Background: General amino acid control (GAAC) is important for cell survival under amino acid starvation.
Results: Absence of fission yeast Cpc2, a homolog of mammalian RACK1, causes defects in eIF2α phosphorylation, induction of amino acid biosynthesis genes, and Gcn2 autophosphorylation.
Conclusion: Cpc2 stimulates the GAAC response by facilitating Gcn2 activation.
Significance: This study provides evidence that RACK1 homolog promotes the GAAC response.
Keywords: Phosphorylation, Signal Transduction, Stress Response, Translation Control, Yeast, Cpc2, Gcn2, General Amino Acid Control
Abstract
General amino acid control (GAAC) is crucial for sensing and adaptation to nutrient availability. Amino acid starvation activates protein kinase Gcn2, which plays a central role in the GAAC response by phosphorylating the α-subunit of eukaryotic initiation factor 2 (eIF2α), leading to the translational switch to stimulate selective expression of stress-responsive genes. We report here that in fission yeast Schizosaccharomyces pombe, Cpc2, a homolog of mammalian receptor for activated C-kinase (RACK1), is important for the GAAC response. Deletion of S. pombe cpc2 impairs the amino acid starvation-induced phosphorylation of eIF2α and the expression of amino acid biosynthesis genes, thereby rendering cells severely sensitive to amino acid limitation. Unlike the Saccharomyces cerevisiae Cpc2 ortholog, which normally suppresses the GAAC response, our findings suggest that S. pombe Cpc2 promotes the GAAC response. We also found that S. pombe Cpc2 is required for starvation-induced Gcn2 autophosphorylation, which is essential for Gcn2 function. These results indicate that S. pombe Cpc2 facilitates the GAAC response through the regulation of Gcn2 activation and provide a novel insight for the regulatory function of RACK1 on Gcn2-mediated GAAC response.
Introduction
Translational control contributes to stress-induced fine-tuning of gene expression patterns (1, 2). Phosphorylation of the α-subunit of eukaryotic translation initiation factor eIF2 (eIF2α) is an important event in translational regulation in response to stress (3). Its molecular mechanism has been extensively studied in the budding yeast Saccharomyces cerevisiae (4), and the phenomenon is known as the general amino acid control (GAAC)3 response, in which depletion of a single amino acid stimulates the expression of many genes involved in the biosynthesis of all amino acids. Stress-induced eIF2α phosphorylation inhibits the formation of an active ternary complex consisting of eIF2, methionyl-tRNA, and GTP, leading to a decrease in global translation initiation efficiency (3, 4). At the same time, translation of a subset of stress-responsive genes, such as S. cerevisiae GCN4 and mammalian ATF4 transcriptional factors, is specifically promoted by a mechanism involving short upstream open reading frames in the 5′-untranslated region of these mRNAs (5–7). Gcn4 and ATF4 induce the expression of a number of genes required for amino acid biosynthesis and stress response (8, 9). Starvation-induced expression of amino acid biosynthesis genes is also observed in the fission yeast Schizosaccharomyces pombe, although the relevant transcriptional factor is not known (10). A single eIF2α kinase, Gcn2, regulates this response in S. cerevisiae, whereas four (GCN2, HRI, PKR, PERK) or three (Gcn2, Hri1, Hri2) eIF2α kinases regulate eIF2α phosphorylation depending on the types of stress in mammals or S. pombe, respectively (3, 11). Because eIF2α kinases are activated in response to diverse forms of stress, eIF2α phosphorylation functions to integrate various stress stimuli into translational controls (12).
Among the eIF2α kinases, Gcn2 (GCN2 in mammals) is a dominant regulator of the GAAC response. Gcn2 binds preferentially to non-aminoacylated (uncharged) tRNA in vitro, through the domain homologous to histidyl-tRNA synthetase (13, 14). Gcn2 forms a dimer, and autophosphorylation within its kinase domain is essential for full activation of Gcn2 (15–17). Moreover, Gcn2 binds to translating ribosomes (18). From these observations and structural analysis of Gcn2, it is predicted that uncharged tRNA binding activates Gcn2 by inducing a conformational change (19) and that ribosomal localization of Gcn2 facilitates the binding of uncharged tRNA in the ribosomal A-site or the interaction with its substrate eIF2α. In addition to tRNA binding, posttranslational modifications or regulatory factors modulate Gcn2 activity (20–23), although the precise mechanism of regulation of Gcn2 activity by these additional inputs is not fully understood.
The receptor for activated C-kinase (RACK1) is a highly conserved protein among eukaryotes (homologs are known as Asc1 in S. cerevisiae (24) and Cpc2 in S. pombe (25)). RACK1 homolog is linked to diverse physiological processes through interactions with numerous signaling molecules (25–28). Also, recent studies have revealed that RACK1 is a stoichiometric component of ribosomes (29, 30). RACK1 homologs facilitate global and selective translation through various regulatory mechanisms (24, 29, 31, 32). Because RACK1 associates with several signaling factors, such as protein kinase C and Src, on ribosomes, it has been thought that RACK1 integrates cellular signals to translational regulation (32, 33).
RACK1 homologs are involved in control of the GAAC response. The absence of S. cerevisiae ASC1 increased the Gcn4-mediated transcription of amino acid biosynthesis genes under nonstarvation conditions (34), presumably by the destabilization of translation initiation complexes on ribosomes (35). ASC1 deletion suppressed the growth defect of gcn2Δ cells under limiting amino acid conditions, indicating that Asc1 negatively regulates the GAAC response in S. cerevisiae. On the other hand, in Neurospora crassa, cpc-2, encoding a RACK1 homolog, was originally isolated as a gene regulating cross-pathway control (36, 37), a similar phenomenon to the GAAC response. The cpc-2 U142 allele impaired the induction of amino acid biosynthesis genes in response to amino acid limitation (36), suggesting a positive effector role of Cpc-2 in the GAAC response. However, the molecular mechanism of action of Cpc-2 in N. crassa has not been determined. Furthermore, it remains unclear how RACK1 homologs regulate the GAAC response in S. pombe and mammals.
Here, we analyzed the function of S. pombe Cpc2 in response to amino acid starvation. We found that S. pombe Cpc2 is required for phosphorylation of eIF2α and the expression of amino acid biosynthesis genes induced by histidine starvation. Moreover, cpc2 deletion caused defects in the autophosphorylation of the eIF2α kinase Gcn2, indicating that S. pombe Cpc2 stimulates Gcn2 activation and the GAAC response.
EXPERIMENTAL PROCEDURES
Yeast Strains and General Techniques
S. pombe strains used in this study are listed in Table 1. Growth media and basic techniques for S. pombe have been described elsewhere (38). For 3-aminotriazole (3AT) treatment, cells were grown in Edinburgh minimal medium (EMM) liquid medium or on EMM plates supplemented with 10 mm 3AT. To construct an epitope-tagged Gcn2 plasmid, nucleotides 1–3,162 of gcn2+ genomic DNA (numbers are relative to the translation start site) were cloned into the plasmid pT7Blue (Merck) to obtain pYTR193. A ura4+ marker cassette was inserted into the EcoRV-HincII sites in pYTR193 to obtain pYTR194. The upstream region of the gcn2+ gene (nucleotides −496 to −1) was cloned into the KpnI-NdeI sites in pYTR194 followed by 5× FLAG or 12× Myc tag insertion at the NdeI site to obtain pYTR198 or pYTR222, respectively. cpc2 ORF was cloned in the plasmid pBlueScriptII SK(−) to obtain pYTR120. Point mutations of gcn2 or cpc2 were introduced into pYTR193 or pYTR120, respectively, using a QuikChange site-directed mutagenesis kit (Agilent Technologies). Fragments containing gcn2 or cpc2 mutations were transformed into Flag:gcn2Δ (YT3648) or cpc2Δ (YT2307) cells, respectively. To construct expression plasmids, cDNA of S. pombe cpc2, S. cerevisiae asc1, and human RACK1 (GNB2L1) were cloned into the NdeI-SmaI site in the plasmid pREP2 (39).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | |
|---|---|---|
| JK317 | h− | leu1-32 ura4-D18 |
| YT2307 | h− | cpc2::ura4+ leu1-32 ura4-D18 |
| YT2313 | h− | cpc2:2HA6His:ura4+ leu1-32 ura4-D18 |
| YT2453 | h− | hri2::ura4+ leu1-32 ura4-D18 |
| YT2459 | h− | hri2::ura4+ gcn2::Kanr leu1-32 ura4-D18 |
| YT2465 | h− | rpS3:Flag:ura4+ leu1-32 ura4-D18 |
| YT2824 | h− | cpc2::ura4+ gcn2::Kanr leu1-32 ura4-D18 |
| YT3033 | h− | leu1-32 |
| YT3173 | h− | hri2::ura4+ cpc2::ura4+ leu1-32 ura4-D18 |
| YT3360 | h− | gcn2::ura4+ leu1-32 ura4-D18 |
| YT3372 | h− | 5Flag:gcn2 leu1-32 ura4-D18 |
| YT3540 | h− | cpc2::Kanr leu1-32 ura4-D18 |
| YT3542 | h− | cpc2_DE leu1-32 ura4-D18 |
| YT3559 | h− | 5Flag:gcn2 cpc2_DE:ura4+ leu1-32 ura4-D18 |
| YT3598 | h− | 5Flag:gcn2 cpc2::ura4+ leu1-32 ura4-D18 |
| YT3648 | h− | 5Flag:gcn2::ura4+ leu1-32 ura4-D18 |
| YT3656 | h− | 5Flag:gcn2_K585R leu1-32 ura4-D18 |
| YT3657 | h− | 5Flag:gcn2_T818/823A leu1-32 ura4-D18 |
| YT4251 | h− | atf1::ura4+ leu1-32 ura4-D18 |
| YT4254 | h− | 5Flag:gcn2 atf1::ura4+ leu1-32 ura4-D18 |
| YT4279 | h+/h− | 5Flag:gcn2/12myc:gcn2 ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 |
| YT4280 | h+/h− | 5Flag:gcn2/12myc:gcn2 cpc2::ura4+/cpc2::ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 |
| YT4281 | h+/h− | 12myc:gcn2/gcn2+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 |
| YT4299 | h− | cpc2_W43* leu1-32 ura4-D18 |
| YT4309 | h− | 5Flag:gcn2 cpc2_W43* leu1-32 ura4-D18 |
| YT4376 | h+/h− | 5Flag:gcn2/gcn2+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 |
| YT4386 | h− | Flag:cpc2 leu1-32 ura4-D18 |
| YT4388 | h− | Flag:cpc2_W43* leu1-32 ura4-D18 |
| YT4389 | h− | cpc2::Kanr:2HA6His:ura4+ leu1-32 ura4-D18 |
| YT4390 | h− | cpc2_W43*:2HA6His:ura4+ leu1-32 ura4-D18 |
RT-PCR
Total RNA was prepared as described previously (40). cDNA was synthesized using an RNA PCR kit (Takara) with random 9-mer primers. Gene expression levels were analyzed by quantitative PCR using a StepOnePlus real-time PCR system (Invitrogen). Information about the primers used in this study is provided in Table 2. The primers for 18 S rRNA are the same as those used previously (41).
