Background: Enzymes mediating chemical redox cycling in the lung are poorly defined.
Results: Sepiapterin reductase was identified as a key mediator of redox cycling and was analyzed using inhibitors and site-directed mutagenesis.
Conclusion: Sepiapterin reductase generates reactive oxygen species during redox cycling in a mechanism distinct from sepiapterin reduction.
Significance: This is the first report demonstrating that sepiapterin reductase mediates chemical redox cycling.
Keywords: Oxidative Stress, Quinones, Reactive Oxygen Species (ROS), Reductase, Tetrahydrobiopterin, Quinone Redox Cycling
Abstract
In the lung, chemical redox cycling generates highly toxic reactive oxygen species that can cause alveolar inflammation and damage to the epithelium, as well as fibrosis. In this study, we identified a cytosolic NADPH-dependent redox cycling activity in mouse lung epithelial cells as sepiapterin reductase (SPR), an enzyme important for the biosynthesis of tetrahydrobiopterin. Human SPR was cloned and characterized. In addition to reducing sepiapterin, SPR mediated chemical redox cycling of bipyridinium herbicides and various quinones; this activity was greatest for 1,2-naphthoquinone followed by 9,10-phenanthrenequinone, 1,4-naphthoquinone, menadione, and 2,3-dimethyl-1,4-naphthoquinone. Whereas redox cycling chemicals inhibited sepiapterin reduction, sepiapterin had no effect on redox cycling. Additionally, inhibitors such as dicoumarol, N-acetylserotonin, and indomethacin blocked sepiapterin reduction, with no effect on redox cycling. Non-redox cycling quinones, including benzoquinone and phenylquinone, were competitive inhibitors of sepiapterin reduction but noncompetitive redox cycling inhibitors. Site-directed mutagenesis of the SPR C-terminal substrate-binding site (D257H) completely inhibited sepiapterin reduction but had minimal effects on redox cycling. These data indicate that SPR-mediated reduction of sepiapterin and redox cycling occur by distinct mechanisms. The identification of SPR as a key enzyme mediating chemical redox cycling suggests that it may be important in generating cytotoxic reactive oxygen species in the lung. This activity, together with inhibition of sepiapterin reduction by redox-active chemicals and consequent deficiencies in tetrahydrobiopterin, may contribute to tissue injury.
Introduction
The lung is highly sensitive to redox-active chemicals, including bipyridyl herbicides such as paraquat, nitroaromatic compounds, and various quinones (1, 2). High doses of these chemicals can induce alveolar inflammation, epithelial cell damage, pneumonia, pulmonary hypertension, and fibrosis (2). Toxicity is thought to result from an accumulation of these chemicals in lung cells and subsequent enzyme-mediated redox cycling. In the redox cycling process, one electron reduction of a redox-active chemical generates radical ions. Under aerobic conditions, these radicals rapidly react with oxygen generating superoxide anion and the parent compound (3–5). Spontaneous and enzyme-mediated dismutation of superoxide anion leads to the formation of hydrogen peroxide (H2O2). In the presence of trace metals, H2O2 generates highly toxic hydroxyl radicals (6). The reaction of superoxide anion with nitric oxide can also generate peroxynitrite, a potent oxidant known to cause tissue injury (7). These reactive nitrogen and reactive oxygen species (ROS)2 can damage many intracellular components, including DNA, lipids, and proteins resulting in nitrosative and oxidative stress and toxicity (7, 8).
The complement of enzymes in the lung that participate in chemical redox cycling has not been clearly established. Several flavin-containing enzymes known to mediate redox cycling are present in lung tissues, including cytochrome P450 reductase, cytochrome b5 reductase, xanthine oxidase, and various forms of nitric-oxide synthase (9–13). These enzymes require NADPH or NADH as the source of reducing equivalents. NADPH-cytochrome P450 reductase, which contains both FAD and FMN as cofactors, is the best characterized enzyme mediating redox cycling (9, 10). It appears that the ability of FAD to accept single electrons from NADPH is critical for redox cycling. This is supported by findings that the redox cycling process is blocked by diphenyleneiodonium, a selective flavoenzyme inhibitor (9, 14). Cytochrome P450 reductase and cytochrome b5 reductase are microsomal enzymes, whereas xanthine oxidase and nitric-oxide synthase are cytoplasmic enzymes; nitric-oxide synthases are also localized in cell membranes (7, 15–18). Localized concentrations of redox-active chemicals in cells, enzymes that mediate their redox cycling, and levels of ROS generated are key factors leading to lung injury (19).
In earlier studies, we demonstrated that the cytoplasm of lung epithelial cells is a rich source of enzymes capable of mediating chemical redox cycling (20). One important enzyme involved in this process is thioredoxin reductase, a homodimeric flavoprotein that catalyzes the reduction of oxidized thioredoxin, as well as other redox-active proteins, and plays a key role in maintaining cellular redox homeostasis (20, 21). In this study, we identified sepiapterin reductase as another highly active cytoplasmic enzyme that mediates redox cycling in lung cells. Sepiapterin reductase is an NADPH-dependent enzyme that catalyzes the formation dihydrobiopterin (BH2), a precursor for tetrahydrobiopterin (BH4), a cofactor critical in aromatic amino acid metabolism and nitric oxide biosynthesis (22–24) (see Fig. 1 for schematic depicting reactions catalyzed by sepiapterin reductase). Using recombinant human enzyme, sepiapterin reductase was found to be highly efficient in mediating chemical redox cycling and generating ROS. Interestingly, redox cycling markedly reduced the ability of the enzyme to generate BH2. Unlike other enzymes that mediate redox cycling, sepiapterin reductase does not contain flavins or other co-factors suggesting that it functions by a unique mechanism. Our results are novel as they identify a flavin-independent pathway mediating chemical redox cycling in lung epithelial cells. Moreover, they demonstrate that redox cycling chemicals can control a key enzyme required for the biosynthesis of a cofactor important in generating mediators that regulate lung function.
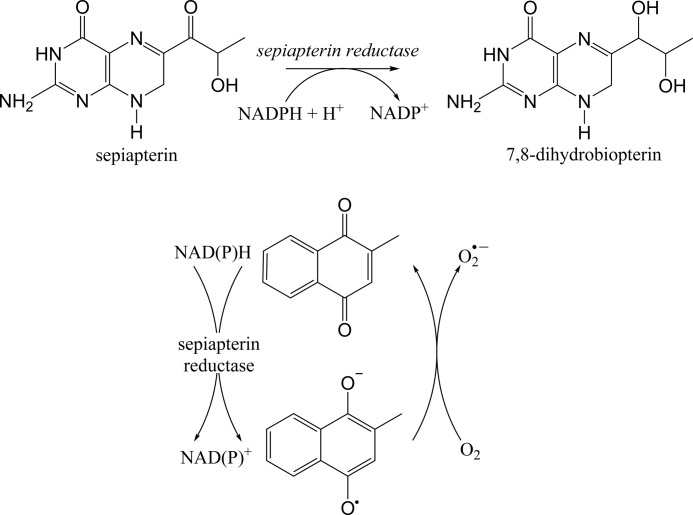
FIGURE 1.
Schematic depicting reactions catalyzed by sepiapterin reductase. Upper panel, reduction of sepiapterin by sepiapterin reductase generates dihydrobiopterin. Additional cellular reductases convert dihydrobiopterin to tetrahydrobiopterin. Lower panel, in the presence of redox cycling chemicals such as menadione, sepiapterin reductase generates reactive oxygen species.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Restriction enzymes were purchased from New England Biolabs (Ipswich, MA). T4 DNA ligase, Amplex Red reagent, and Ni-NTA-agarose were obtained from Invitrogen. A pGEMT TA cloning kit was from Promega (Madison, WI), and Pfu Easy A polymerase was from Stratagene (La Jolla, CA). l-Sepiapterin was from Cayman Chemical (Ann Arbor, MI). Horseradish peroxidase, menadione (2-methylnaphthalene-1,4-dione), NADPH, protease inhibitor mixture 1, and other chemicals were from Sigma unless otherwise indicated. Protease inhibitor mixture 1 contained 4-(2-aminoethyl) benzenesulfonyl fluoride, pepstatin A, E-64 (trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane), bestatin, leupeptin, and aprotinin. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Assays for Sepiapterin Reductase Activity and Redox Cycling
Sepiapterin reductase activity was assayed by measuring decreases in sepiapterin absorbance at 420 nm as described by Katoh (25) with some modifications. Standard reaction mixes contained 100 mm potassium phosphate buffer, pH 6.4, 100 μm NADPH, 50 μm sepiapterin, and 2 μg of enzyme protein in a final volume of 200 μl. Changes in absorbance were monitored using a Spectramax M5 microplate reader (Molecular Devices, Sunnyvale, CA). NADPH, quinones, and cofactors had minimal absorbances at 420 nm and did not interfere with the sepiapterin reductase assay. In some experiments, sepiapterin reductase activity was assayed as described by Ferre and Naylor (26). In the assay, BH2, the reduction product of sepiapterin by sepiapterin reductase, is oxidized by iodine to the highly fluorescent biopterin, which is then quantified by HPLC with fluorescence detection. For the assay, standard reaction mixes were supplemented with an NADPH-regenerating system consisting of 10 mm glucose 6-phosphate and 0.5 units/ml glucose-6-phosphate dehydrogenase, and 0.2 μg of sepiapterin reductase. After incubating the reactions at 37 °C for 1 h in the dark, 25 μl of a mixture of 33 mm iodine and 100 mm KI in 1 m HCl was added. After standing for 10 min at room temperature, the precipitated proteins were removed by centrifugation at 15,000 × g for 3 min at room temperature, and excess iodine was reduced by the addition of 25 μl of 57 mm ascorbic acid to the reaction mix. Biopterin formed in the assay was separated using a Jasco HPLC system (Easton, MD) fitted with a Maxsil-10 250 × 4-mm C18 column (Phenomenex, Torrance, CA). The mobile phase consisted of 5% methanol in water, and the flow rate was set at 1.5 ml/min. Fluorescence was monitored using a Jasco FP-2020 spectrofluorometer with excitation and emission wavelengths set at 362 and 435 nm, respectively. The chromatographic peaks were integrated using the Jasco ChromNAV software.
ROS generated by chemical redox cycling were assayed by measuring the formation of superoxide anion, H2O2, and hydroxyl radicals. Superoxide anion was measured spectrophotometrically by the reduction of acetylated cytochrome c at 550 nm as described by Fussell et al. (27). Typical reaction mixes contained 100 mm potassium phosphate buffer, pH 7.8, 0.05 mm acetylated cytochrome c, 0.2 mm NADPH, 0.5 mm menadione, and 20 μg of sepiapterin reductase in a final volume of 1 ml. H2O2 production was assayed using Amplex-Red/horseradish peroxidase as described previously (20, 28). Standard reactions (0.1 ml) contained 50 mm phosphate buffer, pH 7.8, 25 μm Amplex-Red, 0.1 unit of horseradish peroxidase, 0.2 mm NADPH, 22.5 μg of cell cytosolic proteins, or 1 μg of recombinant sepiapterin reductase and appropriate concentrations of redox-active chemicals. Reactions were initiated by the addition of proteins to the mix. The fluorescent product, resorufin, was recorded as relative fluorescence units using the microplate reader with excitation and emission wavelengths set at 530 and 587 nm, respectively. In experiments using recombinant sepiapterin reductase, the concentration of H2O2 generated in the reactions was calculated from a calibration curve prepared using appropriate standards. H2O2 formation was found to be linear for at least 30 min using 0.1 μg of recombinant sepiapterin reductase. When catalase was added to the redox cycling reactions, it degraded H2O2 as it was formed; it did not alter the ability of the enzyme to reduce sepiapterin. We also found that reduced biopterin did not react directly with quinones such as menadione or 9,10-phenanthrenequinone to generate H2O2.
Hydroxyl radicals were measured by monitoring the formation of 2-hydroxyterephthalate from terephthalate in enzyme assays as described previously (29). In these assays, 4.5 μg of recombinant sepiapterin reductase/100 μl of reaction mix were used. In some experiments, oxygen utilization in enzyme reactions during redox cycling was measured using a Clark-type oxygen electrode as described previously (27). In these assays, 16 μg of recombinant sepiapterin reductase/0.8 ml of reaction mix were used. Kinetic parameters of the enzyme were calculated from linear portions of reaction curves using Lineweaver-Burk plots.
