Abstract
The phloem provides a unique niche for several organisms. Aphids are a large group of Hemipteran insects that utilize stylets present in their mouthparts to pierce sieve elements and drink large volumes of phloem sap. In addition, many aphids also vector viral diseases. Myzus persicae, commonly known as the green peach aphid (GPA), is an important pest of a large variety of plants that includes Arabidopsis thaliana. This review summarizes recent studies that have exploited the compatible interaction between Arabidopsis and GPA to understand the molecular and physiological mechanisms utilized by plants to control aphid infestation, as well as genes and mechanisms that contribute to susceptibility. In addition, recent efforts to identify aphid-delivered elicitors of plant defenses and novel aphid salivary components that facilitate infestation are also discussed.
Keywords: green peach aphid, effectors, Hemiptera, phloem-feeding insect, plant defense mechanisms, susceptibility factors
Introduction
The phloem, which provides a conduit for resource distribution and signaling, also provides a niche for some organisms. However, for these organisms the phloem also provides several challenges in that the phloem sap is under high pressure, has a high C:N ratio and a high osmolarity due to elevated sugar content. Furthermore, when ruptured or punctured the sieve elements are prone to occlusion. Aphids (Hemiptera: Aphididae) constitute a large group of “piercing-sucking” class of insects that have adapted to feeding from sieve elements (Pollard, 1973; Blackman and Eastop, 2000; Walling, 2000). Nearly 250 amongst the ~4000 aphid species that have been described are considered as pests (Dixon, 1998; Blackman and Eastop, 2000). Damage to the plant results from loss of phloem sap and changes in source-sink patterns as a consequence of which nutrient flow to the primary growth zones is reduced (Mittler and Sylvester, 1961; Girousse et al., 2005). Some aphids also vector viral diseases of plants, thereby causing further loss of plant productivity and quality (Kennedy et al., 1962; Matthews, 1991; Dixon, 1998). Viral infection in the host plant can further influence severity of aphid infestation (Ziebell et al., 2011; Gutiérrez et al., 2013).
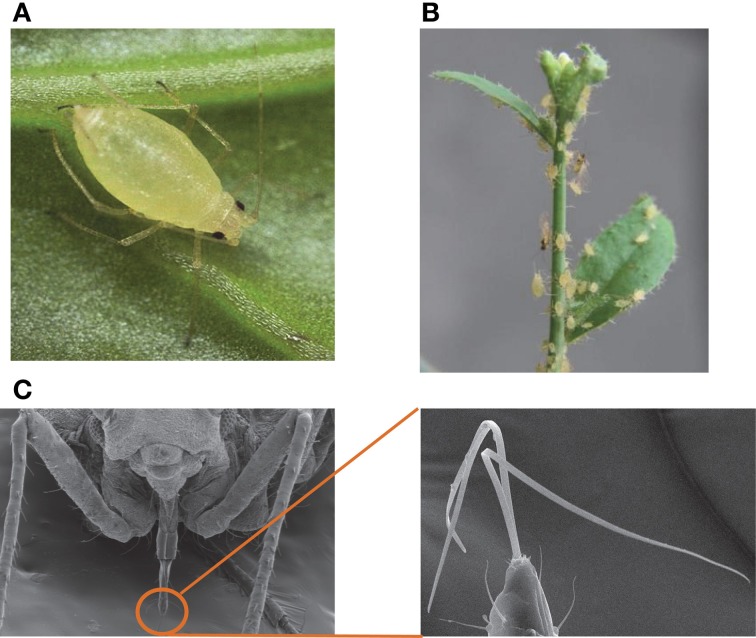
Aphids can be broadly classified as specialists or generalists (Lankau, 2007). Specialists like the cabbage aphid (Brevicoryne brassicae) and the mustard aphid (Lipaphis erysimi) have a limited host range that is restricted to cruciferous plants. In contrast, as described below, a generalist like the green peach aphid (GPA; Myzus persicae Sulzer) (Figures 1A,B) feeds on a large variety of plants belonging to different families (Blackman and Eastop, 2000; Lankau, 2007). The mouthparts of aphids are modified into slender stylets (Figure 1C), which enable penetration of the sieve element to consume phloem sap. Present within each stylet is a salivary canal through which saliva is released into the plant tissue, and a food canal through which the insect uptakes phloem sap. The predominantly intercellular route taken by stylets, combined with the activity of aphid salivary components, minimizes physical damage to the plant tissue, thus averting substantial wounding-related responses from the plant (Miles, 1999; Walling, 2000; Tjallingii, 2006). In addition, the aphid saliva also contains factors that have been suggested to prevent or reverse sieve element occlusion (SEO) (Will et al., 2007, 2009). The aphid stylets occasionally may pierce host cells, seemingly to ingest/sample minute amounts of plant material (Tjallingii, 1990, 2006). These interactions of the aphid stylets with the sieve elements and plant cells also provide an interface for exchange of metabolites and macromolecules between the aphid and the plant that potentially could promote or deter colonization.
