2008) Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood 112, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bergo M. O., Leung G. K., Ambroziak P., Otto J. C., Casey P. J., Gomes A. Q., Seabra M. C., Young S. G. (2001) Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 276, 5841–5845 [DOI] [PubMed] [Google Scholar]
- 24. Lu Q., Harrington E. O., Hai C. M., Newton J., Garber M., Hirase T., Rounds S. (2004) Isoprenylcysteine carboxyl methyltransferase modulates endothelial monolayer permeability: involvement of RhoA carboxyl methylation. Circ. Res. 94, 306–315 [DOI] [PubMed] [Google Scholar]
- 25. Rounds S., Lu Q., Harrington E. O., Newton J., Casserly B. (2008) Pulmonary endothelial cell signaling and function. Trans. Am. Clin. Climatol. Assoc. 119, 155–167; discussion 167–159 [PMC free article] [PubMed] [Google Scholar]
- 26. Jayaram B., Syed I., Singh A., Subasinghe W., Kyathanahalli C. N., Kowluru A. (2011) Isoprenylcysteine carboxyl methyltransferase facilitates glucose-induced Rac1 activation, ROS generation and insulin secretion in INS 832/13 β-cells. Islets 3, 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Backlund P. S., Jr. (1997) Post-translational processing of RhoA. Carboxyl methylation of the carboxyl-terminal prenylcysteine increases the half-life of Rhoa. J. Biol. Chem. 272, 33175–33180 [DOI] [PubMed] [Google Scholar]
- 28. Berg T. J., Gastonguay A. J., Lorimer E. L., Kuhnmuench J. R., Li R., Fields A. P., Williams C. L. (2010) Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holmes R. S., Wright M. W., Laulederkind S. J., Cox L. A., Hosokawa M., Imai T., Ishibashi S., Lehner R., Miyazaki M., Perkins E. J., Potter P. M., Redinbo M. R., Robert J., Satoh T., Yamashita T., Yan B., Yokoi T., Zechner R., Maltais L. J. (2010) Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm. Genome 21, 427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ross M. K., Crow J. A. (2007) Human carboxylesterases and their role in xenobiotic and endobiotic metabolism. J. Biochem. Mol. Toxicol. 21, 187–196 [DOI] [PubMed] [Google Scholar]
- 31. Harrington E. O., Newton J., Morin N., Rounds S. (2004) Barrier dysfunction and RhoA activation are blunted by homocysteine and adenosine in pulmonary endothelium. Am. J. Physiol. Lung Cell Mol. Physiol. 287, L1091–1097 [DOI] [PubMed] [Google Scholar]
- 32. Michaelson D., Ali W., Chiu V. K., Bergo M., Silletti J., Wright L., Young S. G., Philips M. (2005) Postprenylation CAAX Processing Is Required for Proper Localization of Ras but Not Rho GTPases. Mol. Biol. Cell 4, 1606–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cushman I., Casey P. J. (2011) RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh. Migr. 5, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia-Mata R., Boulter E., Burridge K. (2011) The 'invisible hand': regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 12, 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]