Isolation and Characterization of a Redox Cycling Activity from Lung Epithelial Cells
MLE-15 murine lung epithelial cells, kindly provided by Dr. Jacob Finkelstein at the University of Rochester, New York (30), and A549 lung epithelial cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C with 5% CO2 in a humidified incubator. MLE-15 cell cytosolic fractions were prepared by suspending the cells (3 × 108) in 1.0 ml of phosphate buffer (50 mm, pH 7.0) containing protease inhibitors and sonicating on ice. After centrifugation in an Eppendorf 5417R centrifuge at 12,000 × g for 10 min to remove debris, membranes and mitochondrial fractions, supernatants were centrifuged in a Beckman L7–55 ultracentrifuge (100,000 × g, 1 h) to obtain post-microsomal supernatant fractions. NADPH-binding proteins were then purified by affinity chromatography using 2′,5′-ADP-agarose columns as described by Wolff et al. (31). NADPH eluates from the columns were concentrated using an Amicon ultracentrifugal filter (Millipore, Billerica, MA) and then fractionated by size exclusion chromatography on a Superose 12 HR 10/30 column (GE Healthcare) in phosphate-buffered saline containing 0.1% Nonidet P-40, pH 7.4, at a flow rate of 0.3 ml/min. Fractions were monitored for absorbance at 280 nm and redox cycling activity over 30 min using the Amplex-Red/horseradish peroxidase assay. Two activity peaks were collected and analyzed on 10% SDS-polyacrylamide gels using silver staining. An Mr ≈28,000 band, which was detected in both fractions, was cut from the gel of the peak II fraction, subjected to in-gel digestion with trypsin, and analyzed using MALDI-TOF/TOF (Alphalyse, Palo Alto, CA). A combined MS and MS/MS search was performed against the NCBI database using an in-house MASCOT server. Six peptide fragments matched the sequence of mouse sepiapterin reductase.
For metabolism studies, A549 lung epithelial cells, grown to a density of 4 × 105 cells/well in 6-well culture dishes, were pretreated with control medium or medium containing 500 μm menadione. After 2 h, sepiapterin (100 μm final concentration) was added to the cultures. After an additional 2 h, cells were lysed in PBS containing 0.5% Nonidet P40 and analyzed for biopterin content by HPLC as described above (26).
Cloning of the Mouse and Human SPR Genes into a pET28a Expression Vector
Total RNA was isolated from either mouse MLE-15 cells or human A549 lung epithelial cells using the TRIzol reagent (Invitrogen). cDNAs were synthesized using reverse transcription with Superscript II RNase H-RT according to the manufacturer's instructions (Invitrogen). The full-length sepiapterin reductase gene was amplified by PCR using Pfu Easy A polymerase (Stratagene) with the primer pairs hSPR-F and hSPR-R or mSPR-F and mSPR-R (Table 1). Forward primers (hSPR-F and mSPR-F) contain an NdeI restriction site and 15 nucleotides at the 5′-end of sepiapterin reductase, and reverse primers (hSPR-R and mSPR-R) contain a BamHI restriction site and 13 nucleotides at the 3′-end of sepiapterin reductase according to the published human sepiapterin reductase sequence (accession number NM_003124, PubMed) and the mouse sepiapterin reductase sequence (accession number NM_011467, PubMed). The PCR products were purified using a PCR purification kit (Qiagen, Valencia, CA), cloned into the TA vector pGEMT (Stratagene), and transformed into Escherichia coli DH10B cells. The pGEMT-hSPR was isolated using a plasmid miniprep kit (Qiagen), digested with NdeI and BamHI restriction enzymes, and then subcloned into the NdeI and BamHI sites of pET28a, which carries a hexa-histidine tag coding sequence (Novagen, Madison, WI). The positive clones were selected and verified by sequence analysis (DNA Core facility, UMDNJ-Robert Wood Johnson Medical School, Piscataway, NJ).
TABLE 1.
PCR primers used in cloning of SPR and site-directed mutagenesis
| Primer | Sequence |
|---|---|
| hSPR-F | CCGCGCGGCAGCCATATGGAGGGCGGGCTG |
| hSPR-R | CGAATTCGGATCCTTATCATTTGTCATAGAAGTC |
| mSPR-F | CCGCGCGGCAGCCATATGGAGGCAGGCGG |
| mSPR-R | CGAATTCGGATCCTTATCAGTCATAGAAGTCCAC |
| G14S-F | GCTTGCTGACCAGCGCCTCCCGCGGC |
| G14S-R | GCCGCGGGAGGCGCTGGTCAGCAAGC |
| G18D-F | GGCCTCCCGCGACTTCGGCCGGACG |
| G18D-R | CGTCCGGCCGAAGTCGCGGGAGGCC |
| R42G-F | GTCCTTAGCGCCGGCAACGACGAGGC |
| R42G-R | GCCTCGTCGTTGCCGGCGCTAAGGAC |
| N99A-F | GCTTATCAACGCCGCGGGCTCTCTTG |
| N99A-R | CAAGAGAGCCCGCGGCGTTGATAAGC |
| S157A-F | GTTAACATCTCGGCCCTCTGTGCCCTGC |
| S157A-R | GCAGGGCACAGAGGGCCGAGATGTTAAC |
| K174L-F | GTACTGTGCAGGACTGGCTGCTCGTG |
| K174L-R | CACGAG CAGCCAGTCC TGCACAGTAC |
| M205G-F | GGACACAGACGGGCAGCAGTTGGC |
| M205G-R | GCCAACTGCTGCCCGTCTGTGTCC |
| D257H-R | CGAATTCGGATCCTTATCATTTGTCATAGAAGTGCACGTGGGCTCC |
| DEL257-R | CGAATTCGGATCCTTATCACACGTGGGCTCC |
| Y259A-R | CGAATTCGGATCCTTATCATTTGTCAGCGAAGTCCACGTGGGCTCC |
Site-directed Mutagenesis
To generate the human sepiapterin reductase mutants G14S, G18R, R42G, N99A, S157A, K147L, M205G, and D257H, primer-mediated two-step PCR was used because the mutation sites are located in the central region of the gene. Mutation points were designed in the complementary forward and reverse primers. pET-hSPR containing the wild type human sepiapterin reductase gene was used as a template to perform site-directed mutagenesis. In the first round, left arm PCR fragments were obtained using hSPR forward primer and mutation reverse primers (G14S-R, G18R-R, R42G-R, N99A-R, S157A-R, K174L-R, M205G-R, and D257H-R), and right arm PCR fragments were obtained using mutation forward primers (G14S-F, G18R-F, R42G-F, N99A-F, S157A-F, K174L-F, M205G-F, and D257H-F) and hSPR reverse primer. Purified left arm and right arm fragments were mixed to form partial duplex structures in complementary regions that were then used as templates for a second round of PCR with the primers hSPR-F and hSPR-R to generate the full-length mutation PCR fragments. For mutants D257H, Y259A, and Δ257–261, the full-length mutant PCR fragments were produced by one-step PCR using an hSPR-F primer paired with a D257H-R primer, Y259A-R primer, or Δ257–261-R primer, respectively, to produce full-length D257H, Y259A, and Δ257–261 PCR fragments because the mutation points are very close to the C terminus and therefore the mutation points were designed in reverse primers. The full-length mutant PCR products were cloned into the pET28a vector between the NdeI and BamHI restriction sites. The mutations were confirmed by sequence analysis.
Expression and Purification of Recombinant Sepiapterin Reductase
The expression vector containing wild type or mutant sepiapterin reductase genes was transformed into E. coli BL21 (DE3). In this system, the expressed recombinant proteins contain six histidines at the N terminus for nickel affinity (Ni-NTA) purification. The clones were picked and cultured in Terrific Broth medium containing 25 mg/liter kanamycin at 37 °C. The recombinant sepiapterin reductase proteins were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside for 4 h at 37 °C when the absorbance of the cultures reached 0.6 at 600 nm. The cell pellets from low speed centrifugation (6,000 × g for 20 min) were resuspended in buffer A (50 mm NaH2PO4/Na2HPO4, pH 7.0, 300 mm NaCl), sonicated on ice, and then centrifuged at 20,000 × g for 30 min. The recombinant proteins in supernatants were purified using a nickel affinity chromatography column (Invitrogen) under native conditions. Briefly, after loading the crude extracts, the Ni-NTA affinity column was equilibrated with buffer A and then washed with buffer B (buffer A with 20 mm imidazole). Finally, the recombinant enzymes were eluted with buffer C (buffer A containing 150 mm imidazole and 10% glycerol). Eluted fractions were concentrated using VivaSpin 6 column (VWR, West Chester, PA) and analyzed for protein using the DC Protein Assay Reagent (Bio-Rad) with bovine serum albumin as the standard. Recombinant enzymes were analyzed on 12.5% SDS-polyacrylamide gels using Coomassie Brilliant Blue R-25 staining. Recombinant wild type human sepiapterin reductase was 3-fold more active on sepiapterin reduction and redox cycling than recombinant wild type mouse sepiapterin reductase and was used in all further experiments to characterize the enzyme.
RESULTS
Identification of Sepiapterin Reductase as a Mediator of Redox Cycling in Lung Epithelial Cells
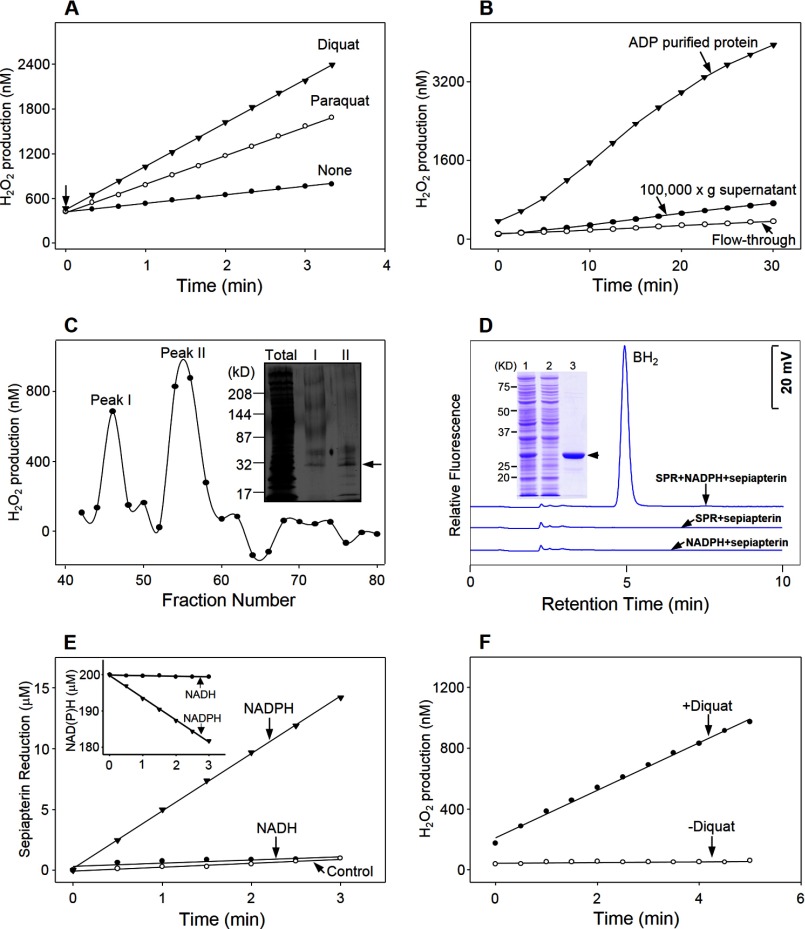
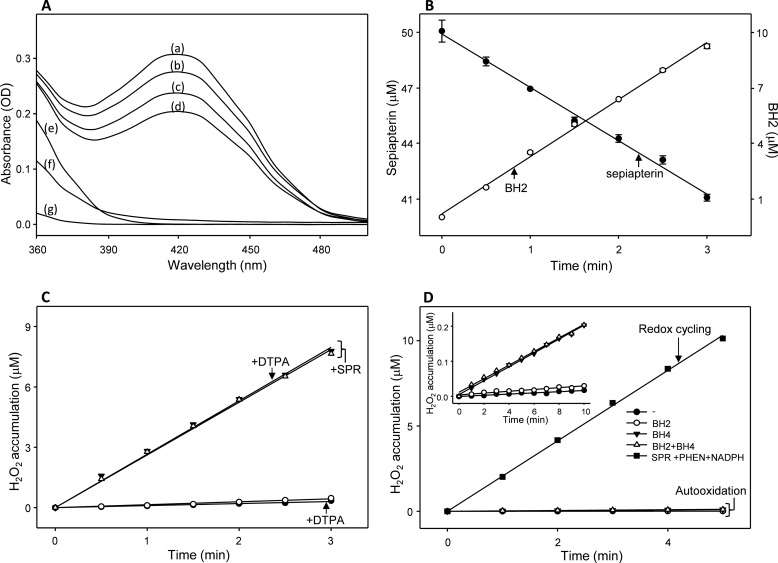
Diquat and paraquat were found to readily stimulate the generation of H2O2 in cytosolic fractions of lung epithelial cells (Fig. 2, panel A). This activity was NADPH-dependent (data not shown); greater than 95% of redox cycling activity could be recovered following ADP-affinity purification (Fig. 2, panel B). Two main peaks of activity (peaks I and II) were obtained following size exclusion chromatography of the affinity-purified material (Fig. 2, panel C). A common distinct protein band (Mr ≈28,000) was detected in both peak fractions following SDS-PAGE (Fig. 2, panel C, inset). The protein band in peak II was cut from the gel, analyzed using MALDI-TOF/TOF MS, and identified as sepiapterin reductase. To confirm that sepiapterin reductase possesses redox cycling activity, the human and mouse genes were cloned into a hexahistidine-tagged vector, expressed in E. coli, and purified by nickel affinity chromatography (Fig. 2, panel D, inset, and data not shown). The enzymes, which appeared as single bands on SDS-polyacrylamide gels (Mr ≈30,000), reduced sepiapterin to BH2, as measured by decreases in absorption of sepiapterin at 420 nm (25) and by the formation of BH2 by HPLC (26). Similar enzyme activity was detected in both assays (Figs. 2, panel D, and 3, panels A and B, and data not shown). In these assays, sepiapterin was reduced in a time-dependent manner with a stoichiometric (1:1) increase in BH2 (Fig. 3, panel B). Using the HPLC assay, the kinetics of the enzymes was measured (Km = 25.4 μm, kcat = 97.0 min−1, kcat/Km = 3.8 min−1 μm−1 for the human enzyme, and Km = 44.2 μm, kcat = 48.4 min−1, kcat/Km = 1.1 min−1μm−1, for the mouse enzyme). For both the human and mouse enzymes, the reaction was NADPH-dependent; NADH did not support enzyme activity (Fig. 2, panels D and E, and data not shown).