TABLE 2.
Primer sets used in quantitative RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| his4 | TAGATCAGGTGCCGACAAAG | CCGAAATAACAACTGCCTGA |
| SPAC56E4.03 | GCTACATCCATGCCGATAAA | CTAGGGTCAACAACGAACCA |
| SPCC364.07 | TCAACTCTCCATACGCCAAC | CACCTTGTTCCACTCACCAC |
| SPAC10F6.13c | TTTGCTGAATGGGAACAAGA | CAACAAGAGAATCGCGAAGA |
| arg3 | GTCATCCCGAGGAAGTGTCT | GTCCATTTGCGGTTCTCTG |
| dld1 | CTGAAGTTGCTTGGGTTGGT | GGCATCCATATTGGTCTTGG |
| lys3 | TCTGGTTATTGGGGCTCTTG | TTAATGTCCCAGCGAAGAATG |
| leu3 | TCTCTCCATCCCCATAACGA | CCAAACAAACAGCCCTCAA |
| SPBC19F5.04 | GCAGCACAGATACCAAAGCA | AACCAAATCATGCACAGAACC |
| cdk9 | CTCTTTGCGGTGCTATTTTG | TTGCTGGATATGGTGGTGTT |
| 18 S rRNA | GGGAACCAGGACTTTTACCTTGA | AACTTGCCTGCTTTGAACACTCTA |
| cpc2 | CTCTGACTGGGTTTCTTGTGTG | GCCATAGTGAGAAGTGCGAAG |
| snoU24b | TATTTGCTACTTCGGAGGCCTTA | GGTGATTTGTTTTGTCTCATCG |
Immunoblotting
For preparation of whole cell extracts, harvested cells were suspended in alkaline lysis buffer (1.85 m NaOH, 7.4% 2-mercaptoethanol) and incubated for 10 min on ice. Samples were combined with an equal volume of 50% trichloroacetic acid, incubated for 10 min, and centrifuged. Precipitated proteins were suspended in 2× sample buffer. For detection of Gcn2-phospho-Thr-818, cell lysates were extracted with zirconia beads and lysis buffer A (25 mm MOPS, pH 7.2, 15 mm EGTA, 150 mm NaCl, 0.1% Nonidet P-40, 1 mm dithiothreitol, 10% glycerol, 50 mm NaF, 1 mm phenylmethylsulfonyl fluoride, Complete protease inhibitor (Roche Applied Science), PhosSTOP (Roche Applied Science)) using a Multi-beads shocker (Yasui Kikai). FLAG-tagged Gcn2 was immunoprecipitated using anti-FLAG M2-agarose resin (Sigma). Anti-Gcn2-phospho-Thr-818 antibody was raised against the following peptide, ADEDL(P)TTGVGC (Medical & Biological Laboratories). Anti-Cpc2 antibody was raised against a recombinant Cpc2 protein. The other antibodies used in this study were anti-phosphorylated eIF2α (44-728G; Invitrogen), anti-FLAG M2 (F-3165; Sigma), anti-c-Myc (Sc-40; Santa Cruz Biotechnology), and anti-Cdc2 (PSTAIRE) (Sc-53; Santa Cruz Biotechnology) antibodies.
Co-immunoprecipitation Assay
Cell lysates were extracted in lysis buffer A as described above. FLAG-tagged Gcn2 was immunoprecipitated using Dynabeads anti-mouse IgG (Dynal Biotech) preconjugated with anti-FLAG M2 antibody in lysis buffer A. After incubation for 2 h, the beads were washed four times with lysis buffer A containing 300 mm NaCl and suspended in 2× sample buffer for immunoblotting.
Sucrose Gradient and Polysome Fractionation
Sucrose gradients were performed as described previously with modifications (29). 100 μg/ml cycloheximide was added prior to harvesting cells. Cells were washed and resuspended in TSM buffer (10 mm Tris-HCl, pH 7.4, 100 mm NaCl, 30 mm MgCl2, 50 μg/ml cycloheximide, 200 μg/ml heparin, 1 mm phenylmethylsulfonyl fluoride, Complete protease inhibitor). After breakage with zirconia beads using a Multi-beads shocker, cell lysates were clarified by centrifugation. Supernatants were loaded on 15–50% (w/v) sucrose gradients in Gradient buffer (7.5 mm Tris-HCl, pH 7.4, 70 mm NH4Cl, 3.9 mm MgOAc) and ultracentrifuged for 3 h at 40,000 rpm in a Beckman SW41 rotor. Samples were fractionated using a gradient station (Biocomp Instruments), and polysome profiles were obtained by monitoring the absorbance at 260 nm along the gradient.
In Vitro Kinase Assay
Cells were cultured to log phase. Cell lysates were extracted with zirconia beads and lysis buffer B (50 mm Tris-HCl, pH 8.0, 5 mm EGTA, 150 mm NaCl, 0.1% Nonidet P-40, 50 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, Complete protease inhibitor) using a Multi-beads shocker. Immunoprecipitation was performed using anti-FLAG M2-agarose resin. After washing twice with lysis buffer B and twice with kinase buffer (20 mm Hepes-KOH, pH 7.5, 20 mm MgCl2, 5 mm EGTA, 2 mm dithiothreitol), the resin was suspended in 30 μl of kinase buffer containing 8.5 μm ATP and 0.2 μl of [γ-32P]ATP (10 mCi/ml) and then incubated for 30 min at 30 °C. Samples were resolved by SDS-PAGE, and autoradiography was performed using a Typhoon 9400 imager and ImageQuant software (GE Healthcare).