Figure 1.
Green peach aphid on Arabidopsis. (A) Green peach aphid on Chinese cabbage (Brassica rapa var chinensis). Images by Nick Sloff. (B) Green peach aphid on Arabidopsis thaliana. (C) Mouthparts of aphid. Left panel: SEM image of showing aphid mouthpart; Right panel: Aphid stylet. Images provided by John Diaz-Montano. The above images were adapted with permission from Louis et al. (2012c) Arabidopsis thaliana—Aphid Interaction. The Arabidopsis Book (First published on May 22, 2012:e0159. doi: 10.1199/tab.0159). Copyright American Society of Plant Biologists (thearabidopsisbook.org).
Green peach aphid
The host range of the GPA comprises over 400 plant species belonging to nearly 50 plant families, including important crops like potato and sugar beet, stone fruits (e.g., peach, almond, and cherry), and horticultural crops in the Brassicaceae, Solanaceae and Cucurbitaceae families (Blackman and Eastop, 2000). More than 100 viral diseases are vectored by GPA, which is the most important virus vector on vegetable crops (Kennedy et al., 1962; Matthews, 1991). These characteristics, combined with the capacity of the GPA population to rapidly increase in size and GPA's resistance to a large number of insecticides (Georghiou and Lagunes-Tejada, 1991; Vasquez, 1995; Devonshire et al., 1998; Silva et al., 2012a,b) has resulted in GPA being categorized amongst the top three agricultural pests in the USA (Klein and Waterhouse, 2000).
GPA is capable of sexual and asexual (parthenocarpic) reproduction. Asexual reproduction is characterized by a telescopic generation in which an adult female contains embryos that themselves contain embryos. In nature, the GPAs life cycle includes primary and secondary hosts. The sexual cycle is completed on the primary host, which comprise stone fruits like peach (Prunus persicae), Canadian plum (P. nigra), black cherry (P. serotina), and dwarf Russian almond (P. tenella) (Blackman and Eastop, 2000). Secondary hosts include a large variety of plants, including potato, tomato, eggplant, lettuce, celery, mustard, cabbage, radish, and squash (Blackman and Eastop, 2000).
The near completion of the GPA genome sequence (http://tools.genouest.org/tools/myzus/; Ramsey et al., 2007) combined with the development of plant-delivered RNA-interference (RNAi) technology for gene silencing in GPA (Pitino et al., 2011; Pitino and Hogenhout, 2013), have made available new genomic resources and powerful reverse-genetic tools that have begun to facilitate understanding the function of GPA genes and their contribution to plant-aphid interaction.
Salivary effectors promote insect performance on plant
Once the aphid initiates feeding on the host plant it delivers salivary secretions into the plant tissue, which potentially allow the aphid to circumvent plant defenses. Aphids produce two kinds of salivary secretions: gelling saliva and watery saliva (Miles, 1999). The gelling saliva, which is secreted when the stylet is penetrating the host tissue but outside the sieve element, forms a sheath around the stylet that likely facilitates stylet movement through the plant tissue and minimizes damage to the stylet tip. In addition, the salivary sheath helps rapidly seal the wound caused by aphid stylet penetration, thus minimizing the likelihood of wounding-response activation by the host. On the other hand, the watery saliva, which is intermittently released in the plant tissue when the insect is feeding, contains factors that enable the aphid to prevent and maybe also reverse phloem occlusion, thus allowing the insect to feed continuously from a single sieve element (Miles, 1999; Will et al., 2007, 2009). The saliva also contain effectors that manipulate host physiology to facilitate colonization (Rodriguez and Bos, 2013). Publicly available salivary gland ESTs have been utilized to identify GPA effectors that promote colonization (Bos et al., 2010). The salivary protein MpC002 when transiently expressed in Nicotiana benthamiana or expressed in transgenic Arabidopsis thaliana was found to enhance GPA colonization (Bos et al., 2010; Pitino and Hogenhout, 2013). By contrast, when MpC002 expression in the GPA was silenced by allowing insects to feed on N. benthamiana leaves that were transiently expressing a dsRNA construct, or in transgenic Arabidopsis stably expressing dsRNA, insect fecundity was significantly reduced (Pitino et al., 2011; Pitino and Hogenhout, 2013), thus indicating that the MpC002 facilitates infestation. Similarly, in pea aphid (Acyrthosiphon pisum) silencing of the homologous C002 gene had detrimental effects on the insect's ability to colonize plants (Mutti et al., 2008). However, unlike MpC002, expression of the pea aphid C002 in Arabidopsis had no effect on GPA fecundity, suggesting specificity in the role of these orthologous genes in promoting aphid infestation (Pitino and Hogenhout, 2013).