FIGURE 2.
Identification of sepiapterin reductase as a mediator of chemical redox cycling. Panel A, redox cycling activity, assessed by the formation of H2O2, was quantified in 100,000 × g supernatant fractions from MLE-15 cells in the presence of 500 μm paraquat or diquat. Arrow indicates initiation of the reaction following the addition of supernatant. Panels B and C, paraquat-stimulated redox cycling activity in cytosolic fractions of MLE-15 cells purified by ADP affinity and size exclusion chromatography, respectively. Panel C, inset, SDS-PAGE analysis of total cell lysate and the two peaks (I and II) of redox cycling activity following size exclusion chromatography. The major band in peak II of SDS-PAGE (shown by the arrow) was analyzed by MALDI-TOF/TOF and identified as sepiapterin reductase. Panel D, dihydrobiopterin (BH2) formation from sepiapterin by human recombinant sepiapterin reductase. Sepiapterin reductase activity required sepiapterin, NADPH, and purified recombinant SPR. Formation of BH2 in enzyme assays was analyzed by HPLC. Inset, SDS-PAGE analysis of recombinant human sepiapterin reductase expressed in E. coli, lane 1, crude extract of E. coli containing sepiapterin reductase induced with 0.5 mm isopropyl β-d-thiogalactopyranoside; lane 2, sample collected from flow-through fraction from the Ni-NTA column; lane 3, sample of imidazole eluted fraction from nickel-affinity column. The arrow indicates the purified enzyme. Panel E, reduction of sepiapterin by purified recombinant sepiapterin reductase was dependent on NADPH, but not NADH. Inset, comparison of NADH and NADPH oxidation by sepiapterin reductase. Enzyme activity was analyzed by changes in absorbance of sepiapterin at 420 nm. Panel F, purified recombinant sepiapterin reductase mediates redox cycling of diquat.
FIGURE 3.
Assays for sepiapterin reduction and redox cycling activity of sepiapterin reductase. Panel A, changes in the absorbance spectra of sepiapterin in the sepiapterin reductase assay. Note the decrease in absorption of sepiapterin over time. Standard reaction mixes in a total volume of 0.2 ml contained 50 mm phosphate buffer, pH 7.8, 200 μm NADPH, 50 μm sepiapterin, and 1 μg/ml sepiapterin reductase and were analyzed after 0 min (curve a), 1 min (curve b), 2 min (curve c), and 3 min (curve d). Controls contained the standard reaction mix without the enzyme and sepiapterin (curve e), with the standard reaction mix buffer plus 250 μm menadione (curve f), or with the standard reaction mix buffer plus 50 μm BH2 (curve g). Panel B, comparison of sepiapterin consumption and BH2 production in sepiapterin reductase enzyme assays. Both sepiapterin reduction reactions were run under the conditions indicated above. Sepiapterin reduction was measured by decreases in absorbance at 420 nm and was presented as micromolar concentrations of the substrate remaining in the assay over time. BH2 was measured using HPLC as described under “Experimental Procedures” and was presented as micromolar concentrations of the product formed in the assay over time. Panel C, effects of DTPA on H2O2 formation by sepiapterin reductase. Redox cycling was run in standard reaction mixes without sepiapterin and supplemented with 5 μm 9,10-phenanthrenequinone without (open triangles) and with 250 μm DTPA (closed triangles). In control experiments, sepiapterin reductase was left out of reaction mixes without (open circles) and with DTPA (closed circles). Panel D, comparison of H2O2 production by sepiapterin reductase redox cycling and autooxidation of BH2 and/or BH4. H2O2 production in the redox cycling reaction was compared with autooxidation of 50 μm BH2, 50 μm BH4, or the combination of BH2 and BH4. Autooxidation reactions were run in the buffer of the standard reaction mix. Inset, enlarged scale for autooxidation reactions run over 10 min. Note that the redox cycling reaction generated much greater amounts of H2O2 when compared with the autooxidation reactions.
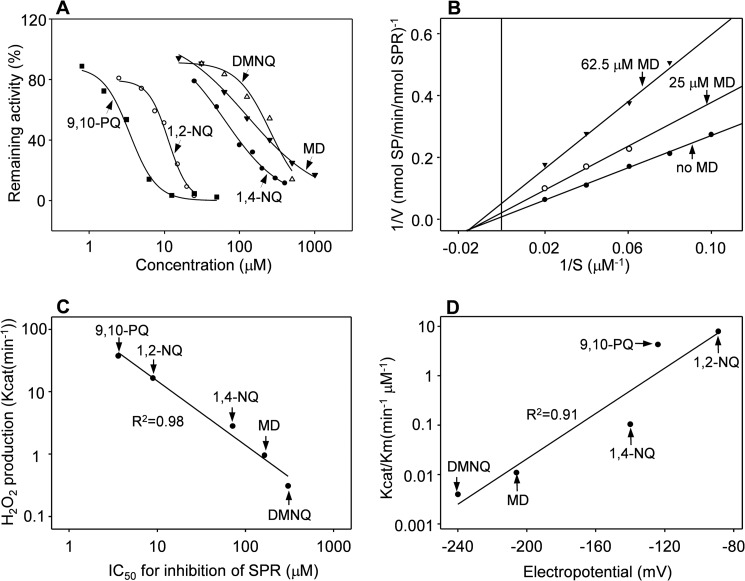
In further studies, we characterized sepiapterin reductase-mediated redox cycling using human recombinant enzyme. Recombinant sepiapterin reductase was found to readily mediate redox cycling of diquat (Fig. 2, panel F), as well as a number of quinones known to redox cycle, including 9,10-phenanthrenequinone, menadione, 2,3-dimethoxy-1,4-naphthoquinone, 1,2-naphthoquinone, and 1,4-naphthoquinone (Fig. 4, panel A, and Table 2), as measured by the formation of H2O2 in enzyme assays. The reaction of each of these redox cycling chemicals was time-, concentration-, and NADPH-dependent (Fig. 4, panel B, and data not shown). The most active redox cycling quinone based on kcat/Km was 1,2-naphthoquinone followed by 9,10-phenanthrenequinone, 1,4-naphthoquinone, menadione, and 2,3-dimethoxy-1,4-naphthoquinone (Table 2), which is generally consistent with previous reports on the redox potentials of these compounds (32–34). In contrast, two related quinones, p-benzoquinone (1,4-benzoquinone) and phenylquinone (2-phenyl-1,4-benzoquinone), did not redox cycle with sepiapterin reductase (Fig. 4, panel C, inset).
FIGURE 4.
Characterization of redox cycling by human recombinant sepiapterin reductase. Panel A, redox cycling of different quinones by sepiapterin reductase. H2O2 formation was measured in the absence or presence of 5 μm 9,10-phenanthrenequinone, 500 μm menadione, or 500 μm dimethoxy-1,4-naphthoquinone. Data are the average of three independent measurements. SPR was added at the indicated time. Panel B, effects of increasing concentrations of NADPH on menadione redox cycling; inset, effects of increasing concentrations of menadione on redox cycling activity. Panel C, inhibition of menadione redox cycling by 100 μm benzoquinone or phenylquinone; inset, inability of benzoquinone and phenylquinone to redox cycle with sepiapterin reductase. Panel D, effects of benzoquinone and phenylquinone on sepiapterin reductase activity. Enzyme activity was analyzed by changes in absorbance of sepiapterin at 420 nm.
TABLE 2.
Redox cycling activity of quinones by sepiapterin reductase
The mouse enzyme also mediated redox cycling, although less efficiently that the human enzyme (Km = 8.8 μm, kcat = 37.7 min−1, kcat/Km = 4.3 min−1 μm−1 for the human enzyme, and Km = 6.3 μm, kcat = 13.3 min−1, kcat/Km = 2.1 min−1 μm−1, for the mouse enzyme, using 9,10-phenanthrenequinone as the redox cycling chemical).
Because the buffers and recombinant enzymes used in our studies contain trace redox metals that could contribute to the formation of H2O2 in enzyme assays, we analyzed redox cycling in enzyme assays containing diethylenetriaminepentaacetic acid (DTPA). We found that DTPA had no effect on the rate of H2O2 formation, indicating that there was no or only a minimal contribution of trace metals to the sepiapterin reductase redox cycling reaction (Fig. 3, panel C).
It is also possible that oxidation of BH2 or BH4 contributes to the formation of H2O2 by sepiapterin reductase. Fig. 3 (panel D) compares H2O2 formation generated by chemical redox cycling and via oxidation of the biopterin cofactors. Under our assay conditions, BH4 was found to generate H2O2, whereas only minimal amounts were generated from BH2 (Fig. 3, panel D, inset). Much greater amounts of H2O2 were generated by the chemical redox cycling reaction (Fig. 3, panel D). These data indicate that oxidation of BH2 and BH4 do not contribute significantly to H2O2 formed in the chemical redox cycling assays.
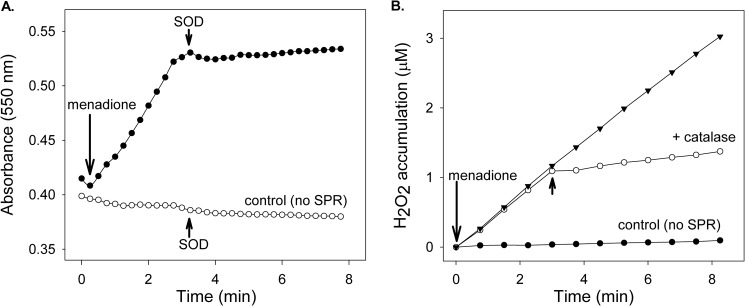
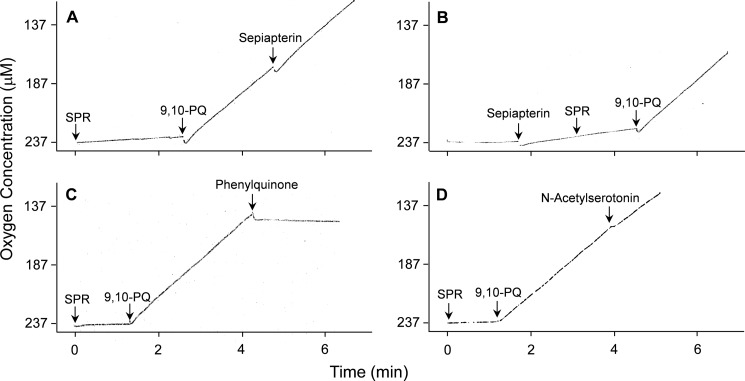
During redox cycling, one electron reduction of a redox-active chemical generates radical ions. Under aerobic conditions, these radicals are rapidly oxidized back to the parent compounds generating superoxide anion. Spontaneous and enzyme-supported dismutation of superoxide anion generates H2O2. Using menadione, we found that redox cycling by recombinant human sepiapterin reductase readily generated superoxide anion, as measured by the reduction of acetylated cytochrome c; the accumulation of this ROS was inhibited by superoxide dismutase (Fig. 5, panel A). The formation of superoxide anion in reaction mixes during redox cycling was associated with an accumulation of H2O2; this was inhibited by catalase (Fig. 5, panel B). In the presence of iron, redox cycling by sepiapterin reductase also generated hydroxyl radicals (data not shown). The accumulation of these ROS was inhibited by DMSO, a scavenger of hydroxyl radicals. The addition of 9,10-phenanthrenequinone, but not sepiapterin, to reaction mixes readily increased oxygen utilization (Fig. 6, panels A and B). This is consistent with the fact that sepiapterin reduction by sepiapterin reductase is not an oxygen-requiring reaction.