RESULTS
Cpc2 Plays an Important Role in the Induction of Amino Acid Biosynthesis Genes in S. pombe
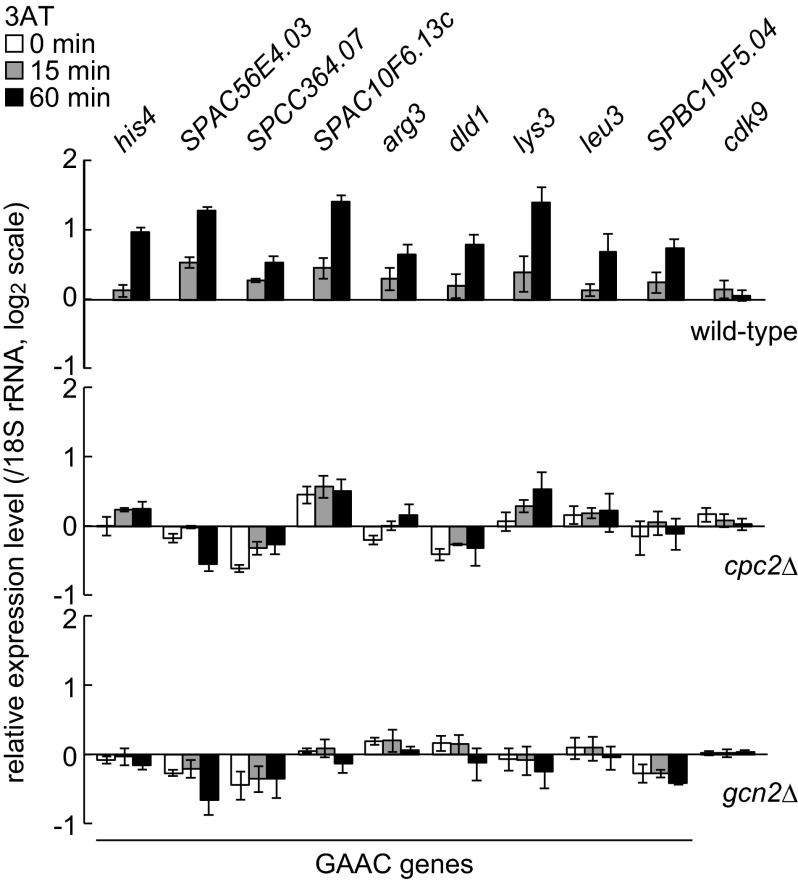
To investigate whether S. pombe Cpc2 is involved in the response to amino acid starvation, wild-type, gcn2Δ and cpc2Δ cells were exposed to 3AT, which inhibits histidine biosynthesis and induces the GAAC response in S. pombe. A recent analysis revealed that 3AT treatment induces expression of about 40% of the genes that are predicted to function in amino acid biosynthesis in S. pombe (10). We selected nine genes from those genes as the GAAC genes (his4+, SPAC56E4.03, SPCC364.07, SPAC10F6.13c, arg3+, dld1+, lys3+, leu3+, SPBC19F5.04) and checked their expression levels by quantitative RT-PCR. The addition of 3AT significantly increased the expression of all nine genes in wild-type cells, but not in gcn2Δ cells (Fig. 1), as reported in a previous microarray analysis (10). Furthermore, we found that deletion of cpc2 abolished 3AT-induced expression of most of these amino acid biosynthesis genes (Fig. 1). The expression of some genes such as SPAC10F6.13c and lys3 was detectable even in cpc2Δ cells. However, in the absence of cpc2, 3AT-induced up-regulation of SPAC10F6.13c was not observed, and the fold increase of lys3 expression upon 3AT addition was significantly smaller than that observed in wild-type cells. These data indicate that S. pombe Cpc2 is important for the precise regulation of gene expression in the GAAC response.
FIGURE 1.
Cpc2 is important for induction of amino acid biosynthesis genes. Gene expression levels of nine GAAC genes and cdk9+ (control) were analyzed by quantitative RT-PCR from wild-type (YT3033), cpc2Δ (YT2307), and gcn2Δ (YT3360) cells treated with 10 mm 3-aminotriazole (3AT) for 0 min (white bar), 15 min (gray bar), and 60 min (black bar). All data are normalized to the expression level of 18 S rRNA and shown as the relative fold to those at time 0 in wild-type cells with S.E. (n = 3). GAAC genes are: his4+, imidazoleglycerol-phosphate synthase (predicted); SPAC56E4.03, aromatic aminotransferase (predicted); SPCC364.07, d-3 phosphoglycerate dehydrogenase (predicted); SPAC10F6.13c, aspartate aminotransferase (predicted); arg3+, ornithine carbamoyltransferase; dld1+, dihydrolipoamide dehydrogenase; lys3+, saccharopine dehydrogenase; leu3+, 2-isopropylmalate synthase; SPBC19F5.04, aspartate kinase (predicted).
Cpc2 Is Required for Gcn2-mediated eIF2α Phosphorylation and Survival under Amino Acid Starvation
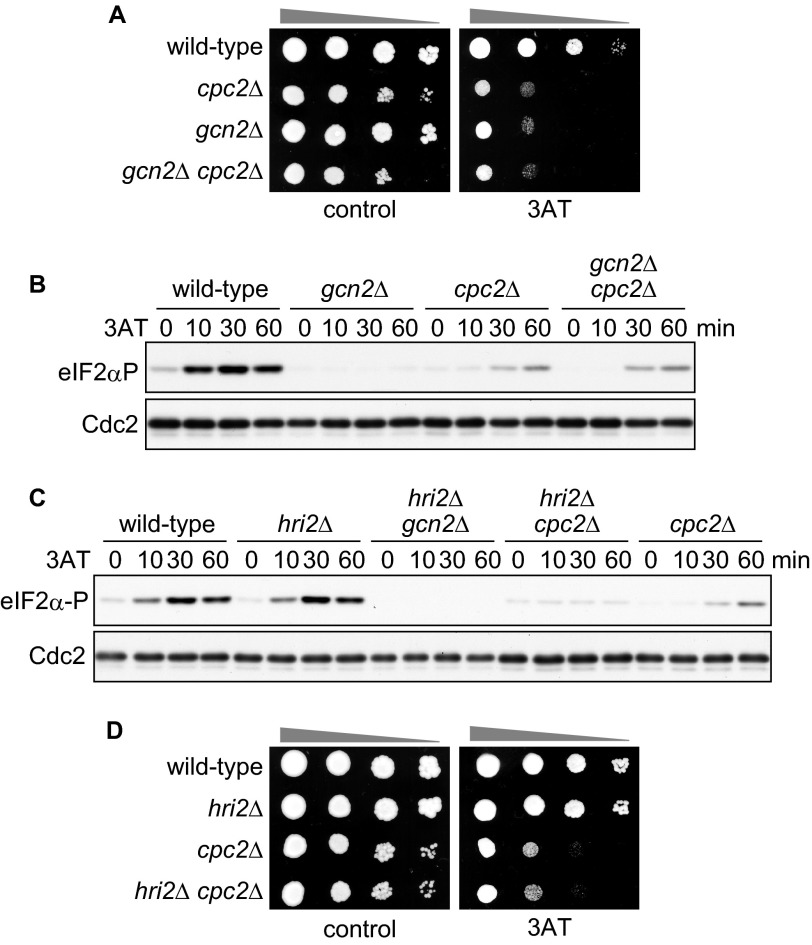
Mutant cpc2Δ cells exhibited a severe growth defect in the presence of 3AT (Fig. 2A), which is consistent with a defect in induction of amino acid biosynthesis genes, as described above. Moreover, 3AT sensitivity was not significantly different between cpc2Δ and gcn2Δ cpc2Δ double-mutant cells (Fig. 2A). These data suggest that S. pombe Cpc2 plays a role in the GAAC response through a common mechanism with Gcn2. Treatment of wild-type cells with 3AT induces phosphorylation of eIF2α in a Gcn2-dependent manner (Fig. 2B), which is a key step in the GAAC response. To examine whether Cpc2 is required for eIF2α phosphorylation, we checked the phosphorylation pattern after the addition of 3AT in cpc2Δ cells. Loss of cpc2 resulted in a marked decrease in the level of eIF2α phosphorylation when compared with wild-type cells, indicating that Cpc2 is important for eIF2α phosphorylation under amino acid starvation (Fig. 2B, wild-type and cpc2Δ).
FIGURE 2.
Cpc2 is required for Gcn2-mediated eIF2α phosphorylation (eIF2αP) and survival under amino acid starvation. A, 10-fold serial dilutions of wild-type (YT3033), cpc2Δ (YT2307), gcn2Δ (YT3360), and gcn2Δ cpc2Δ (YT2824) cells were spotted on EMM plates ± 10 mm 3AT. B, immunoblots of phosphorylated eIF2α and Cdc2 (control) from wild-type (YT3033), cpc2Δ (YT2307), gcn2Δ (YT3360), and gcn2Δ cpc2Δ (YT2824) cells treated with 10 mm 3AT. C, immunoblots of phosphorylated eIF2α and Cdc2 (control) from wild-type (YT3033), hri2Δ (YT2453), hri2Δ gcn2Δ (YT2459), hri2Δ cpc2Δ (YT3173), and cpc2Δ (YT2307) cells treated with 10 mm 3AT. D, 10-fold serial dilutions of wild-type (YT3033), hri2Δ (YT2453), cpc2Δ (YT2307), and hri2Δ cpc2Δ (YT3173) cells were spotted on EMM plates ± 10 mm 3AT.