PIntO1 (Progeny Increase to Overexpression 1; also known as Mp1) and PIntO2 are two other putative salivary protein-encoding GPA genes that have been suggested to facilitate GPA colonization on Arabidopsis. Compared to non-transgenic plants, the number of nymphs produced was larger on transgenic plants expressing PIntO1 or PIntO2 from the phloem-specific AtSUC2 promoter (Pitino and Hogenhout, 2013). However, GPA colonization was not impacted when the pea aphid homologs (ApPIntO1 and ApPIntO2) were similarly expressed in Arabidopsis (Pitino and Hogenhout, 2013), thus suggesting that the GPA PIntO1 and PIntO2 proteins specifically target an Arabidopsis factor/mechanism that is involved in promoting GPA fecundity. Plant-delivered RNAi silencing of PIntO2 expression in GPA had a detrimental effect on the insect's ability to replicate on Arabidopsis, further indicating that PIntO2, which is delivered into the plant by the GPA, is essential for promoting insect multiplication on Arabidopsis. Although the biochemical function of MpC002, PIntO1, and PIntO2 are not known, the above studies indicate that the GPA saliva contains factors that likely manipulate host physiology, thus allowing the insect to better adapt to the host plant and promote reproduction.
The arabidopsis thaliana-green peach aphid pathosystem: a model system for understanding plant defense and susceptibility against aphids
Arabidopsis has been used as a model plant by researchers to study plant growth, development and response to stress (Koornneef and Meinke, 2010). Advantages offered by Arabidopsis for molecular-genetic studies include its small size, short generation time, completely sequenced genome and the ease with which it can be transformed (Meinke et al., 1998; Koornneef and Meinke, 2010). The compatible interaction between Arabidopsis and the GPA (Figure 1B) has been successfully utilized to characterize plant response against phloem-feeding insects and to identify plant genes and mechanisms that contribute to defense and susceptibility to these phloem sap-consuming insects (Table 1 and Figure 2) (Louis et al., 2012c). In addition, this pathosystem has also been utilized to study natural genetic variation amongst Arabidopsis accessions to identify quantitative trait loci that influence this interaction (Cabrera y Poch et al., 1998).
Table 1.
Arabidopsis mutants that impact green peach aphid colonization.
| AtG No. | Mutant | Name/function | References |
|---|---|---|---|
| DEFENSE SIGNALING | |||
| At5g05170 | cev1 | constitutive expression of VSP1a | Ellis et al., 2002a |
| At2g39940 | coi1 | coronatine-insensitive1 | Ellis et al., 2002a |
| At5g03280 | ein2 | ethylene-insensitive 2 | Kettles et al., 2013 |
| At1g66340 | etr1 | ethylene response 1 | Mewis et al., 2005, 2006; Kettles et al., 2013 |
| At3g23250 | mby15 | Myb domain protein | Liu et al., 2010 |
| At3g28910 | myb30 | Myb domain protein | Liu et al., 2010 |
| At5g67300 | myb44 | Myb domain protein | Liu et al., 2010 |
| At1g18570 | mby51 | Myb domain protein | Liu et al., 2010 |
| At4g37260 | myb73 | Myb domain protein | Liu et al., 2010 |
| At3g06490 | myb108 | Myb domain protein | Liu et al., 2010 |
| At1g64280 | npr1 | non-expresser of PR genes1 | Mewis et al., 2005 |
| At3g52430 | pad4 | phytoalexin-deficient4b | Pegadaraju et al., 2005, 2007; Louis et al., 2012a |
| At5g13330 | rap2.6L | AP2 domain protein | Liu et al., 2010 |