FIGURE 5.
Ability of human recombinant sepiapterin reductase to generate ROS. Reaction mixes contained sepiapterin reductase, 200 μm NADPH, and 500 μm menadione. Panel A, menadione stimulated superoxide anion production by sepiapterin reductase. Menadione was added to reaction mixes to stimulate superoxide anion formation as indicated by the arrow. Superoxide anion was measured spectrophotometrically by monitoring superoxide dismutase inhibitable changes in absorbance of acetylated cytochrome c at 550 nm. Reactions were run in the absence and presence of SPR. SOD (40 units), which dismutates superoxide anion, was added to the reactions as indicated by the arrowheads. Panel B, menadione stimulated H2O2 production by sepiapterin reductase. Menadione was added to reaction mixes to stimulate H2O2 production. Catalase (2000 units), which breaks down H2O2, was added to the reactions as indicated by the arrowheads.
FIGURE 6.
Oxygen consumption by human recombinant sepiapterin reductase during chemical redox cycling. A Clark-type oxygen electrode was used to quantify oxygen consumption during redox cycling. The reaction was run in the presence of 200 μm NADPH and an NADPH-regenerating system. After establishing a stable base line, SPR or sepiapterin was added. Panels A and B, 5 μm 9,10-phenanthrenequinone (9,10-PQ), but not sepiapterin (200 μm), initiated oxygen consumption by sepiapterin reductase. Panels C and D, 200 μm phenylquinone, but not 200 μm N-acetylserotonin, inhibited 9,10-phenanthrenequinone-induced oxygen consumption.
As indicated above, the reduction of sepiapterin by sepiapterin reductase required NADPH, but not NADH. Unexpectedly, we found that redox cycling by human recombinant sepiapterin reductase also utilized NADH. The Km and kcat values for NADPH in the reaction was 30.2 μm and 0.74 min−1, respectively, and for NADH was 110 μm and 0.2 min−1, respectively. Thus, NADPH was significantly more efficient in supporting the reaction (kcat/Km = 0.025 min−1 μm−1 for NADPH versus 0.002 min−1 μm−1 for NADH). In these studies, redox cycling was measured by quantifying the formation of H2O2 in enzyme assays containing 0.5 mm menadione and increasing concentrations of NADPH or NADH.
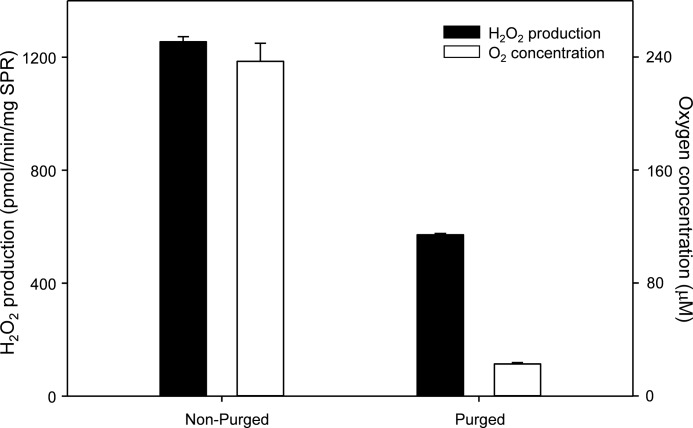
The measured ratio of NADPH utilization to oxygen consumption to H2O2 formation in our assays was found to be 2.0:2.6:1.2 (Table 3). H2O2 formation was significantly different from NADPH utilization and oxygen consumption. These data are close to the theoretical stoichiometry of 1:1:0.5 or 2:2:1 for the redox cycling reaction. It is important to note that redox cycling in our assays was measured under normoxic conditions (∼240 μm oxygen in solution in vitro). Levels of oxygen are often 10 times lower in vivo, which lowers ROS formation during redox cycling. Using our in vitro assay, we found that decreasing oxygen tension 10-fold resulted in an ∼50% decrease in H2O2 formation during redox cycling (Fig. 7). These data indicate that oxygen is in excess in the redox cycling reaction and that high levels of ROS can be formed even at reduced tissue oxygen concentrations. It is important to point out, however, that the one electron reduction of redox-active chemicals by sepiapterin reductase is not an oxygen-requiring reaction. At low oxygen tension, radical ions generated by this process are stabilized. They are by themselves highly reactive and can contribute to toxicity (35).
TABLE 3.
Stoichiometry of the chemical redox cycling reaction of sepiapterin reductase
| Stoichiometry | NADPH consumeda | Oxygen consumed | H2O2 formed |
|---|---|---|---|
| Enzyme activity (μmol/min/mg protein) | 1.8 ± 0.2 (11.3) | 2.3 ± 0.4 (14.5) | 1.1 ± 0.1 (7.1) |
| Measured ratio | 2.0 ± 0.2 | 2.6 ± 0.3b | 1.2 ± 0.1d |
| Theoretical ratioc | 2 | 2 | 1 |
a Reactions contained 5 μm 9,10-phenanthrenequinone as the redox cycling chemical. The number in parentheses is the μm concentration of NADPH or oxygen consumed or H2O2 formed after the first 3 min of incubation. Each value represents the mean ± S.E. (n = 3 for NADPH and oxygen consumption and 7 for H2O2 formation).
b Not significantly different from NADPH consumption; significantly different (p ≤ 0.05) from H2O2 formation (t test).
c The theoretical stoichiometry is based on the fact that two oxygen molecules (O2) and two electrons from two NADPH molecules (each of which acts as a one-electron donor) are needed to generate one H2O2 molecule.
d Significantly different (p ≤ 0.05) from NADPH and oxygen consumption (t-test).
FIGURE 7.
Effects of oxygen on H2O2 production by 9,10-phenanthrenequinone-induced redox cycling with sepiapterin reductase. Reactions were run in Oxygraph reaction cells in a total volume of 0.6 ml and contained 50 mm phosphate buffer, pH 7.8, 5 μm 9,10-phenanthrenequinone, and 200 μm NADPH. To generate reduced oxygen levels, the reaction cell was purged with 100% nitrogen gas, and levels of oxygen were monitored with the Oxygraph. To initiate the redox cycling reactions, 0.3 μg of sepiapterin reductase was injected into the reaction cells. Samples were removed at different time points; the reactions were stopped by the addition of acetonitrile at a final concentration of 30%, and the H2O2 concentration was determined by the Amplex red assay.
Effects of Inhibitors on the Reduction of Sepiapterin and Redox Cycling by Sepiapterin Reductase
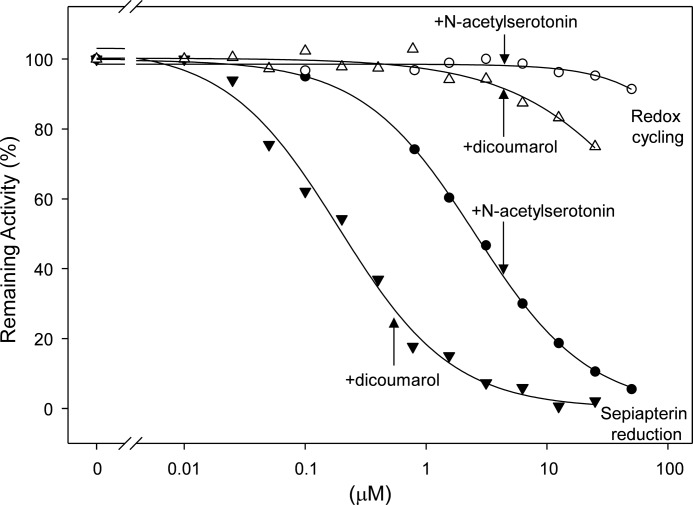
A number of aldo-keto reductase inhibitors have been reported to block the activity of purified rat sepiapterin reductase, including dicoumarol, indomethacin, ethacrynic acid, and the flavonoid glycoside rutin, as well as the indoleamine, N-acetylserotonin (36, 37). Each of these compounds was found to inhibit sepiapterin reduction mediated by recombinant human sepiapterin reductase. The most potent inhibitor was dicoumarol followed by N-acetylserotonin, indomethacin, ethacrynic acid, and rutin (Fig. 8 and Table 4). Whereas dicoumarol, ethacrynic acid, and rutin were noncompetitive inhibitors of the enzyme, N-acetylserotonin and indomethacin were competitive inhibitors (Ki values = 0.9 and 2.7 μm, respectively, see Table 4). In contrast, none of these inhibitors affected sepiapterin reductase-mediated redox cycling (Table 4 and Fig. 6, panel D). These data suggest that the reduction of sepiapterin and chemical redox cycling occur by distinct mechanisms. This is supported by our findings that sepiapterin, at concentrations >1 mm, also failed to inhibit redox cycling mediated by sepiapterin reductase with any of the redox-active quinones (data not shown). Diphenyleneiodonium, an inhibitor of flavin-containing oxidoreductases, which has been reported to inhibit redox cycling by NADPH-cytochrome P450 reductase and thioredoxin reductase (9, 20), was also unable to inhibit either the reduction of sepiapterin or redox cycling by sepiapterin reductase (Table 4 and data not shown), a finding consistent with the fact that the enzyme does not require flavin cofactors for activity.
FIGURE 8.
Effects of N-acetylserotonin and dicoumarol on sepiapterin reduction and redox cycling by human recombinant sepiapterin reductase. Sepiapterin reductase activity was measured by decreases in sepiapterin absorbance at 420 nm. Redox cycling was measured by the formation of superoxide anion in enzyme assays in the presence of 100 μm menadione. Note that both N-acetylserotonin and dicoumarol inhibit sepiapterin reduction (IC50 = 2.6 and 0.2 μm, respectively) but not redox cycling activity. Sepiapterin reduction was analyzed by changes in absorbance of sepiapterin at 420 nm.
TABLE 4.
Ability of various compounds to inhibit sepiapterin reduction and redox cycling by sepiapterin reductase
| Inhibitor | Sepiapterin reductiona |
Redox cycling |
|||
|---|---|---|---|---|---|
| Inhibition type | IC50 | Ki | Inhibition type | IC50 | |
| μm | μm | μm | |||
| Non-redox cycling agents | |||||
| Dicoumarol | Noncompetitive | 0.2 | NIb | ||
| N-Acetylserotonin | Competitive | 2.6 | 0.9 | NI | |
| Indomethacin | Competitive | 8.1 | 2.7 | NI | |
| Ethacrynic acid | Noncompetitive | 22.9 | NI | ||
| Rutin | Noncompetitive | 24.0 | NI | ||
| Diphenyleneiodonium | NIc | NIc | |||
| Benzoquinone | Competitive | 2.8 | 0.9 | Noncompetitive | 3.4 |
| Phenylquinone | Competitive | 1.6 | 0.5 | Noncompetitive | 3.2 |
| Redox cycling agents | |||||
| 9,10-Phenanthrenequinone | Noncompetitive | 3.6 | |||
| 1,2-Naphthoquinone | Noncompetitive | 8.9 | |||
| 1,4-Naphthoquinone | Noncompetitive | 71.4 | |||
| Menadione (2-methyl-1,4-naphthoquinone) | Noncompetitive | 164.3 | |||
| 2,3-Dimethoxyl-1,4-naphthoquinone | Noncompetitive | 304.6 | |||
a Sepiapterin reduction was measured by decreases on absorbance of sepiapterin at 420 nm as described under “Experimental Procedures.”
b NI means not inhibitory at concentrations up to 100-fold higher than the IC50 for sepiapterin reduction inhibition.
c NI means not inhibitory at concentrations up to 1 mm.