We observed a relatively weak and delayed eIF2α phosphorylation signal in cpc2Δ cells at 60 min after 3AT addition (Fig. 2B, wild-type and cpc2Δ). This signal pattern was similar in gcn2Δ cpc2Δ double-mutant cells (Fig. 2B), supporting the idea that Gcn2-dependent eIF2α phosphorylation is abrogated in cpc2Δ cells. On the other hand, these results raise the possibility that the delayed eIF2α phosphorylation in cpc2Δ cells is independent of Gcn2. Among three eIF2α kinases (Gcn2, Hri1, and Hri2) in S. pombe, Gcn2 plays a major role in phosphorylation of eIF2α under amino acid starvation (10, 11), as shown in the data for gcn2Δ cells (Fig. 2B, gcn2Δ). To test the possibility that other eIF2α kinases are responsible for the delayed eIF2α phosphorylation in cpc2Δ cells, the effects of deletions of other eIF2α kinases were examined. We found that although the eIF2α phosphorylation kinetics in hri2Δ cells were comparable with wild-type cells, the hri2 deletion in a cpc2Δ background caused the loss of delayed eIF2α phosphorylation (Fig. 2C). These data suggest that prolonged 3AT treatment in cpc2Δ cells activates Hri2 and leads to Hri2-mediated delayed eIF2α phosphorylation. However, the 3AT sensitivity of hri2Δ cpc2Δ double-mutant cells was comparable with cpc2Δ cells (Fig. 2D), suggesting that delayed phosphorylation does not contribute substantially to cell viability under this condition. Collectively, these results indicate that S. pombe Cpc2 facilitates survival under amino acid starvation, presumably by promoting Gcn2-mediated eIF2α phosphorylation.
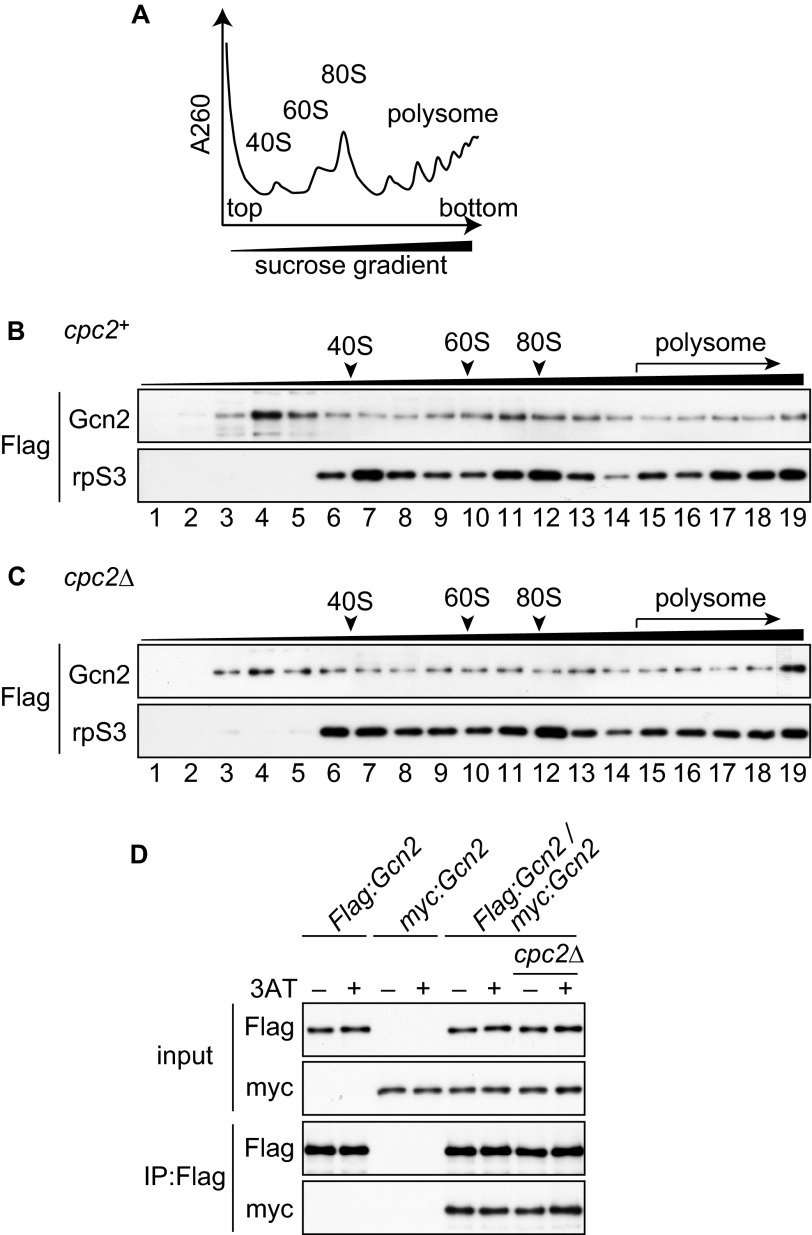
Cpc2 Is Dispensable for Ribosomal Association and Dimerization of Gcn2
Cpc2 and Gcn2 exhibited similar defects in the amino acid starvation response in S. pombe (Figs. 1 and 2). These observations raised the possibility that S. pombe Cpc2 modulates the Gcn2-mediated stress response. S. cerevisiae Gcn2 associates with the ribosome via its carboxyl-terminal domain, and appropriate ribosomal localization potentiates Gcn2 function in translational control (18). This carboxyl-terminal domain is also important for dimerization, which is essential for S. cerevisiae Gcn2 function in vivo (15, 16). Because Cpc2 is also a ribosomal binding protein (29), we first asked whether Cpc2 affects ribosomal binding of Gcn2 in S. pombe. To analyze the association of Gcn2 with ribosomes, we constructed cells in which both Gcn2 and rpS3, a component of the 40 S ribosomal subunit, were tagged with FLAG. Fractionation of whole cell extracts using sucrose gradient centrifugation yields ribosomes distributed along the gradient depending on their sedimentation coefficient (Fig. 3A). We detected a significant amount of Gcn2 in ribosomal fractions (fraction numbers 6–19) from wild-type cells (Fig. 3B), consistent with the idea that S. pombe Gcn2 interacts with ribosomes, as observed in S. cerevisiae. Ribosomal association of Gcn2 remained largely unchanged in cpc2Δ cells (Fig. 3C), suggesting that Gcn2 does not require Cpc2 to interact with ribosomes. We next examined the effect of Cpc2 on Gcn2 dimerization using heterozygous diploid strains expressing Gcn2 tagged with FLAG or Myc from each gcn2 allele. Immunoprecipitation assays revealed that FLAG-tagged Gcn2 associates with Myc-tagged Gcn2 (Fig. 3D), suggesting that S. pombe Gcn2 formed a dimer, as is the case in S. cerevisiae. This interaction was independent of Cpc2 (Fig. 3D). These results indicate that S. pombe Cpc2 does not regulate ribosomal localization or dimerization of Gcn2.
FIGURE 3.
Cpc2 is dispensable for ribosomal binding and dimerization of Gcn2. A, schematic diagram of distribution of ribosomes after fractionation using sucrose gradient centrifugation. The predicted absorbance curve at 260 nm is shown along the gradient. B and C, immunoblots of gradient fractions to detect FLAG-Gcn2 and FLAG-rpS3. Whole cell extracts were prepared from Flag:gcn2 rpS3:Flag (YT3650) (B) or Flag:gcn2 rpS3:Flag cpc2Δ (YT3658) (C) cells, and both Gcn2 and rpS3 were detected using anti-FLAG antibody. D, co-immunoprecipitation (IP) assay of Gcn2 dimerization from Flag:gcn2/gcn2+ diploid (YT4376), myc:gcn2/gcn2+ diploid (YT4281), Flag:gcn2/myc:gcn2 diploid (YT4279), and Flag:gcn2/myc:gcn2 cpc2Δ/cpc2Δ diploid (YT4280) cells treated with 10 mm 3AT 15 min (+) or not (−). Immunoblots were performed using anti-FLAG and anti-Myc antibodies.