| At1g67030 | zfp6 | Zinc-finger protein | Liu et al., 2010 |
| GLUCOSINOLATE METABOLISM | |||
| At5g60890 | atr1D | altered tryptophan regulation1 | Kim et al., 2008 |
| At4g39950 At2g22330 | cyp79B2 cyp79B3 | Double mutant is deficient in indole-glucosinolates | Kim et al., 2008 |
| At5g57220 | cyp81F2 | cytochrome P450 monooxygenase | Pfalz et al., 2009 |
| At3g09710 | iqd1 | IQ-Domain1 | Levy et al., 2005 |
| LIPID METABOLISM | |||
| At3g01420 | α-dox1 | α-dioxygenase1 | Avila et al., 2013 |
| At3g11170 | fad7 | fatty acid desaturase7 | Avila et al., 2012 |
| At3g22400 | lox5 | lipoxygenase 5 (9-lipoxygenase) | Nalam et al., 2012 |
| At5g14180 | mpl1 | Myzus persicae-induced lipase1 | Louis et al., 2010a |
| At2g43710 | ssi2 | stearoyl-ACP desaturase | Pegadaraju et al., 2005 |
| CARBOHYDRATE METABOLISM | |||
| At2g18700 | tps11 | trehalose-6-phosphate synthase11 | Singh et al., 2011 |
| At1g10550 | xth33 | xyloglucan:xyloglucosyl transferase33 | Divol et al., 2007 |
| SENESCENCE AND OXIDATIVE BURST | |||
| At5g64930 | cpr5 | constitutive expression of PR genes5 | Pegadaraju et al., 2005 |
| At5g47910 | rbohd | respiratory burst oxidase homolog D | Miller et al., 2009 |
| PHLOEM FUNCTION | |||
| At4g19840 | pp2-A1 | phloem protein 2A1 | Zhang et al., 2011 |
| SMALL RNA GENE SILENCING PATHWAY | |||
| At1g01040 | dcl1 | dicer-like1 | Kettles et al., 2013 |
| At1g09700 | hyl1 | hyponastic leaves 1 | Kettles et al., 2013 |
| At4g20910 | hen1 | hua enhancer1 | Kettles et al., 2013 |
| At3g05040 | hst | hasty | Kettles et al., 2013 |
| At2g27100 | se | serrate | Kettles et al., 2013 |
| At1g48410 | ago1 | argonaute1 | Kettles et al., 2013 |
CEV1 is involved in cellulose metabolism. JA and ethylene signaling are hyperactive in the cev1 mutant. JA signaling is required for the enhanced resistance phenotype of the cev1 mutant.
Although PAD4 is associated with SA signaling and camalexin metabolism, PAD4's involvement in controlling GPA colonization is independent of SA signaling and camalexin metabolism.
Figure 2.
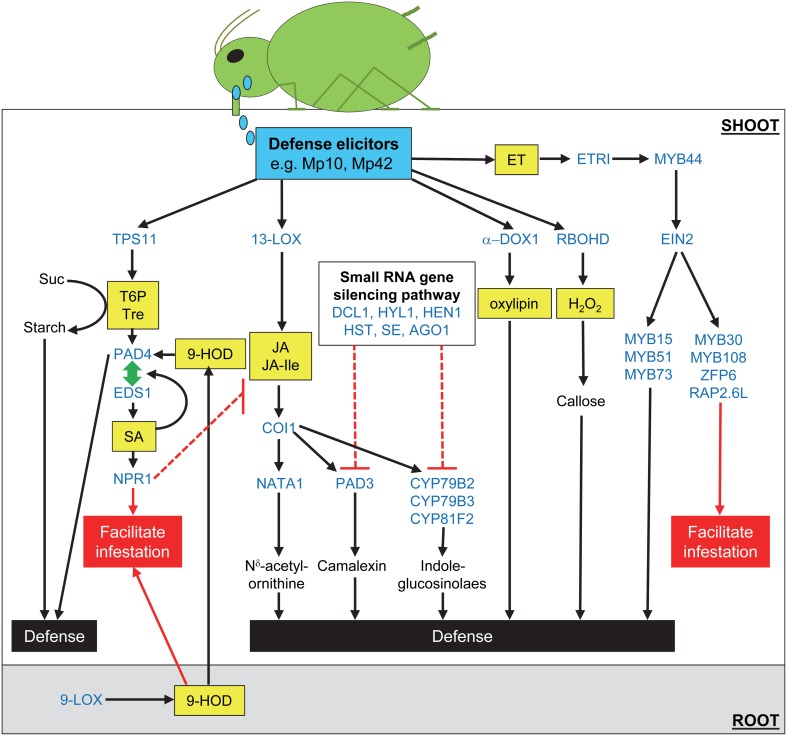
Model depicting relationship between genes and mechanisms that influence Arabidopsis interaction with the green peach aphid. Green peach aphid (GPA) salivary secretions contain effectors that promote infestation, as well as elicitors (e.g., Mp10 and Mp42) that are recognized by the host to turn on defense responses. GPA infestation on the shoot results in the induction of LOX5 expression in roots and a concomitant increase in the levels of LOX5-derived oxylipins (e.g., 9-HOD). LOX5 expression is likely induced by a GPA infestation-induced factor that is translocated from the leaves to the roots. The LOX5-derived oxylipins are transported from the roots to the shoots where one or more of these oxylipins stimulate expression of the defense regulatory gene, PAD4. A PAD4-dependent mechanism adversely impacts GPA settling, feeding and fecundity on Arabidopsis. PAD4 expression is further stimulated by the trehalose (Tre) metabolic pathway. GPA infestation results in the elevated expression of TPS11, which encodes an enzyme with Trehalose-6-phosphate (T6P) synthase and T6P phosphatase activities that is required for promoting PAD4 expression in GPA-infested plants. TPS11 also promotes accumulation of starch at the expense of sucrose (Suc), which is a major feeding stimulant, thereby generating