Effects of Redox Cycling Compounds on Sepiapterin Reductase Activity
Each of the quinones that redox cycles with sepiapterin reductase was found to inhibit the ability of the enzyme to reduce sepiapterin (Fig. 9, panel A, and Table 4). 9,10-Phenanthrenequinone was the most potent inhibitor followed by 1,2-naphthoquinone, 1,4-naphthoquinone, menadione, and 2,3-dimethoxy-1,4-naphthoquinone. The relative inhibitory effect of these quinones was proportional to their kcat values (Table 2 and Fig. 9, panel C). All of the inhibitors were noncompetitive with respect to sepiapterin (Fig. 9, panel B, and Table 4). Interestingly, p-benzoquinone and phenylquinone, which did not redox cycle with sepiapterin reductase (Fig. 4, panel C, inset), inhibited both the ability of sepiapterin reductase to reduce sepiapterin and to mediate redox cycling (Fig. 4, panels C and D, and Table 4), as measured by the production of ROS and oxygen utilization (Fig. 6, panel C, and data not shown). p-Benzoquinone and phenylquinone were competitive inhibitors of sepiapterin reductase-mediated reduction of sepiapterin but noncompetitive inhibitors of redox cycling (Table 4). These data indicate that redox cycling is not required for quinones to inhibit sepiapterin reduction and further support the idea that mechanisms underlying sepiapterin reduction and redox cycling by sepiapterin reductase are distinct.
FIGURE 9.
Effects of quinones on human recombinant sepiapterin reductase activity. Panel A, effects of quinones (9,10-phenanthrenequinone (9,10-PQ), 1,2-naphthoquinone (1,2-NQ), 1,4-naphthoquinone (1,4-NQ), menadione (MD), and dimethoxy-1,4-naphthoquinone (DMNQ)) on sepiapterin (SP) reduction by human recombinant sepiapterin reductase. Panel B, Lineweaver-Burk analysis of menadione inhibition of sepiapterin reduction. Note that menadione is a noncompetitive inhibitor. Panels A and B, sepiapterin reduction was measured by changes in absorbance of sepiapterin at 420 nm. Panel C, correlation between the kcat for redox cycling, as measured by H2O2 production, and the ability to inhibit reduction of sepiapterin. Data are presented as the IC50 value for inhibition of sepiapterin reduction. Panel D, correlation between kcat/Km for redox cycling by sepiapterin reductase and redox potential for various quinones.
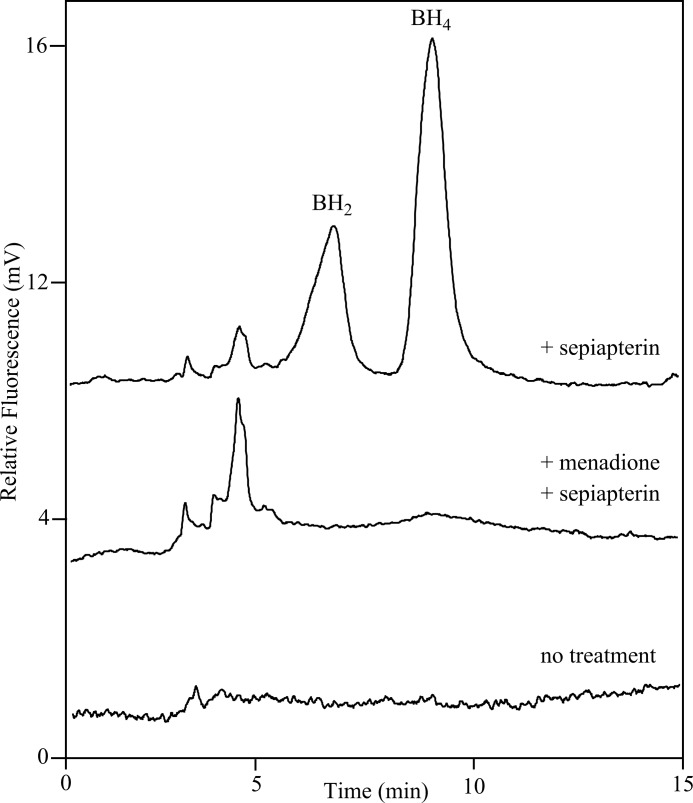
Because redox cycling inhibited reduction of sepiapterin by purified recombinant sepiapterin reductase, we next determined whether sepiapterin reductase could be inhibited in vivo by redox cycling chemicals. Fig. 10 shows that in A549 human lung epithelial cells, treatment with sepiapterin readily generates BH2 and BH4, a finding that confirms that these cells express sepiapterin reductase. Menadione completely prevented the formation of BH2 and BH4. These data demonstrate that menadione redox cycling in vivo can suppress sepiapterin reductase activity.
FIGURE 10.
Inhibition of dihydrobiopterin and tetrahydrobiopterin formation by menadione in A549 lung epithelial cells. Cells were pretreated with control medium or medium containing 500 μm menadione. After 2 h, sepiapterin (100 μm final concentration) was added to the cultures. After an additional 2 h, cells were extracted and analyzed for BH2 and BH4 content by HPLC. The upper and middle tracings were from cells treated with sepiapterin alone or sepiapterin and menadione. The lower tracing is from control cells that were not treated with sepiapterin or menadione.
Site-directed Mutagenesis Defines Distinct Sites on Sepiapterin Reductase for the Reduction of Sepiapterin and Redox Cycling
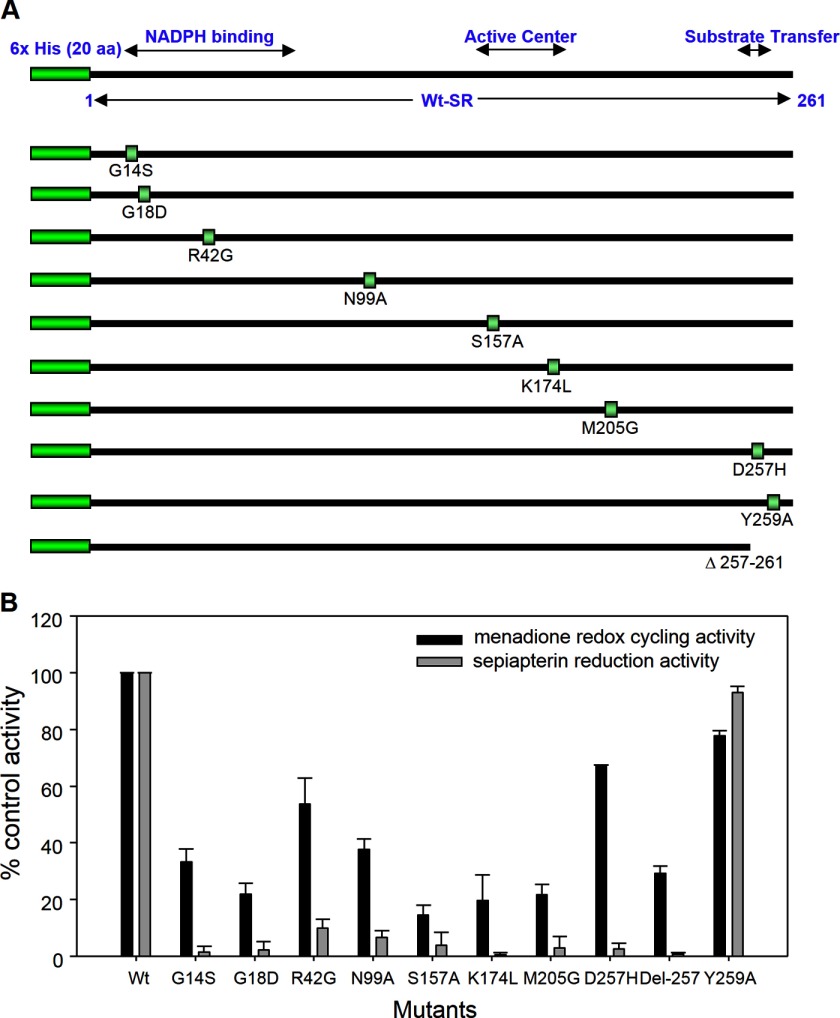
Previous studies have shown that sepiapterin reductase contains an active center conserved region important for NADPH binding and a substrate transfer site (38–41). In further studies we used site-directed mutagenesis to investigate the role of the different functional domains of the enzyme in its sepiapterin reduction and redox cycling activities. In the human enzyme, the catalytic center has been defined by a conserved Ser-157–Tyr-170–Lys-174 triad (38). We found that mutations in Ser-157 or Lys-174 in this region reduced both sepiapterin reduction and redox cycling activity by 80–95% (Fig. 11). The catalytic efficiencies (kcat/Km) for sepiapterin reduction of S157A and K174L decreased to 1.8 and 0.8% of wild type sepiapterin reductase, respectively, and for redox cycling to 6.8 and 1.4%, respectively (Table 5). Similarly, mutations in Gly-14 and Gly-18 in the NADPH-binding motif of sepiapterin reductase resulted in almost complete loss of the ability to reduce sepiapterin and a 65–75% decrease in redox cycling (Fig. 11, panel B). For both of these mutations, the catalytic efficiencies for redox cycling decreased to 0.2% of wild type sepiapterin reductase (Table 5). Mutation in Asp-42, which is thought to be important in the selectivity of the enzyme for NADPH (42), led to a 90% reduction in sepiapterin reduction activity and a 50% reduction in redox cycling activity (Fig. 11, panel B). The catalytic efficiencies for this mutant decreased to 2 and 7% of wild type sepiapterin reductase for sepiapterin reduction and redox cycling, respectively (Table 5). These data confirm the importance of the catalytic center and the NADPH binding regions in mediating the activities of sepiapterin reductase. Residues Asn-99 and Met-205 are conserved amino acids in human, mouse, and rat sepiapterin reductases and may play important roles in maintaining the structure of the enzyme (40), and mutations in these amino acids also caused marked reductions in the activities of both sepiapterin reduction and redox cycling (Fig. 11, panel B). The catalytic efficiency of N99A and M205G for sepiapterin reduction decreased to ∼1 and 5%, respectively, and for redox cycling, 5 and 25%, respectively, when compared with the wild type enzyme (Table 5).
FIGURE 11.
Site-directed mutagenesis of human recombinant sepiapterin reductase. Panel A, schematic drawing of recombinant sepiapterin reductase and mutant enzymes. The human recombinant sepiapterin reductase used in this investigation contains a hexahistidine tag at the N terminus and the 261 original full-length amino acids. The NADPH-binding motif, active center, and substrate transfer motif are indicated. The mutated positions are labeled separately. Panel B, sepiapterin reduction and redox cycling activity of sepiapterin reductase and mutant enzymes were assayed as described under “Experimental Procedures.” Data are presented as percentage of the wild type control enzyme activity. Sepiapterin reduction was assayed in the presence of 50 μm sepiapterin and measured by changes in absorbance of sepiapterin at 420 nm. Redox cycling was assayed in the presence of 500 μm menadione.
TABLE 5.
Kinetic parameters of wild type and mutant sepiapterin reductases
| SPR |
Km |
kcat |
kcat/Km |
|||
|---|---|---|---|---|---|---|
| SPRa | Redoxb | SPR | Redox | SPR | Redox | |
| μm | min−1 | min−1 μm−1 | ||||
| Wt-hSPR | 25.4 | 8.8 | 97.0 | 37.67 | 3.82 | 4.28 |
| G14S | NDc | 50.1 | ND | 0.34 | ND | 0.01 |
| G18D | ND | 464.1 | ND | 2.90 | ND | 0.01 |
| R42G | 45.7 | 50.1 | 3.2 | 5.90 | 0.07 | 0.32 |
| N99A | 94.1 | 126.0 | 3.3 | 17.40 | 0.04 | 0.14 |
| S157A | 50.3 | 3.2 | 3.5 | 0.92 | 0.07 | 0.29 |
| K174L | 169.8 | 27.7 | 5.4 | 1.79 | 0.03 | 0.06 |
| M205G | 133.8 | 13.4 | 25.7 | 13.45 | 0.19 | 1.08 |
| D257H | 60.6 | 1.9 | 9.4 | 6.44 | 0.16 | 3.39 |
| Δ257–261 | ND | 116.0 | ND | 0.35 | ND | <0.01 |
| Y259A | 25.5 | 19.6 | 36.2 | 48.30 | 1.42 | 2.46 |
a Sepiapterin reduction activity was assayed by the formation of BH2 from sepiapterin by HPLC as described under “Experimental Procedures.”
b Redox cycling was assayed using 9,10-phenanthrenequinone.
c ND means not determined.