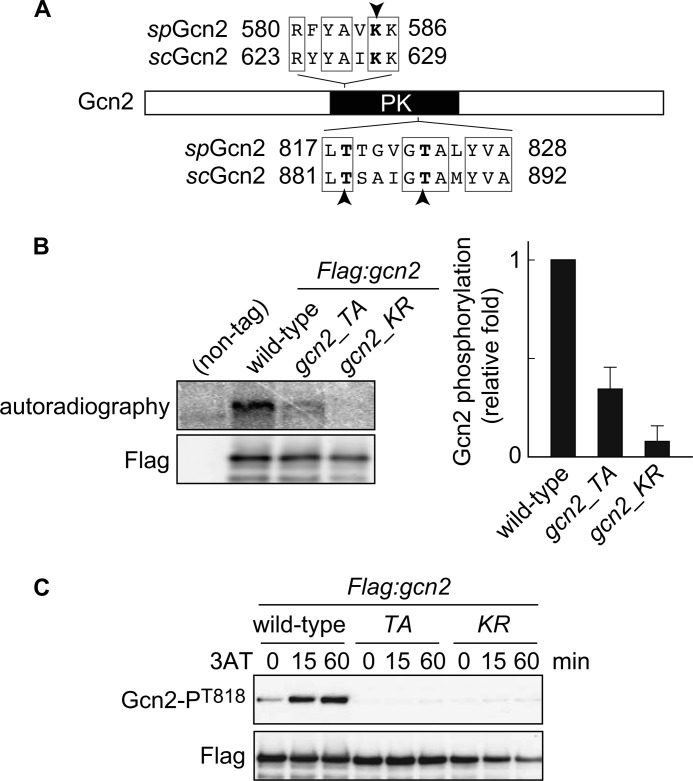
Cpc2 Is Required for Gcn2 Autophosphorylation
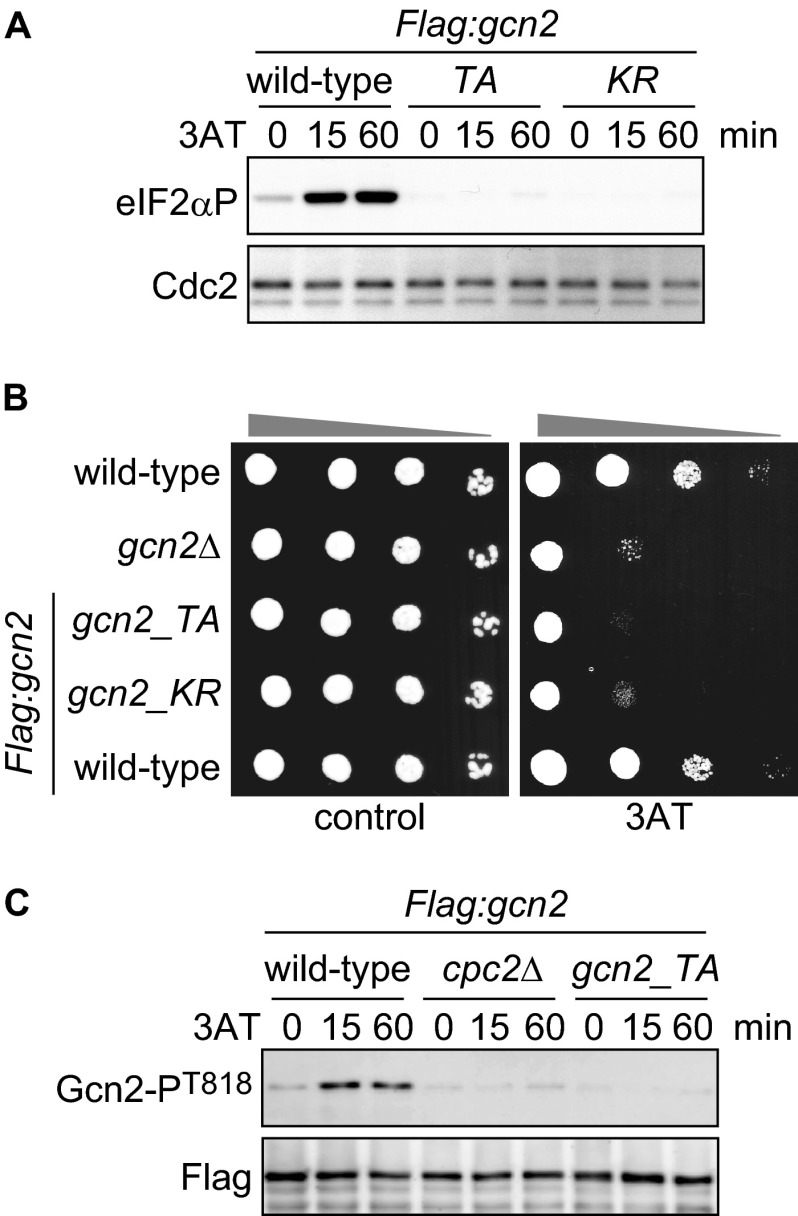
To further investigate how Cpc2 regulates Gcn2-dependent eIF2α phosphorylation, we next focused on autophosphorylation of Gcn2. In S. cerevisiae, autophosphorylation of two threonine residues (Thr-882 and Thr-887) in Gcn2 protein is essential for its full kinase activity (17). To examine its relevance in S. pombe, we generated gcn2_TA cells carrying alanine substitutions at predicted autophosphorylation sites (Thr-818 and Thr-823) corresponding to Thr-882 and Thr-887 in S. cerevisiae Gcn2 (Fig. 4A). As a control, a kinase-dead mutant (gcn2_KR), in which a lysine residue (Lys-585) essential for kinase activity was replaced by arginine, was also created (Fig. 4A). We performed in vitro kinase assays using FLAG-tagged Gcn2 immunoprecipitated from wild-type and mutant S. pombe cells and observed a Gcn2 phosphorylation signal that was dependent on its kinase activity (Fig. 4B, wild-type and gcn2_KR). Moreover, this experiment revealed that the T818A and T823A mutations largely abolished the phosphorylation signal of Gcn2, suggesting that Thr-818 and/or Thr-823 of S. pombe Gcn2 are major target residues that are autophosphorylated in vitro, as shown in S. cerevisiae (17) (Fig. 4B, gcn2_TA). To assess the significance of the phosphorylation in vivo, we raised an anti-Gcn2-phospho-Thr-818 antibody. We detected an increased phosphorylation signal at Thr-818 of Gcn2 in wild-type cells upon 3AT treatment, whereas no phosphorylation was observed in gcn2_TA or gcn2_KR mutant cells (Fig. 4C), supporting the idea that Thr-818 is an autophosphorylation site of S. pombe Gcn2. We also found that the increase in eIF2α phosphorylation under amino acid starvation was lost in gcn2_TA and gcn2_KR mutant cells (Fig. 5A). Concordantly, these mutant cells were highly sensitive to 3AT treatment (Fig. 5B). These results indicate that autophosphorylation is essential for Gcn2 activity and cell survival during amino acid starvation in S. pombe. To investigate whether Cpc2 is required for autophosphorylation of Gcn2, we examined Gcn2 Thr-818 phosphorylation in cpc2Δ cells. Intriguingly, the Gcn2 phospho-Thr-818 signal was virtually abrogated by the cpc2 deletion (Fig. 5C), indicating that S. pombe Cpc2 is required for Gcn2 activation. Taken together, these results demonstrate that S. pombe Cpc2 positively regulates the GAAC response by facilitating Gcn2 autophosphorylation.
FIGURE 4.
Gcn2 is autophosphorylated in vitro and in vivo. A, domain structure of Gcn2 protein. Lys-628 and threonines Thr-882 and Thr-887, indicated by arrowheads, in S. cerevisiae Gcn2 (scGcn2) are essential sites for kinase activity and autophosphorylation, respectively. The corresponding amino acids in S. pombe Gcn2 (spGcn2) are also shown. PK, protein kinase domain. B, in vitro kinase assay using FLAG-Gcn2 immunoprecipitated from cells harboring FLAG-tagged Gcn2 (Flag:gcn2, YT3372) and nontagged control cells (JK317). Autoradiography and immunoblot probed with anti-FLAG antibody are shown. Flag:gcn2_TA cells (YT3657) have alanine substitution at predicted autophosphorylation sites. Flag:gcn2_KR cells (YT3656) are kinase-dead mutants. Quantified Gcn2 phosphorylation signals (Gcn2P/Gcn2) are shown as the relative fold (S.E. (n = 3)) with respect to the level of Flag:gcn2 cells. C, immunoblots of phosphorylated Gcn2 and FLAG (control) from Flag:gcn2 (YT3372), Flag:gcn2_TA (YT3657), and Flag:gcn2_KR (YT3656) cells treated with 10 mm 3AT. FLAG-Gcn2 was immunoprecipitated before immunoblots.
FIGURE 5.
Cpc2 is required for starvation-induced Gcn2 activation. A, immunoblots of phosphorylated eIF2α and Cdc2 (control) from Flag:gcn2 (YT3372), Flag:gcn2_TA (YT3657), and Flag:gcn2_KR (YT3656) cells treated with 10 mm 3AT. B, wild-type (JK317), gcn2Δ (YT3360), Flag:gcn2 (YT3372), Flag:gcn2_TA (YT3657), and Flag:gcn2_KR (YT3656) cells were 10-fold serially diluted and spotted on EMM plates ± 10 mm 3AT. C, immunoblots of anti-FLAG-Gcn2 immunoprecipitates from Flag:gcn2 (YT3372), Flag:gcn2 cpc2Δ (YT3598), and Flag:gcn2_TA (YT3657) cells treated with 10 mm 3AT, probed with anti-phospho-Gcn2 and anti-FLAG (control) antibodies.