a secondary sink that is detrimental to the insect's ability to colonize Arabidopsis. TPS11 and PAD4 are also required for accumulation of an antibiosis factor in the petiole exudates that limits insect fecundity. However, the GPA has evolved mechanisms that over time spent on the plant suppress this TPS11/PAD4-determined antibiosis activity. The GPA has also evolved to utilize one or more of the 9-LOX-derived oxylipins, or products thereof, as cues to stimulate feeding from phloem and xylem, and enhance fecundity. These oxylipins, which are consumed by the insect from the plant, likely induce changes in the GPA gene expression/physiology, thus allowing the insect to overcome and/or bypass plant defenses and adapt to the host plant. Salicylic acid (SA) signaling through NPR1 is also stimulated in GPA-infested plants. In plant-pathogen interaction, the PAD4 protein functions along with its interacting partner EDS1 in an amplification loop that promotes SA synthesis, leading to activation of SA dependent defenses. SA in turn amplifies PAD4 and EDS1 expression, thus resulting in positive amplification of this PAD4/EDS1-SA loop in plant defense against pathogens. Although EDS1 expression and SA signaling are activated in GPA-infested Arabidopsis, genetic studies confirm that SA and EDS1 are not required for controlling GPA infestation on Arabidopsis. Quite to the contrary, SA by antagonizing the jasmonic acid (JA; active form is JA-Isoleucine [JA-Ile]) signaling mechanism likely facilitates GPA infestation. JA, which is synthesized by the 13-LOX pathway, is required for controlling severity of GPA infestation. JA promotes the accumulation of Nδ-acetylornithine, camalexin and indole-glucosinolates, which are detrimental to GPA. Expression of PAD3, which is involved in camalexin synthesis and some genes involved in glucosinolate synthesis (e.g., CYP79B2 and CYP81F2) are negatively regulated by the small RNA gene-silencing mechanism involving DCL1, HYL1, HENT1, HST, SE, and AGO1. Oxylipins synthesized by the α DOX1 pathway and reactive oxygen species (ROS) produced by the NADPH oxidase RBOHD are also involved in controlling GPA infestation. H2O2 promotes callose deposition and thus likely contributes to phloem occlusion and plant defense against GPA. ROS's could also impinge on other signaling/defense mechanisms. Ethylene signaling through ETR1 and EIN2 has also been implicated in Arabidopsis defense against GPA. The ethylene inducible MYB44 gene is required for controlling GPA infestation. MYB44 is required for promoting EIN2 expression in response to harpin treatment, which also induces resistance against GPA in Arabidopsis. The ethylene- and harpin-inducible MYB15, MYB51, and MYB73 genes were required for harpin-induced resistance against GPA. By contrast, since mutations in the ethylene- and harpin-inducible MYB30, MYB108, ZFP6, and RAP2.6L genes enhanced the effect of harpin on controlling GPA infestation, these genes are shown as factors that facilitate GPA infestation. The relationship between many of these different pathways/mechanisms remains to be studied. All genes/proteins are in blue and signaling molecules are in yellow boxes. Red lines/arrows indicate steps/mechanisms that facilitate GPA infestation, while black lines indicate steps that contribute to defense. Lines ending with a perpendicular bar are indicative of a repressive effect.