The C-terminal region of sepiapterin reductase has been reported to be critical for binding of sepiapterin to the enzyme (40). Deletion of the C-terminal five amino acids almost completely eliminated enzyme activity (Fig. 11, panel B). For redox cycling, the catalytic efficacy decreased to less than 1% of the wild type enzyme (Table 5). Interestingly, mutation of Asp-257 to histidine eliminated sepiapterin reduction activity but had minimal effects on redox cycling activity (Fig. 11, panel B). Thus, the catalytic efficiency of D257H for sepiapterin reduction and redox cycling decreased to ∼4 and 80% of wild type enzyme activity, respectively (Table 5). Phosphorylation has also been reported to regulate sepiapterin reductase activity (43). Mutation of Tyr-259, a unique potential phosphorylation site in the C-terminal substrate transfer motif, had no major effects on sepiapterin reduction and redox cycling activity (Fig. 11, panel B, and Table 5).
DISCUSSION
This study demonstrates that sepiapterin reductase is a mediator of chemical redox cycling in lung epithelial cells. Active substrates include bipyridinium herbicides, as well as various redox-active quinones. The one electron reduction of these substrates by sepiapterin reductase readily generates ROS, including superoxide anion and H2O2, at the expense of a reduced nicotinamide adenine dinucleotide cofactor. The redox cycling reaction consumes oxygen and utilizes either NADPH or NADH; NADPH is significantly more efficient than NADH in supplying electrons to the reaction. This is in contrast to enzyme-mediated reduction of sepiapterin, an obligate NADPH-dependent reaction. The phosphate group in the adenine nucleotide is presumably important in cofactor recognition by the enzyme during sepiapterin reduction; in contrast, the redox cycling reaction also allows electrons from NADH to mediate the one electron reduction of redox-active substrates, possibly due to less stringent requirements for binding of the cofactor to the enzyme (44). Our findings that NADH can mediate redox cycling are in accord with reports that other enzymes catalyzing this process, including cytochrome b5 reductase and ubiquinone oxidoreductase, preferentially utilize NADH (44, 45).
Sepiapterin reductase is widely distributed in tissues, including the lung, kidneys, and brain (25). Thus, chemical redox cycling by sepiapterin reductase and consequent ROS generation may contribute to tissue injury in a manner dependent on localized concentrations of the enzyme, reduced pyridine nucleotide cofactors, oxygen, and redox-active chemicals. It should be noted that chemical redox cycling by sepiapterin reductase is flavin cofactor-independent. For flavin-containing enzymes such as cytochrome P450 reductase, cytochrome b5 reductase, xanthine oxidase, and various forms of nitric-oxide synthase, the flavin cofactor is thought to participate in chemical redox cycling because of its ability to accept single electrons from the pyridine nucleotide cofactor, which are presumably used for the one electron reduction of the redox-active chemicals. This reaction is blocked by the flavin inhibitor diphenyleneiodonium (14). The fact that sepiapterin reductase is not a flavin-containing enzyme and is not blocked by diphenyleneiodonium suggests that there are multiple mechanisms by which enzymes can mediate chemical redox cycling. Recent studies have shown that aldo-keto reductases also mediate chemical redox cycling in the absence of a flavin cofactor (46, 47). At the present time, the precise mechanism mediating flavin-independent redox cycling is not known. It is possible that binding of redox-active chemicals in an appropriate orientation in proximity to the pyridine nucleotide cofactor is sufficient to allow single electron transfers. Further studies are needed to explore this possibility.
Redox-active quinones were found to vary in their activity with respect to redox cycling with sepiapterin reductase with 1,2-naphthoquinone displaying the greatest activity, followed by 9,10-phenanthrenequinone, 1,4-naphthoquinone, menadione, and 2,3-dimethoxy-1,4-naphthoquinone. The catalytic activity of these quinones was tightly correlated (R2 = 0.91) with their redox potentials. Of note is our finding that each of the redox cycling agents was an effective noncompetitive inhibitor of sepiapterin reduction. These data indicate that there are distinct mechanisms underlying sepiapterin reduction and redox cycling. The efficient inhibition of sepiapterin reduction by the quinones is likely due to their ability to divert electron flux from sepiapterin; this is supported by the observation that the IC50 values of the quinones for inhibition of sepiapterin reductase were correlated with their Km values for NADPH oxidation. A similar mechanism for quinone inhibition of disulfide substrate reduction has been suggested for thioredoxin reductase (48). Earlier studies have identified a number of substrates of purified rat erythrocyte sepiapterin reductase, including dicarbonyl compounds, quinones, aldehydes, and ketones in assays measuring the NAD(P)H oxidase activity of the enzyme (36). Although this activity was ascribed to the “carbonyl reductase” or “aldo-keto reductase” activity of sepiapterin reductase, one cannot exclude the possibility that these enzyme activities were due, at least in part, to redox cycling. Additional studies are required to identify specific enzyme products and reaction mechanisms that could distinguish non-sepiapterin reduction and non-redox cycling activities of sepiapterin reductase with different chemical substrates.
Earlier work has identified inhibitors of sepiapterin reductase largely based on structural similarities with natural substrates for the enzyme (i.e. sepiapterin or pyruvoyltetrahydropterin), in particular, N-acetylserotonin, N-acetyldopamine, and N-acetyl-m-tyramine (37). These compounds were all competitive inhibitors of the enzyme substrates. Similarly, using human recombinant enzyme, we found that N-acetylserotonin was a competitive inhibitor of sepiapterin utilization. These data are consistent with crystallographic studies showing similarities in the structures of substrates and inhibitors bound to sepiapterin reductase (40). Sueoka and Katoh (36) showed that aldo-keto reductase inhibitors, including indomethacin, ethacrynic acid, rutin, and dicoumarol, were inhibitors of rat erythrocyte sepiapterin reductase. Our findings that these compounds are potent inhibitors of human recombinant sepiapterin reductase are in accord with this report. Notably, these compounds blocked sepiapterin reduction by the enzyme, but not redox cycling, further supporting the idea that the two catalytic enzyme activities are distinct. Indomethacin, like N-acetylserotonin, was found to be a competitive inhibitor, whereas dicoumarol, ethacrynic acid, and rutin were noncompetitive inhibitors; these data indicate that the various inhibitors of sepiapterin reduction also function by distinct mechanisms. In general, inhibitors of sepiapterin reductase are structurally diverse; further studies are needed to better define the mechanisms by which they selectively block substrate reduction by sepiapterin reductase.
Of interest was our finding that benzoquinone and phenylquinone, two non-redox cycling quinones, inhibited the reduction of sepiapterin, as well as quinone redox cycling by sepiapterin reductase. Although both compounds were competitive inhibitors of sepiapterin reduction, they were noncompetitive inhibitors of redox cycling, providing additional evidence that the active sites on the enzyme for sepiapterin reduction and redox cycling are unique. At present, it is not known if the inhibitory activities of the quinones are due to binding to different or overlapping sites on sepiapterin reductase. Both reactions require the adenine nucleotide co-factor suggesting that their active sites on the enzyme are in close proximity. The fact that benzoquinone and phenylquinone inhibit sepiapterin reduction without redox cycling indicates that the production of superoxide anion is not required for inhibition of sepiapterin reduction.
Our data demonstrate that A549 lung epithelial cells contain sepiapterin reductase and that the enzyme is active and readily generates BH2 and BH4 from sepiapterin. Of note was our observation that menadione, a highly effective redox cycling chemical, inhibited this process. Thus, chemical redox cycling can suppress the formation of BH4 in intact cells. These data suggest that chemical redox cycling can inhibit sepiapterin reductase in vivo.
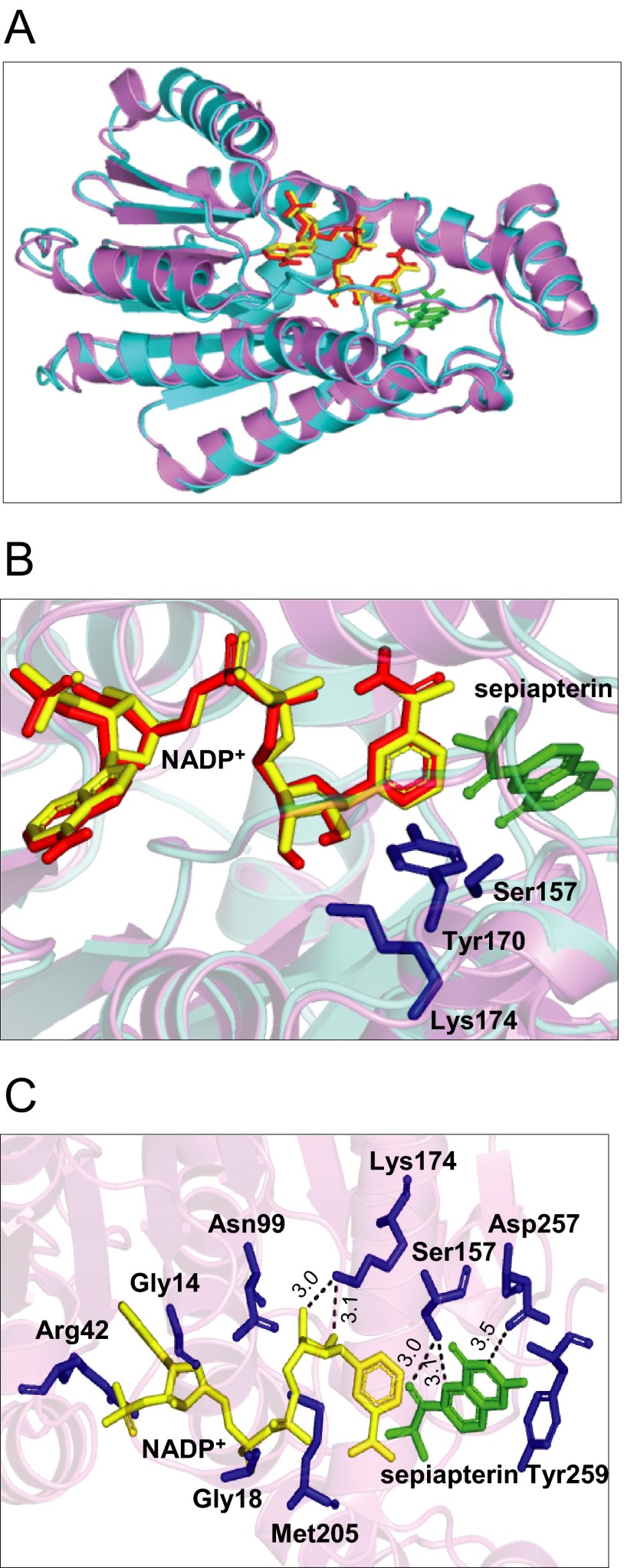
Based on sequence alignment showing a conserved N-terminal motif (15GXXXGXG21 in the mouse enzyme and 14GXXXGXG20 in the human enzyme) for NADPH binding, referred to as a “Rossmann-fold,” and an active site motif (YXXXK), sepiapterin reductase has been classified as a member of the short chain-dehydrogenase reductase family (38, 49). This has been confirmed by crystallographic studies with the mouse enzyme in which sepiapterin reductase was identified as a homodimeric protein with each monomer containing an NADPH and a sepiapterin-binding site in close proximity (see Fig. 12 for structure). These latter studies showed that Arg-43 (Arg-42 in the human enzyme) functions as a critical anchoring site for the adenine moiety in NADPH (40, 41). This amino acid is thought to exist only in short chain-dehydrogenase reductase family members that preferentially utilize NADPH (42). In contrast, Met-206 in the mouse enzyme (Met-205 in the human enzyme) functions to maintain a critical hydrophobic pocket in proximity to the nicotinamide moiety of NADPH (40). To further understand the role of these amino acids in sepiapterin reductase functioning, we performed site-directed mutational analysis. Initially, we analyzed mutations in the human enzyme NADPH binding domain, including G14S, G18D, R42G, and M205G (see Figs. 11 and 12 for summaries). Mutations in glycine add more polar structure to the enzyme at positions where adenine binds in the NADPH binding pocket and can decrease interactions of the protein with adenine. A similar change in adenine binding can result from the arginine mutation, which prevents hydrogen bonding with adenine, adding a less polar, more neutral amino acid to the NADPH binding pocket. In contrast, the methionine mutation significantly decreases hydrophobic interactions with nicotinamide, which can disrupt electron transfer to sepiapterin. We found that these point mutations caused major losses (90–98%) in the ability of sepiapterin reductase to reduce sepiapterin confirming that in the wild type enzyme these amino acids bind tightly to NADPH, generating a unique structural conformation that is essential for substrate reduction. These data are also consistent with the requirement for the adenine nucleotide for optimal enzyme activity (25). Interestingly, NADPH-binding site mutant sepiapterin reductase enzymes still retained 25–55% of their redox cycling activity. Although each of the point mutations appear to alter the alignment of the nicotinamide with respect to sepiapterin, a process that limits electron transfer to the substrate, NADPH appears to remain in close enough proximity to the redox cycling chemicals allowing for partial activity. These data are in accord with our inhibitor studies and indicate that redox cycling occurs at a site on the enzyme distinct from sepiapterin reduction. Presumably, NADPH remains bound to the Rossmann-fold containing pocket in the mutant enzymes. Thus, as long as redox cycling chemicals are in proximity to the nicotinamide moiety, redox cycling reactions occur, although with reduced efficiency. However, further studies are needed to rule out the possibility of altered binding of redox cycling chemicals to the mutant enzymes, which may also reduce enzyme activity.