Cpc2, but Not snoU24b, Is Important for the GAAC Response
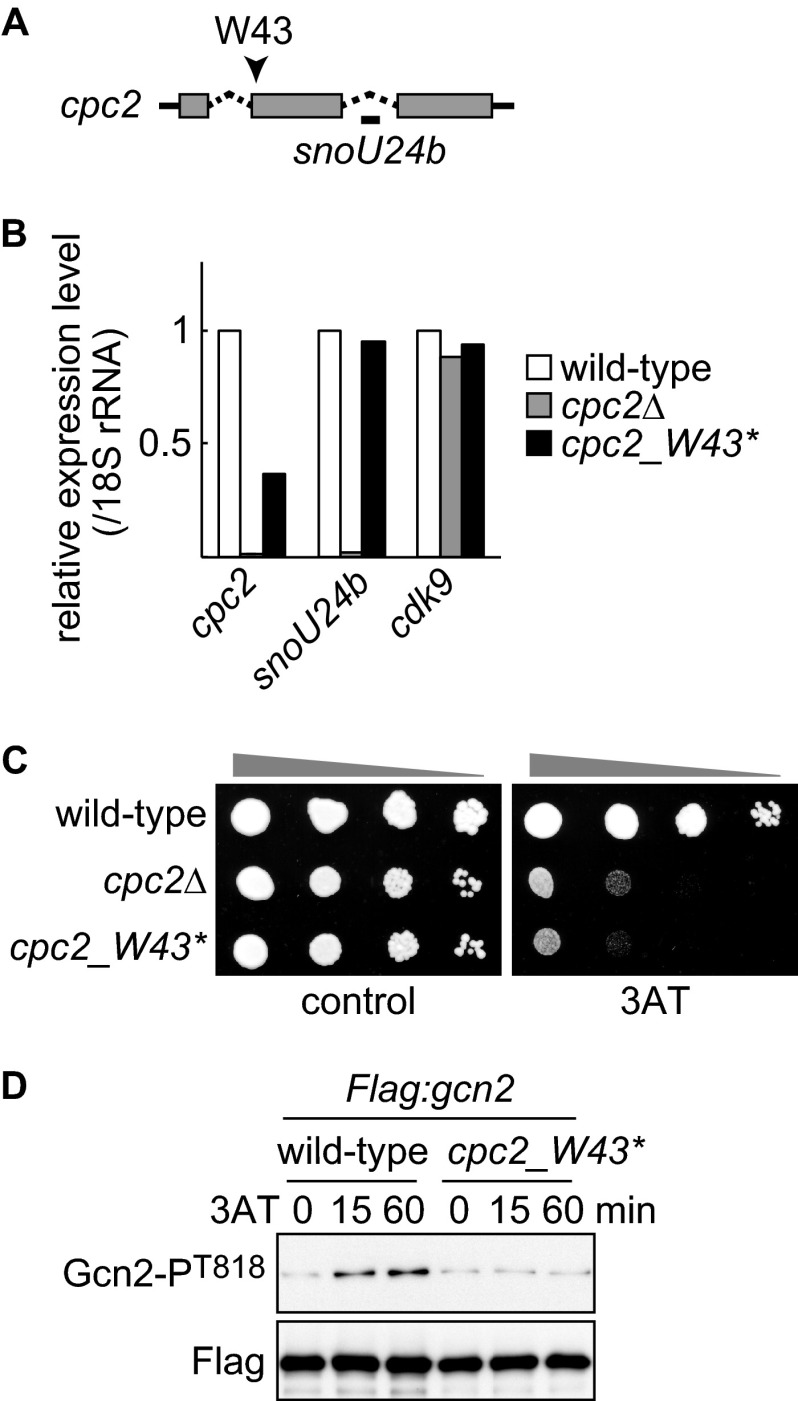
An intron of the S. pombe cpc2 gene contains snoU24b, encoding a C/D box U24 small nucleolar RNA (Fig. 6A). The particular cpc2Δ strain that we used was one in which the cpc2 gene was disrupted by replacement of the first exon with a marker gene (supplemental Fig. S1A). As expected, this strain did not express Cpc2 protein (supplemental Fig. S1, A and B), but it also lost snoU24b expression (Fig. 6B, cpc2Δ). Although most previous studies have not taken this point into account, it has been recently reported that in S. cerevisiae, the ribosome assembly defect (half-mer polysomes) observed in the cells deleted within the asc1-containing genomic region is ascribed to the loss of a small nucleolar RNA carried in an intron of the ASC1 gene (35), underlining the importance of testing whether defective snoU24b is responsible for phenotypes observed in cpc2Δ cells. To that end, we generated cells (cpc2_W43*) harboring a nonsense mutation in place of Trp43 in Cpc2 (Fig. 6A). No Cpc2 protein was detected in cpc2_W43* cells (supplemental Fig. S1, A–C). As expected, the expression level of snoU24b in cpc2_W43* cells was comparable with that in wild-type cells (Fig. 6B). A decrease in cpc2 mRNA abundance is presumably caused by nonsense-mediated mRNA decay. cpc2_W43* cells exhibited a severe 3AT sensitivity similar to that in cpc2Δ cells (Fig. 6C). Moreover, 3AT-induced Gcn2 autophosphorylation was abrogated in cpc2_W43* cells (Fig. 6D), indicating that Cpc2, but not snoU24b, contributes to regulation of Gcn2 and the GAAC response.
FIGURE 6.
Cpc2, but not snoU24b RNA, is important for Gcn2 regulation. A, small nucleolar RNA U24b is encoded within the intron of cpc2 mRNA. Gray box, coding sequence; dashed line, intron; solid line, untranslated region. The position encoding Trp43 is indicated by an arrowhead. B, gene expression levels of cpc2, snoU24b, and cdk9+ (control) were analyzed by quantitative RT-PCR from wild-type (YT3033), cpc2Δ (YT2307), and cpc2_W43* (YT4299) cells. All data are normalized to the expression level of 18 S rRNA, and the average expression level of two independent experiments is shown as the relative fold to those in wild-type cells. C, 10-fold serial dilutions of wild-type (JK317), cpc2Δ (YT3540), and cpc2_W43* (YT4299) cells were spotted on EMM plates ± 10 mm 3AT. D, immunoblots of anti-FLAG-Gcn2 immunoprecipitates from Flag:gcn2 (YT3372) and Flag:gcn2 cpc2_W43* (YT4309) cells treated with 10 mm 3AT, probed with anti-phospho-Gcn2 and anti-FLAG (control) antibodies.
Ribosome-free Cpc2 Has a Role in Regulating Gcn2
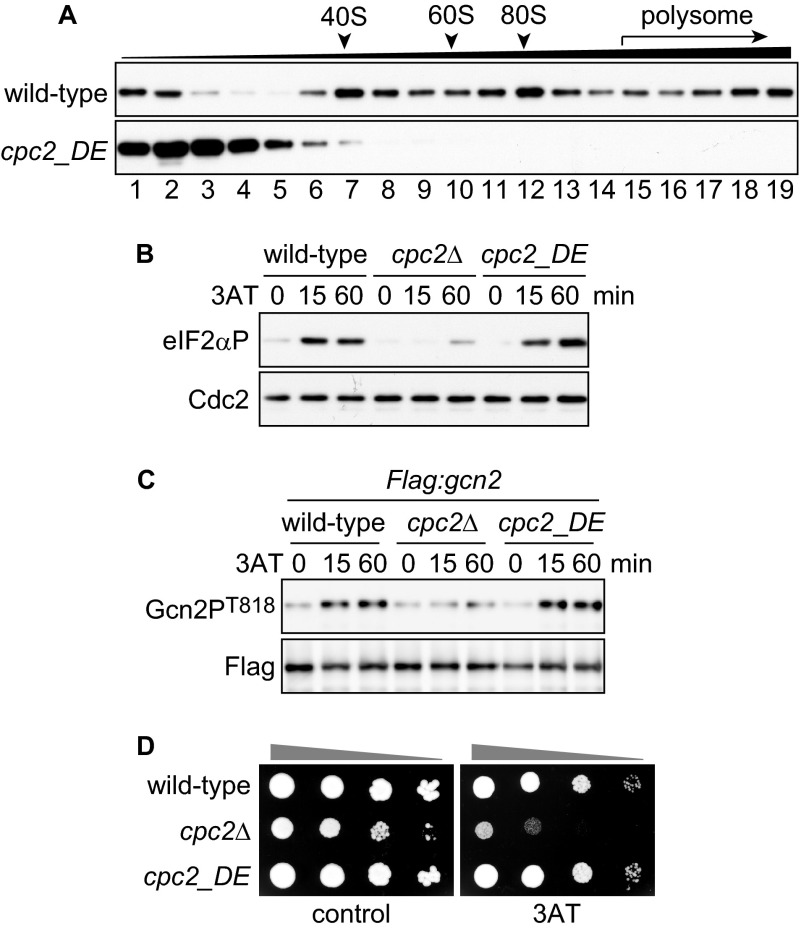
Although Cpc2 does not affect the ribosomal association of Gcn2 (Fig. 3), the fact that both Cpc2 and Gcn2 bind to ribosomes prompted us to investigate whether the association of Cpc2 with ribosomes is a prerequisite for Gcn2 activation. To further examine this point, we prepared ribosome-unbound cpc2_DE mutant cells, in which two amino acids (Arg-36 and Lys-38) responsible for ribosome binding were substituted by aspartic acid and glutamic acid, respectively (30, 42). As reported previously (42), distribution of Cpc2_DE was observed in ribosome-free fractions after fractionation using sucrose gradient centrifugation (Fig. 7A). We examined whether eIF2α phosphorylation and Gcn2 autophosphorylation were abrogated in cpc2_DE mutant cells and found no significant difference in the kinetics or intensities of the phosphorylation signals between wild-type and cpc2_DE mutant cells (Fig. 7, B and C). These results suggest that Cpc2 retains an ability to regulate Gcn2 without ribosomal association. Moreover, cpc2_DE mutant cells exhibited no sensitivity to 3AT treatment (Fig. 7D), supporting the notion that ribosome-unbound Cpc2 is functional in Gcn2 regulation and the GAAC response.