Mechanisms that limit GPA infestation on arabidopsis
Both constitutive and inducible factors/mechanisms contribute to plant defense against aphids. In general, plant resistance mechanisms against aphids can be broadly classified as antixenosis and antibiosis (Painter, 1951; Kogan and Ortman, 1978). Antixenosis is used to describe mechanisms that result in a plant either not serving as a host, or given the option the insect preferring an alternate host. Antixenotic defenses could influence insect feeding behavior, for example adversely impacting its ability to find sieve elements. In contrast, antibiosis results from defenses that impact insect physiology leading to impairment of aphid growth, development, reproduction and/or survival (Smith, 2005). In some cases, antibiosis could also result from limited availability of nutrients required by an aphid (Pedigo, 1999). Severe cases of antibiosis could impact insect feeding behavior thus contributing to antixenosis, as well. Tolerance is another phenomenon that results in the plant withstanding or recovering from the infestation despite supporting an insect population that is comparable to that which causes damage on a susceptible variety (Painter, 1951). Thus, tolerance does not adversely impact the insect, but rather is an adaptation that benefits the plant. As discussed below, Arabidopsis engages both antibiotic and antixenotic defenses to control GPA infestation.
Perception of aphids
In plant-microbe interaction, immune receptors have been suggested to facilitate recognition of specific pathogen-derived effectors or infection-associated elicitors, leading to the activation of defenses that limit infection (Boller and Felix, 2009; Thomma et al., 2011; Gassmann and Bhattacharjee, 2012). Similar surveillance mechanisms likely allow plants to recognize aphid infestation (Smith and Clement, 2012). For example, in tomato the Mi-1.2-encoded nucleotide binding site (NBS) leucine-rich-repeat (LRR) protein confers resistance against certain biotypes of Macrosiphum euphorbiae (potato aphid) and Meloidogyne sp. (root-knot nematodes) (Milligan et al., 1998; Rossi et al., 1998; Vos et al., 1998), and in melon the Vat-encoded NBS-LRR protein confers resistance against Aphis gossypii (melon and cotton aphid) (Pauquet et al., 2004). Analogous to the involvement of immune receptors in plant immunity against pathogens, it is plausible that Mi-1.2 and Vat likely help recognize effectors delivered into the plant by the aphid or elicitors produced in planta in response to aphid infestation. Loci controlling resistance against aphids have been identified in other plants, as well. In lettuce resistance against Nasonovia ribisnigri (lettuce aphid) is conferred by the Nr gene, in soybean resistance against Aphis glycines (soybean aphid) is conferred by the RAG1 and RAG2 genes, in apple the Sd1 gene confers resistance against some biotypes of Dysaphis devecta (rosy leaf curling aphid) (Roche et al., 1997), and ten loci (Dn1-Dn9 and Dnx) have been identified in rye, wheat or Tausch's goatgrass that confer resistance against Diuraphis noxia (Russian wheat aphid) (Helden et al., 1993; Hill et al., 2006; Kim et al., 2010; Bouhssini et al., 2011; Smith and Clement, 2012).
Whether a similar immune receptor-mediated mechanism is utilized by plants against a generalist like the GPA is not known. However, a few recently concluded studies have demonstrated that GPA saliva contains factors that elicit defense responses in Arabidopsis. De Vos and Jander (2009) showed that saliva from the GPA when infiltrated into Arabidopsis leaves resulted in reduced GPA population size on the treated leaves. The resistance induced by saliva did not require salicylic acid (SA), jasmonic acid (JA) and ethylene signaling, which are involved in controlling infestation by insects and pathogens. The resistance inducing elicitor was sensitive to boiling and proteinase K treatment, suggesting that it is a protein. Size fractionation experiments indicated a 3–10 kD molecular range for this elicitor from GPA saliva. Expression levels of several Arabidopsis genes, including those associated with defense, signal transduction and senescence were altered in response to saliva application (De Vos and Jander, 2009), suggesting that the resistance enhancing effect of this proteinaceous elicitor is likely due to the activation of plant defense mechanisms. Mp10 and Mp42 are two salivary proteins from the GPA, which were recently shown to elicit host defenses and reduce insect fecundity when transiently over-expressed in N. benthamiana leaves (Bos et al., 2010). Mp10, which shows homology to insect protein olfactory segment D2-like protein (OS-D2-like protein), when over-expressed in N. benthamiana resulted in chlorosis. In addition Mp10 cross-talk with defense signaling mediated by the bacterial flg22 peptide resulted in the attenuation of flg22-induced ROS production (Bos et al., 2010). These results suggest that aphid salivary components, or products thereof, are likely recognized by Arabidopsis cells, leading to activation of defenses.