FIGURE 12.
Molecular models of sepiapterin reductase complexed with NADP+ and sepiapterin. Panel A, superposition of human and mouse sepiapterin reductase. Human sepiapterin reductase is shown in violet (Protein Data Bank code 1Z6Z) and mouse sepiapterin reductase in cyan (Protein Data Bank code 1SEP). The NADP cofactor and sepiapterin substrate complexed with mouse sepiapterin reductase are shown in red and green, respectively. NADP bound with human sepiapterin reductase is shown in yellow. Panel B, close-up of the active site of the NADP-sepiapterin-sepiapterin reductase complex. Key residues in the active site are represented as blue sticks. Panel C, interactions of sepiapterin and NADP with the active site of human sepiapterin reductase. Protein residues selected for site-directed mutagenesis are shown as blue sticks. Hydrogen bonds are shown by broken lines, and the corresponding distances (Å) are indicated.
Another highly conserved sequence motif in sepiapterin reductase is the catalytic site. In mouse sepiapterin reductase, this site is composed of several hydrophobic amino acids, including Leu-105, Leu-159, Tyr-165, Trp-168, Tyr-171, Met-206, and Cys-160 (40). Tyr-171 is a key active site residue; the phenyl ring hydroxyl group of Tyr-171 is situated in an orientation for proton transfer between the C4′N of NADPH and the C1′-carbonyl function of the substrate. Arg-178, which is outside the active site cavity, is thought to act in concert with Lys-175, to facilitate proton transfer from the hydroxyl function of Tyr-171 to the substrate's carbonyl oxygen, although Ser-158 stabilizes the orientation of the substrate (42, 50, 51). Thus, a critical triad pocket in sepiapterin reductase composed of Ser-158, Tyr-171, and Lys-175 is formed and serves a key function in stabilizing the protein structure, maintaining cofactor/substrate proximity, and proton transfer (40). Three highly conserved asparagine residues (Asn-100, Asn-128, and Asn-155) are also located within hydrogen bonding distance to the guanidinium moiety of Arg-178 and are thought to be important for maintaining this amino acid in an orientation required for functional interactions with Tyr-171 (40). Previous site-directed mutagenesis studies with rat sepiapterin reductase have confirmed that Ser-158, Tyr-171, and Lys-175 play key roles in the function of the enzyme (38, 39). We found that mutations in Ser-157 and Lys-174 in the human enzyme (corresponding to Ser-158 and Lys-175 in the mouse enzyme) resulted in an 80–85% loss in the ability of sepiapterin reductase to mediate reduction of sepiapterin. This is likely due to the fact that hydrogen bonds cannot form between the substituted amino acids and the substrate in the mutated enzymes. Interestingly, low levels (15–20%) of redox cycling activity remained. These data indicate that, despite mutations at critical functional sites, the active site on the enzyme mediating redox cycling is close to the active site triad, allowing for low level activity. As observed with mutants in the NAD(P)H binding pocket, active site triad mutants are able to position redox cycling chemicals close enough to NAD(P)H to permit low levels of redox cycling. Similar decreases in sepiapterin reduction and chemical redox cycling were observed with the Asn-99 mutant (corresponding to Asn-100 in the mouse enzyme). These data further support the idea that this amino acid is critical for maintaining the functional activity of the active site triad.
Analysis of the crystal structure of mouse sepiapterin reductase reveals that the pterin substrate is positioned in the active site anchored with its guanidine moiety to Asp-258 (40). This amino acid, which is located at the C terminus of the enzyme, is thought to play a key role in positioning the pterin substrate side chain C1′-carbonyl group near the hydroxyl group in Tyr-171 and NADPH C4′N in the active site of the enzyme (40). This study shows that mutation in this aspartate residue (D257H) in the human enzyme (corresponding to Asp-258 in the mouse enzyme) caused a loss of sepiapterin reduction activity, with minimal effects on its chemical redox cycling activity. It appears that decreases in sepiapterin reduction are due to a reduced affinity for the substrate resulting in lower catalytic efficiency. However, with respect to redox cycling, the D257H substitution, which changes the amino acid residue from negatively charged to positively charged, caused an increase in Km and a decrease in kcat for the reaction. Thus, although the net result did not alter apparent redox cycling activity, this substitution changed the kinetics of the reaction, possibly due to conformational changes in the active site of the enzyme. These results indicate that Asp-257 is important in generating BH2; however, substitutions do allow redox cycling. These data also support the idea that sepiapterin reduction and redox cycling by sepiapterin reductase occur by distinct mechanisms.
Deletion of the last five amino acids in the C terminus (Δ257–261) of the enzyme abolished sepiapterin reductase activity and decreased redox cycling by ∼70%. These findings confirm a role for Asp-257 in sepiapterin reduction and suggest that the C terminus of the enzyme is required for optimal chemical redox cycling.
Katoh et al. (43) showed that serine/threonine phosphorylation of rat sepiapterin reductase by Ca2+/calmodulin-dependent protein kinase II or protein kinase C modified the kinetic properties of the enzyme. The C terminus of human sepiapterin reductase contains a single tyrosine residue (Tyr-259) in proximity to the active site that has the potential to control enzyme activity. We found that mutation of this amino acid (Y259A) had no major effects on sepiapterin reduction or on chemical redox cycling indicating that this tyrosine residue is not required for enzyme activity. It remains to be determined whether phosphorylation of this or other tyrosine residues and/or serine or threonine residues in sepiapterin reductase is important in controlling its enzymatic activities.
BH4 is a key cofactor for a number of aromatic amino acid hydroxylases important in synthesizing catecholamines, neurotransmitters, and indoleamines, as well as for nitric-oxide synthases. Deficiencies in BH4 result in movement disorders such as dystonia and Parkinson disease, Alzheimer disease, and atypical phenylketonuria (53–55). Specific mutations in sepiapterin reductase, which compromise its ability to generate BH4, are associated with these diseases. For example, the mutation K251X, which causes the deletion of C-terminal amino acids, including the critical Asp-257, results in delayed psychomotor development and a complex movement disorder (56, 57). Early onset Parkinson disease and other neurological deficits have also been described in patients with point mutations in sepiapterin reductase (e.g. R150G), deletion mutations (e.g. Q119X), as well as splicing mutations (IVS2–2A>G) (57, 58). That sepiapterin reductase is crucial for neuronal functioning has been highlighted recently by findings that sepiapterin reductase knock-out mice have reduced levels of neurotransmitters and that this is associated with a variety of movement disorders (59, 60).
Our data showing that redox cycling chemicals can inhibit sepiapterin reductase suggest a mechanism leading to deficiencies in BH4 and consequent disease pathologies. Sepiapterin reductase transfers electrons from NADPH to sepiapterin. Both endogenous and exogenous redox cyclers preferentially utilize these electrons, at the expense of their supply to sepiapterin, a process that effectively blocks the formation of BH4. In many tissues, including the brain and lung, this process could compromise BH4-containing enzymes and lead to neurological deficits and/or aberrant pulmonary functioning. In this regard, epidemiological studies have demonstrated that exposure to paraquat, which we have shown redox cycles with sepiapterin reductase, leads to increased risk of Parkinson disease (61). Moreover, neonatal exposure of mice to paraquat results in dopaminergic cell loss and the development of a Parkinson disease phenotype (52). In the lung, altered levels of BH4 can lead to pulmonary hypertension and fibrosis (2). However, it should be noted that exposure to redox cycling chemicals may only be for short periods of time, and this may not be sufficient to reduce BH4 levels below those needed to sustain aromatic amino acid hydroxylases and nitric-oxide synthases. Because chemical redox cycling by sepiapterin reductase also generates cytotoxic ROS, which can contribute to altered cell functioning, this may be a more important short term toxic mechanism. Additional studies are needed to determine whether exposure to paraquat or other redox cycling chemicals can lead to BH4 deficiencies and the production of cytotoxic concentrations of ROS in human tissues.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM034310, R01ES004738, R01CA132624, U54AR055073, and P30ES005022.
- ROS
- reactive oxygen species
- SPR
- sepiapterin reductase
- m
- mouse
- h
- human
- Ni-NTA
- nickel-nitrilotriacetic acid
- BH4
- tetrahydrobiopterin
- BH2
- dihydrobiopterin
- DTPA
- diethylenetriaminepentaacetic acid
- F
- forward
- R
- reverse.