FIGURE 7.
Ribosome-free Cpc2 is able to regulate Gcn2. A, immunoblots of Cpc2 from fractionated samples prepared from Flag:gcn2 (YT3372) and Flag:gcn2 cpc2_DE (YT3559) cells. B, immunoblots of phosphorylated eIF2α and Cdc2 (control) from wild-type (JK317), cpc2Δ (YT3540), and cpc2_DE (YT3542) cells treated with 10 mm 3AT. C, immunoblots of anti-FLAG-Gcn2 immunoprecipitates from Flag:gcn2 (YT3372), Flag:gcn2 cpc2Δ (YT3598), and Flag:gcn2 cpc2_DE (YT3559) cells treated with 10 mm 3AT, probed with anti-phospho-Gcn2 and anti-FLAG (control) antibodies. D, 10-fold serial dilutions of wild-type (JK317), cpc2Δ (YT3540), and cpc2_DE (YT3542) cells were spotted on EMM plates ± 10 mm 3AT.
RACK1 Homologs Compensate for the Defects in the Regulation of Gcn2 in S. pombe cpc2 Mutant Cells
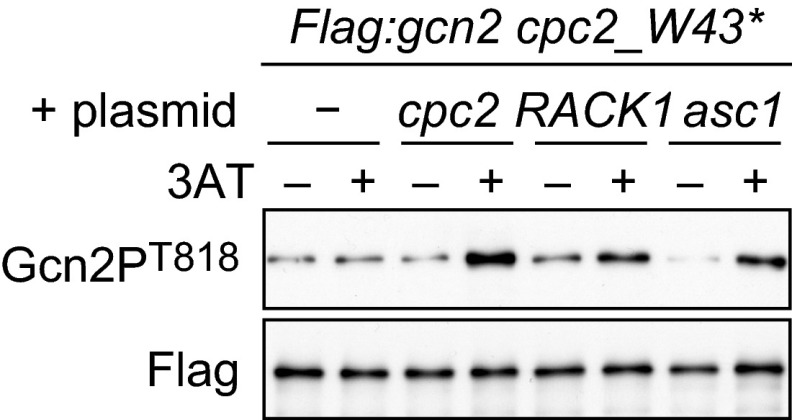
As mentioned above, S. pombe Cpc2 facilitates the Gcn2-mediated GAAC response. By contrast, deletion of S. cerevisiae Asc1 promotes the GAAC response (34). To address this discrepancy between the two species, we investigated whether S. cerevisiae Asc1 and human RACK1 rescue the Gcn2 defects in S. pombe cpc2Δ cells. We cloned cDNA of cpc2, asc1, and RACK1 genes in an expression plasmid and introduced them individually into cpc2 mutant cells. The expression levels of Asc1 and RACK1 proteins were similar to the level of Cpc2 (supplemental Fig. S2). Immunoblotting of the phosphorylated Gcn2 in the cells harboring the expression plasmids revealed that either asc1 or RACK1 expression restored 3AT-induced Gcn2 phosphorylation in cpc2 mutant cells (Fig. 8), indicating that Asc1 and RACK1 play the same role as Cpc2 in the regulation Gcn2 in S. pombe. These observations suggest that the phenotypic difference between S. pombe and S. cerevisiae lies in other components in the system regulating the GAAC response.
FIGURE 8.
RACK1 homologs compensate for the defects in the regulation of Gcn2. Flag:gcn2 cpc2_W43* (YT4309) cells harboring an empty plasmid (−) or the expression plasmids (cpc2, RACK1, or asc1) were treated with 10 mm 3AT 60 min (+) or not (−). Immunoblots of anti-FLAG-Gcn2 immunoprecipitates were probed with anti-phospho-Gcn2 and anti-FLAG (control) antibodies.
DISCUSSION
It has been reported that RACK1 homologs are involved in the GAAC response in S. cerevisiae and N. crassa. However, the molecular mechanisms of action of the RACK1 homologs in the GAAC response have not been fully elucidated. In this study, we have focused on S. pombe Cpc2 in the GAAC response. Our findings demonstrate that deletion of S. pombe cpc2 impairs 3AT-induced eIF2α phosphorylation and subsequent expression of amino acid biosynthesis genes and that cpc2Δ cells are highly sensitive to 3AT treatment. Moreover, S. pombe Cpc2 is required for autophosphorylation of Gcn2. Thus, S. pombe Cpc2 positively contributes to the GAAC response through the activation of eIF2α kinase Gcn2.
Amino acid sequences among RACK1 homologs (mammalian RACK1, S. pombe Cpc2, S. cerevisiae Asc1, and N. crassa Cpc-2) are highly conserved, and they show >70% similarity (25, 34). Indeed, ectopic expression of mammalian RACK1 compensated several defects observed in S. pombe cpc2Δ cells (25) and S. cerevisiae asc1Δ cells (43). Moreover, both S. pombe and N. crassa devoid of cpc2 (cpc-2) exhibit defects in fertility (25, 37), supporting the idea that RACK1 homologs phylogenetically share a similar function. On the other hand, the requirement of RACK1 homologs in the GAAC response seems to differ among species. In S. pombe, Cpc2 promotes the GAAC response as shown in this work. By contrast, S. cerevisiae Asc1 has an apparently opposite effect on the GAAC response as Asc1 physically interacts with the eIF3 complex to facilitate efficient translation initiation, which suppresses the GAAC response (35). In N. crassa, the cpc-2 U142 mutation causes defects in amino acid deprivation-induced gene expression (36), suggesting that N. crassa Cpc-2 contributes to the GAAC response in a similar fashion to S. pombe Cpc2. However, because ectopic expression of the N. crassa cpc-2 U142 allele in S. cerevisiae asc1Δ cells causes the defect in the GAAC response (34), one cannot exclude the possibility that cpc-2 U142 produces an allele-specific effect. Our rescue experiment revealed that the ectopic expression of S. cerevisiae asc1 and human RACK1 restored 3AT-induced Gcn2 phosphorylation in cpc2 mutant cells. This observation suggests that the discrepancy between the function of RACK1 homologs in S. cerevisiae and S. pombe is not due to different structures intrinsic to each of the RACK1 homologs, but rather to Gcn2 or other upstream factors mediating the activation signal of Gcn2. Thus, although RACK1 homologs have a conserved function, additional factor(s) may modify their function in the GAAC response in different species.
In S. pombe cpc2Δ cells, but not in gcn2Δ cells, Hri2-dependent delayed phosphorylation was observed. Hri2 is activated in response to oxidative stress (11). Prolonged 3AT treatment causes the accumulation of reactive oxygen species in the cells, and Gcn2 counters this by inducing the expression of antioxidant genes (44). Moreover, it is known that Cpc2 is also involved in the defense against oxidative stress by regulating the amount of Atf1 protein (42), which is an important transcriptional factor for stress-activated MAP kinase-dependent induction of stress-responsive genes. These results raised the possibility that defects in both Gcn2-mediated and Atf1-mediated responses in cpc2Δ cells cause oxidative stress in the cells, thence leading to the activation of Hri2. However, the delayed phosphorylation was not observed in gcn2Δ atf1Δ double-mutant cells,4 suggesting that Cpc2 may suppress the delayed eIF2α phosphorylation through yet unknown mechanisms.
RACK1 homologs across many eukaryotic species generally locate at the head of the 40 S ribosomal subunit near the mRNA exit site on 80 S ribosomes (30). Ribosome-localized RACK1 promotes translational initiation (31, 35) or mediates translation of specific mRNAs through the interaction with RNA-binding protein (33). Indeed, S. pombe cpc2 deletion causes the reduction of steady-state levels of several proteins, some of which are regulated at the translational level by ribosome-bound Cpc2 (29, 42). On the other hand, RACK1 homologs also have ribosome localization-independent roles (33, 45). In our study of cpc2_DE mutant cells, ribosomal association of Cpc2 was dispensable for regulation of Gcn2 and survival under amino acid starvation. This result suggests that the defect in Gcn2-mediated responses observed in cpc2Δ cells is not simply caused by the reduction of steady-state translation. Rather, it is conceivable that Cpc2 modulates an activity or interaction with regulatory proteins of Gcn2 to promote Gcn2 activation under stress conditions.