The tug-O-War for resources
Source-sink patterns are altered in aphid-infested plants resulting in the diversion of nutrient flow from the natural sinks to the aphid-infested organs (Mittler and Sylvester, 1961; Larson and Whitham, 1991; Dixon, 1998; Girousse et al., 2005). In Arabidopsis, GPA infestation resulted in changes in expression of genes involved in resource partitioning and sugar signaling (Moran and Thompson, 2001; Pegadaraju, 2005). GPA-infestation also resulted in increase in sucrose content in the aphid-infested organs (Singh et al., 2011). This increase in sucrose was relatively rapid, beginning as early as 6–12 h post infestation. Increase in sucrose content occurred even when the infested plants were kept in the dark, suggesting that changes in photosynthetic rate are not the likely cause of this increase. Increased turnover of other molecules could be a potential source for this increase in sucrose. Blockage in export of resources from the infested organ could also potentially contribute to this build-up of sucrose in the infested organ. Sucrose, which is responsible for the high osmolarity of the phloem sap is also the major feeding stimulant encountered by aphids while feeding on phloem sap (Douglas, 2006). However, aphids expend a lot of energy to counter the high osmolarity of the phloem sap they consume (Spiller et al., 1990; Pompon et al., 2010). For example, sucrases present in the insect gut hydrolyze sucrose to hexoses, which are then polymerized into oligosaccharides that have a lower contribution to osmotic pressure than hexoses and sucrose (Wilkinson et al., 1997; Ashford et al., 2000). Furthermore, these oligosaccharides are expelled out in the honeydew. Water consumption from the xylem has also been suggested as a means utilized by aphids to dilute the sugar content in the gut and thus contribute to osmoregulation (Spiller et al., 1990; Pompon et al., 2010). Thus, the infestation-associated increase in sucrose content in plant tissues could potentially be detrimental to the aphid. Genetic studies with the Arabidopsis tps11 (trehalose-6-phosphate synthase 11) mutants suggest that this increase in sucrose in GPA-infested leaves is likely not detrimental to GPA. Quite to the contrary, GPA population was larger on the tps11 mutant, despite sucrose content being 40% higher in the GPA-infested leaves of the tps11 mutant compared to the wild type (WT) plants (Singh et al., 2011). Further studies are required to determine if this increase in sucrose in GPA-infested leaves is in the phloem sap consumed by the aphid, and/or is within the mesophyll cells of the infested leaves. In potato, antisense-mediated silencing of the StSUT1 gene, which encodes a sucrose transporter, resulted in a reduction in sucrose content compared to the non-transgenic plants and simultaneously resulted in poor performance of potato aphid (Pescod et al., 2007). Taken together, these studies with tps11 mutant and potato StSUT1-silenced lines suggest that aphids require an optimal level of sucrose and/or osmolarity and likely target sucrose accumulation to facilitate infestation.
GPA infestation also results in an increase in starch content in Arabidopsis leaves (Singh et al., 2011; Singh, 2012). This increase in starch was observed even when the plants were kept in complete darkness during the course of the experiment. Expression of the Arabidopsis APL3 gene, which encodes a subunit of AGPase that synthesizes ADP-glucose, the donor of glucosyl moieties to the growing starch chain (Geigenberger, 2011), was also upregulated in GPA-infested plants (Singh, 2012). In Arabidopsis the PGM1 gene encodes a phosphoglucomutase that synthesizes glucose-1-phosphate, which is the precursor for starch synthesis. pgm1 mutants fail to accumulate starch (Yu et al., 2000). GPA population size was larger on the pgm1 mutant compared to the WT plant (Singh et al., 2011; Singh, 2012). GPA numbers were also higher on a gbss1 mutant that lacks a plastid-localized amylose synthesizing starch synthase activity (Singh, 2012). In contrast, GPA numbers were lower on the Arabidopsis ssIII (starch synthase III) mutant (Singh, 2012), which hyper-accumulates starch (Zhang et al., 2005) compared to the WT plant. Singh et al. (2011) have suggested that an increase in starch likely functions as a “secondary sink” that is associated with plant defense against the GPA, presumably by redirecting C into starch. Starch was previously shown to have an inhibitory effect on GPA feeding (Campbell et al., 1986). Hence, it is plausible that starch accumulation makes Arabidopsis leaves less desirable to the GPA.