REFERENCES
- 1. Kovacic P., Somanathan R. (2009) Pulmonary toxicity and environmental contamination: radicals, electron transfer, and protection by antioxidants. Rev. Environ. Contam. Toxicol. 201, 41–69 [DOI] [PubMed] [Google Scholar]
- 2. Kimbrough R. D., Gaines T. B. (1970) Toxicity of paraquat to rats and its effect on rat lungs. Toxicol. Appl. Pharmacol. 17, 679–690 [DOI] [PubMed] [Google Scholar]
- 3. Rashba-Step J., Cederbaum A. I. (1994) Generation of reactive oxygen intermediates by human liver microsomes in the presence of NADPH or NADH. Mol. Pharmacol. 45, 150–157 [PubMed] [Google Scholar]
- 4. Bonneh-Barkay D., Reaney S. H., Langston W. J., Di Monte D. A. (2005) Redox cycling of the herbicide paraquat in microglial cultures. Mol. Brain Res. 134, 52–56 [DOI] [PubMed] [Google Scholar]
- 5. Dicker E., Cederbaum A. I. (1991) NADH-dependent generation of reactive oxygen species by microsomes in the presence of iron and redox cycling agents. Biochem. Pharmacol. 42, 529–535 [DOI] [PubMed] [Google Scholar]
- 6. Winterbourn C. C. (1995) Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 82, 969–974 [DOI] [PubMed] [Google Scholar]
- 7. Pacher P., Beckman J. S., Liaudet L. (2007) Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radak Z., Zhao Z., Goto S., Koltai E. (2011) Age-associated neurodegeneration and oxidative damage to lipids, proteins, and DNA. Mol. Aspects Med. 32, 305–315 [DOI] [PubMed] [Google Scholar]
- 9. Wang Y., Gray J. P., Mishin V., Heck D. E., Laskin D. L., Laskin J. D. (2008) Role of cytochrome P450 reductase in nitrofurantoin-induced redox cycling and cytotoxicity. Free Radic. Biol. Med. 44, 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Gray J. P., Mishin V., Heck D. E., Laskin D. L., Laskin J. D. (2010) Distinct roles of cytochrome P450 reductase in mitomycin C redox cycling and cytotoxicity. Mol. Cancer Ther. 9, 1852–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marín A., López de Cerain A., Hamilton E., Lewis A. D., Martinez-Peñuela J. M., Idoate M. A., Bello J. (1997) DT-diaphorase and cytochrome b5 reductase in human lung and breast tumours. Br. J. Cancer 76, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osman A. M., van Noort P. C. (2003) Evidence for redox cycling of lawsone (2-hydroxy-1,4-naphthoquinone) in the presence of the hypoxanthine/xanthine oxidase system. J. Appl. Toxicol. 23, 209–212 [DOI] [PubMed] [Google Scholar]
- 13. Day B. J., Patel M., Calavetta L., Chang L. Y., Stamler J. S. (1999) A mechanism of paraquat toxicity involving nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 96, 12760–12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Donnell B. V., Tew D. G., Jones O. T., England P. J. (1993) Studies on the inhibitory mechanism of iodonium compounds with special reference to neutrophil NADPH oxidase. Biochem. J. 290, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Backes W. L., Kelley R. W. (2003) Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol. Ther. 98, 221–233 [DOI] [PubMed] [Google Scholar]
- 16. Kubota S., Yoshida Y., Kumaoka H. (1977) Studies on the microsomal electron-transport system of anaerobically grown yeast. IV. Purification and characterization of NADH-cytochrome b5 reductase. J. Biochem. 81, 187–195 [DOI] [PubMed] [Google Scholar]
- 17. Agarwal A., Banerjee A., Banerjee U. C. (2011) Xanthine oxidoreductase: a journey from purine metabolism to cardiovascular excitation-contraction coupling. Crit. Rev. Biotechnol. 31, 264–280 [DOI] [PubMed] [Google Scholar]
- 18. Fulton D., Gratton J. P., Sessa W. C. (2001) Post-translational control of endothelial nitric oxide synthase: why isn't calcium/calmodulin enough? J. Pharmacol. Exp. Ther. 299, 818–824 [PubMed] [Google Scholar]
- 19. Park H. S., Kim S. R., Lee Y. C. (2009) Impact of oxidative stress on lung diseases. Respirology 14, 27–38 [DOI] [PubMed] [Google Scholar]
- 20. Gray J. P., Heck D. E., Mishin V., Smith P. J., Hong J. Y., Thiruchelvam M., Cory-Slechta D. A., Laskin D. L., Laskin J. D. (2007) Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H:paraquat oxidoreductase: identification of the enzyme as thioredoxin reductase. J. Biol. Chem. 282, 7939–7949 [DOI] [PubMed] [Google Scholar]
- 21. Holmgren A., Lu J. (2010) Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem. Biophys. Res. Commun. 396, 120–124 [DOI] [PubMed] [Google Scholar]
- 22. Thöny B., Auerbach G., Blau N. (2000) Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347, 1–16 [PMC free article] [PubMed] [Google Scholar]
- 23. Werner E. R., Blau N., Thöny B. (2011) Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem. J. 438, 397–414 [DOI] [PubMed] [Google Scholar]
- 24. Crabtree M. J., Channon K. M. (2011) Synthesis and recycling of tetrahydrobiopterin in endothelial function and vascular disease. Nitric Oxide 25, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh S. (1971) Sepiapterin reductase from horse liver: purification and properties of the enzyme. Arch. Biochem. Biophys. 146, 202–214 [DOI] [PubMed] [Google Scholar]
- 26. Ferre J., Naylor E. W. (1988) Sepiapterin reductase in human amniotic and skin fibroblasts, chorionic villi, and various blood fractions. Clin. Chim. Acta 174, 271–282 [DOI] [PubMed] [Google Scholar]
- 27. Fussell K. C., Udasin R. G., Gray J. P., Mishin V., Smith P. J., Heck D. E., Laskin J. D. (2011) Redox cycling and increased oxygen utilization contribute to diquat-induced oxidative stress and cytotoxicity in Chinese hamster ovary cells overexpressing NADPH-cytochrome P450 reductase. Free Radic. Biol. Med. 50, 874–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou M., Diwu Z., Panchuk-Voloshina N., Haugland R. P. (1997) A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 253, 162–168 [DOI] [PubMed] [Google Scholar]
- 29. Mishin V. M., Thomas P. E. (2004) Characterization of hydroxyl radical formation by microsomal enzymes using a water-soluble trap, terephthalate. Biochem. Pharmacol. 68, 747–752 [DOI] [PubMed] [Google Scholar]
- 30. Barrett E. G., Johnston C., Oberdörster G., Finkelstein J. N. (1998) Silica-induced chemokine expression in alveolar type II cells is mediated by TNF-α. Am. J. Physiol. 275, L1110–L1119 [DOI] [PubMed] [Google Scholar]
- 31. Wolff D. J., Lubeskie A., Li C. (1997) Inactivation and recovery of nitric oxide synthetic capability in cytokine-induced RAW 264.7 cells treated with “irreversible” NO synthase inhibitors. Arch. Biochem. Biophys. 338, 73–82 [DOI] [PubMed] [Google Scholar]
- 32. Butler J., Hoey B. M. (1993) The one-electron reduction potential of several substrates can be related to their reduction rates by cytochrome P-450 reductase. Biochim. Biophys. Acta 1161, 73–78 [DOI] [PubMed] [Google Scholar]
- 33. Roginsky V. A., Barsukova T. K., Stegmann H. B. (1999) Kinetics of redox interaction between substituted quinones and ascorbate under aerobic conditions. Chem. Biol. Interact. 121, 177–197 [DOI] [PubMed] [Google Scholar]
- 34. Ham S. W., Choe J. I., Wang M. F., Peyregne V., Carr B. I. (2004) Fluorinated quinoid inhibitor: possible “pure” arylator predicted by the simple theoretical calculation. Bioorg. Med. Chem. Lett. 14, 4103–4105 [DOI] [PubMed] [Google Scholar]
- 35. Cohen G. M., d'Arcy Doherty M. (1987) Free radical mediated cell toxicity by redox cycling chemicals. Br. J. Cancer Suppl. 8, 46–52 [PMC free article] [PubMed] [Google Scholar]
- 36. Sueoka T., Katoh S. (1985) Carbonyl reductase activity of sepiapterin reductase from rat erythrocytes. Biochim. Biophys. Acta 843, 193–198 [DOI] [PubMed] [Google Scholar]
- 37. Smith G. K., Duch D. S., Edelstein M. P., Bigham E. C. (1992) New inhibitors of sepiapterin reductase. Lack of an effect of intracellular tetrahydrobiopterin depletion upon in vitro proliferation of two human cell lines. J. Biol. Chem. 267, 5599–5607 [PubMed] [Google Scholar]
- 38. Fujimoto K., Hara M., Yamada H., Sakurai M., Inaba A., Tomomura A., Katoh S. (2001) Role of the conserved Ser-Tyr-Lys triad of the SDR family in sepiapterin reductase. Chem. Biol. Interact. 130, 825–832 [DOI] [PubMed] [Google Scholar]
- 39. Fujimoto K., Ichinose H., Nagatsu T., Nonaka T., Mitsui Y., Katoh S. (1999) Functionally important residues tyrosine-171 and serine-158 in sepiapterin reductase. Biochim. Biophys. Acta 1431, 306–314 [DOI] [PubMed] [Google Scholar]
- 40. Auerbach G., Herrmann A., Gütlich M., Fischer M., Jacob U., Bacher A., Huber R. (1997) The 1.25-Å crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters. EMBO J. 16, 7219–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Supangat S., Seo K. H., Choi Y. K., Park Y. S., Son D., Han C. D., Lee K. H. (2006) Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of l-threo-tetrahydrobiopterin. J. Biol. Chem. 281, 2249–2256 [DOI] [PubMed] [Google Scholar]
- 42. Tanaka N., Nonaka T., Nakanishi M., Deyashiki Y., Hara A., Mitsui Y. (1996) Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 Å resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 43. Katoh S., Sueoka T., Yamamoto Y., Takahashi S. Y. (1994) Phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein kinase C of sepiapterin reductase, the terminal enzyme in the biosynthetic pathway of tetrahydrobiopterin. FEBS Lett. 341, 227–232 [DOI] [PubMed] [Google Scholar]
- 44. Yubisui T., Takeshita M. (1980) Characterization of the purified NADH-cytochrome b5 reductase of human erythrocytes as a FAD-containing enzyme. J. Biol. Chem. 255, 2454–2456 [PubMed] [Google Scholar]
- 45. King M. S., Sharpley M. S., Hirst J. (2009) Reduction of hydrophilic ubiquinones by the flavin in mitochondrial NADH:ubiquinone oxidoreductase (complex I) and production of reactive oxygen species. Biochemistry 48, 2053–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsunaga T., Shinoda Y., Inoue Y., Shimizu Y., Haga M., Endo S., El-Kabbani O., Hara A. (2011) Aldo-keto reductase 1C15 as a quinone reductase in rat endothelial cell: its involvement in redox cycling of 9,10-phenanthrenequinone. Free Radic. Res. 45, 848–857 [DOI] [PubMed] [Google Scholar]
- 47. Shultz C. A., Quinn A. M., Park J. H., Harvey R. G., Bolton J. L., Maser E., Penning T. M. (2011) Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase, and carbonyl reductases to redox-cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-o-quinone. Chem. Res. Toxicol. 24, 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mau B. L., Powis G. (1990) Inhibition of thioredoxin reductase (E.C. 1.6.4.5.) by antitumor quinones. Free Radic. Res. Commun. 8, 365–372 [DOI] [PubMed] [Google Scholar]
- 49. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 50. Tanaka N., Nonaka T., Tanabe T., Yoshimoto T., Tsuru D., Mitsui Y. (1996) Crystal structures of the binary and ternary complexes of 7-α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35, 7715–7730 [DOI] [PubMed] [Google Scholar]
- 51. Oppermann U. C., Filling C., Berndt K. D., Persson B., Benach J., Ladenstein R., Jörnvall H. (1997) Active site-directed mutagenesis of 3β/17β-hydroxysteroid dehydrogenase establishes differential effects on short-chain dehydrogenase/reductase reactions. Biochemistry 36, 34–40 [DOI] [PubMed] [Google Scholar]
- 52. Nisticò R., Mehdawy B., Piccirilli S., Mercuri N. (2011) Paraquat- and rotenone-induced models of Parkinson's disease. Int. J. Immunopathol. Pharmacol. 24, 313–322 [DOI] [PubMed] [Google Scholar]
- 53. Curtius H. C., Niederwieser A., Levine R., Müldner H. (1984) Therapeutic efficacy of tetrahydrobiopterin in Parkinson's disease. Adv. Neurol. 40, 463–466 [PubMed] [Google Scholar]
- 54. Tani Y., Fernell E., Watanabe Y., Kanai T., Långström B. (1994) Decrease in 6R-5,6,7,8-tetrahydrobiopterin content in cerebrospinal fluid of autistic patients. Neurosci. Lett. 181, 169–172 [DOI] [PubMed] [Google Scholar]
- 55. Blau N., Hennermann J. B., Langenbeck U., Lichter-Konecki U. (2011) Diagnosis, classification, and genetics of phenylketonuria and tetrahydrobiopterin (BH4) deficiencies. Mol. Genet. Metab. 104, S2–S9 [DOI] [PubMed] [Google Scholar]
- 56. Verbeek M. M., Willemsen M. A., Wevers R. A., Lagerwerf A. J., Abeling N. G., Blau N., Thöny B., Vargiami E., Zafeiriou D. I. (2008) Two Greek siblings with sepiapterin reductase deficiency. Mol. Genet. Metab. 94, 403–409 [DOI] [PubMed] [Google Scholar]
- 57. Bonafé L., Thöny B., Penzien J. M., Czarnecki B., Blau N. (2001) Mutations in the sepiapterin reductase gene cause a novel tetrahydrobiopterin-dependent monoamine-neurotransmitter deficiency without hyperphenylalaninemia. Am. J. Hum. Genet. 69, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Farrugia R., Scerri C. A., Montalto S. A., Parascandolo R., Neville B. G., Felice A. E. (2007) Molecular genetics of tetrahydrobiopterin (BH4) deficiency in the Maltese population. Mol. Genet. Metab. 90, 277–283 [DOI] [PubMed] [Google Scholar]
- 59. Yang S., Lee Y. J., Kim J. M., Park S., Peris J., Laipis P., Park Y. S., Chung J. H., Oh S. P. (2006) A murine model for human sepiapterin-reductase deficiency. Am. J. Hum. Genet. 78, 575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Homma D., Sumi-Ichinose C., Tokuoka H., Ikemoto K., Nomura T., Kondo K., Katoh S., Ichinose H. (2011) Partial biopterin deficiency disturbs postnatal development of the dopaminergic system in the brain. J. Biol. Chem. 286, 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mandel J. S., Adami H. O., Cole P. (2012) Paraquat and Parkinson's disease: an overview of the epidemiology and a review of two recent studies. Regul. Toxicol. Pharmacol. 62, 385–392 [DOI] [PubMed] [Google Scholar]