The observation that ribosome-free Cpc2 is functional for regulation of Gcn2 is intriguing because most of the Cpc2 and Gcn2 in the cell interact with ribosomes, as shown in our experiments. There are two plausible explanations for Gcn2 regulation by ribosome-unbound Cpc2. In one scenario, free Cpc2 may modulate the activity of Gcn2 at extra-ribosomal sites prior to ribosomal loading of Gcn2. Alternatively, Cpc2 may regulate Gcn2 on ribosomes. It has been thought that Gcn2 associates with the 60 S ribosomal subunit near the decoding A-site, which is distant from the Cpc2-binding site. If Cpc2 interacts with Gcn2, it seems likely that Cpc2 transiently dissociates from its binding site to access Gcn2 on ribosomes. Posttranslational modifications modulate Gcn2 activity. TOR (target of rapamycin), an important kinase for nutrient response, indirectly regulates phosphorylation of Ser-577 of S. cerevisiae Gcn2, which inhibits Gcn2 activation (20, 46). Moreover, Snf1, an ortholog of mammalian AMP-activated kinase, promotes Gcn2 activation under amino acid starvation by an unknown mechanism in S. cerevisiae (23). These observations indicate that, besides uncharged tRNA, other signals including posttranslational modifications modulate Gcn2 activity. RACK1 homologs function as a scaffold in signal transduction (26, 32). Thus, although it is unknown whether Cpc2 and Gcn2 interact directly in S. pombe, Cpc2 may transmit an activating signal from an upstream factor to Gcn2 in ribosomal or extra-ribosomal contexts. Precisely how and where Cpc2 regulates Gcn2 in the GAAC response will be explored in future analyses.
Supplementary Material
Acknowledgments
We are grateful to M. Kitabatake and M. Ohno for polysome fractionation, T. Nakamura, P. Russell, and T. Inada for materials and advice, J. Hejna for critical reading of the manuscript, and laboratory members for support.
This work was supported by a Kyoto University start-up grant-in-aid for young scientists (to Y. T.) and a grant-in-aid for cancer research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to F. I.).

This article contains supplemental Figs. S1 and S2.
Y. Tarumoto, J. Kanoh, and F. Ishikawa, unpublished data.
- GAAC
- general amino acid control
- RACK1
- receptor for activated C-kinase
- 3AT
- 3-aminotriazole
- EMM
- Edinburgh minimal medium.
REFERENCES
- 1. Sonenberg N., Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spriggs K. A., Bushell M., Willis A. E. (2010) Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40, 228–237 [DOI] [PubMed] [Google Scholar]
- 3. Holcik M., Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 4. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 5. Mueller P. P., Hinnebusch A. G. (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45, 201–207 [DOI] [PubMed] [Google Scholar]
- 6. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., Hinnebusch A. G., Marton M. J. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell Biol. 21, 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ameri K., Harris A. L. (2008) Activating transcription factor 4. Int. J. Biochem. Cell Biol. 40, 14–21 [DOI] [PubMed] [Google Scholar]
- 10. Udagawa T., Nemoto N., Wilkinson C. R., Narashimhan J., Jiang L., Watt S., Zook A., Jones N., Wek R. C., Bähler J., Asano K. (2008) Int6/eIF3e promotes general translation and Atf1 abundance to modulate Sty1 MAPK-dependent stress response in fission yeast. J. Biol. Chem. 283, 22063–22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhan K., Narasimhan J., Wek R. C. (2004) Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics 168, 1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 13. Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6, 269–279 [DOI] [PubMed] [Google Scholar]
- 14. Wek S. A., Zhu S., Wek R. C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell Biol. 15, 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu H., Garcia-Barrio M. T., Hinnebusch A. G. (1998) Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell Biol. 18, 2697–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narasimhan J., Staschke K. A., Wek R. C. (2004) Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 279, 22820–22832 [DOI] [PubMed] [Google Scholar]
- 17. Romano P. R., Garcia-Barrio M. T., Zhang X., Wang Q., Taylor D. R., Zhang F., Herring C., Mathews M. B., Qin J., Hinnebusch A. G. (1998) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell Biol. 18, 2282–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu S., Wek R. C. (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J. Biol. Chem. 273, 1808–1814 [DOI] [PubMed] [Google Scholar]
- 19. Padyana A. K., Qiu H., Roll-Mecak A., Hinnebusch A. G., Burley S. K. (2005) Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2α protein kinase GCN2. J. Biol. Chem. 280, 29289–29299 [DOI] [PubMed] [Google Scholar]
- 20. Garcia-Barrio M., Dong J., Cherkasova V. A., Zhang X., Zhang F., Ufano S., Lai R., Qin J., Hinnebusch A. G. (2002) Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2α kinase activities of GCN2. J. Biol. Chem. 277, 30675–30683 [DOI] [PubMed] [Google Scholar]
- 21. Sattlegger E., Swanson M. J., Ashcraft E. A., Jennings J. L., Fekete R. A., Link A. J., Hinnebusch A. G. (2004) YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 279, 29952–29962 [DOI] [PubMed] [Google Scholar]
- 22. Sattlegger E., Hinnebusch A. G. (2005) Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 280, 16514–16521 [DOI] [PubMed] [Google Scholar]
- 23. Cherkasova V., Qiu H., Hinnebusch A. G. (2010) Snf1 promotes phosphorylation of the α subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell Biol. 30, 2862–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baum S., Bittins M., Frey S., Seedorf M. (2004) Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 380, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McLeod M., Shor B., Caporaso A., Wang W., Chen H., Hu L. (2000) Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell Biol. 20, 4016–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 27. Robles M. S., Boyault C., Knutti D., Padmanabhan K., Weitz C. J. (2010) Identification of RACK1 and protein kinase Cα as integral components of the mammalian circadian clock. Science 327, 463–466 [DOI] [PubMed] [Google Scholar]
- 28. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 29. Shor B., Calaycay J., Rushbrook J., McLeod M. (2003) Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 278, 49119–49128 [DOI] [PubMed] [Google Scholar]
- 30. Sengupta J., Nilsson J., Gursky R., Spahn C. M., Nissen P., Frank J. (2004) Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 11, 957–962 [DOI] [PubMed] [Google Scholar]
- 31. Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhäuser N., Marchisio P. C., Biffo S. (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 32. Nilsson J., Sengupta J., Frank J., Nissen P. (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coyle S. M., Gilbert W. V., Doudna J. A. (2009) Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell Biol. 29, 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann B., Mösch H. U., Sattlegger E., Barthelmess I. B., Hinnebusch A., Braus G. H. (1999) The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 31, 807–822 [DOI] [PubMed] [Google Scholar]
- 35. Kouba T., Rutkai E., Karásková M., Valášek L. (2012) The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res. 40, 2683–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krüger D., Koch J., Barthelmess I. B. (1990) cpc-2, a new locus involved in general control of amino acid synthetic enzymes in Neurospora crassa. Curr. Genet. 18, 211–215 [DOI] [PubMed] [Google Scholar]
- 37. Müller F., Krüger D., Sattlegger E., Hoffmann B., Ballario P., Kanaan M., Barthelmess I. B. (1995) The cpc-2 gene of Neurospora crassa encodes a protein entirely composed of WD-repeat segments that is involved in general amino acid control and female fertility. Mol. Gen. Genet. 248, 162–173 [DOI] [PubMed] [Google Scholar]
- 38. Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 39. Basi G., Schmid E., Maundrell K. (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131–136 [DOI] [PubMed] [Google Scholar]
- 40. Kanoh J., Sadaie M., Urano T., Ishikawa F. (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 15, 1808–1819 [DOI] [PubMed] [Google Scholar]
- 41. Gómez E. B., Espinosa J. M., Forsburg S. L. (2005) Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol. Cell Biol. 25, 8887–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Núñez A., Franco A., Madrid M., Soto T., Vicente J., Gacto M., Cansado J. (2009) Role for RACK1 orthologue Cpc2 in the modulation of stress response in fission yeast. Mol. Biol. Cell 20, 3996–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerbasi V. R., Weaver C. M., Hill S., Friedman D. B., Link A. J. (2004) Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell Biol. 24, 8276–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nemoto N., Udagawa T., Ohira T., Jiang L., Hirota K., Wilkinson C. R., Bähler J., Jones N., Ohta K., Wek R. C., Asano K. (2010) The roles of stress-activated Sty1 and Gcn2 kinases and of the protooncoprotein homologue Int6/eIF3e in responses to endogenous oxidative stress during histidine starvation. J. Mol. Biol. 404, 183–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Warner J. R., McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cherkasova V. A., Hinnebusch A. G. (2003) Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17, 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.