Premature leaf senescence characterized by chlorophyll loss and upregulation of a subclass of SENESCENCE ASSOCIATED GENES (SAG) is observed in GPA-infested Arabidopsis (Pegadaraju et al., 2005; Louis et al., 2010b, 2012a). Expression of the SAG genes was induced within 24 h of infestation. As mentioned above, the GPA salivary protein Mp10 when expressed in leaves results in chlorosis (Bos et al., 2010). Thus, Mp10 is a likely elicitor of premature leaf senescence in GPA-infested Arabidopsis. Leaf senescence results in the export of nutrients from the senescing leaves and thus could potentially counter the ability of aphids to increase the sink strength of infested leaves. Indeed, transient expression of Mp10 in N. benthamiana also resulted in reduction of GPA fecundity (Bos et al., 2010). In support of a role for senescence in Arabidopsis defense, Pegadaraju et al. (2005) noted that the hyper-senescent ssi2 and cpr5 mutants exhibited enhanced resistance to GPA. Furthermore, mutation in the PAD4 (PHYTOALEXIN-DEFICIENT4) gene, which resulted in the attenuation of the GPA infestation-induced premature leaf senescence phenotype, was accompanied by improved performance of the GPA. Senescence is accompanied by changes in redox status (Khanna-Chopra, 2012). Indeed, GPA infestation is accompanied by an increase in H2O2 content in potato leaves (Kerchev et al., 2012). Analysis of GPA performance on the Arabidopsis rbohd mutant, which is defective in ROS production, confirmed an important role for ROS in Arabidopsis defense against the GPA (Miller et al., 2009). The GPA attained a larger population size on the rbohd mutant than the WT plant (Miller et al., 2009). Thus, senescence associated physiological and developmental changes are presumably engaged by Arabidopsis to counter the ability of GPA to alter resource allocation and thereby control severity of aphid infestation. It is equally possible that senescence is accompanied by the production of factors that are toxic to the GPA.
Defenses in the phloem
Phloem occlusion. Phloem, the site of aphid feeding, provides an ideal location to pack defense metabolites as well as activate mechanisms that promote phloem occlusion (Walz et al., 2004; Will and van Bel, 2006; Gaupels et al., 2008). Comparison of insect feeding behavior with an Electrical Penetration Graph (EPG) set-up (Tjallingii, 1990; Tjallingii and Esch, 1993; Reese et al., 2000; Walker, 2000) demonstrated that Arabidopsis attempts to control GPA feeding from sieve elements (Pegadaraju et al., 2007; Singh et al., 2011). GPA spent more time feeding from the sieve elements on the pad4 and tps11 mutants than the WT plant (Pegadaraju et al., 2007; Singh et al., 2011). In contrast, time spent feeding from sieve elements was reduced when feeding on transgenic plants overexpressing PAD4 (Pegadaraju et al., 2007), thus suggesting that PAD4 and TPS11-mediated mechanisms are required for controlling GPA feeding from sieve elements. However, the involvement of SEO in PAD4 and TPS11-dependent defense against GPA remains to be experimentally tested.
Callose deposition and phloem protein aggregation are mechanisms that contribute to SEO (Will and van Bel, 2006). Although the role of callose in Arabidopsis defense against GPA is not clear, studies with other plant-hemipteran interactions have suggested that callose is an important factor in defense against hemipteran insects. For example, callose deposition in rice is associated with resistance against the brown planthopper (Nilaparvata lugens) (Hao et al., 2008), and in Arabidopsis, expression of the CALS1 gene, which encodes a callose synthase, was up-regulated and callose accumulation enhanced in response to Bemisia tabaci (silverleaf whitefly) infestation (Kempema et al., 2007).
Dispersion of forisomes, which appear to be composed of multiple SEO proteins, contributes to SEO in Fabaceae. In Arabidopsis, the AtSEOR1 and AtSEOR2 genes are homologs of the Fabaceae SEO protein-encoding genes (Anstead et al., 2012). However, loss of AtSEOR1 or AtSEOR2 function in the atseor1 and atseor2 mutants, respectively, did not adversely impact basal resistance against GPA (Anstead et al., 2012). Quite to the contrary, the number of nymphs produced was higher on the WT than the atseor1 and atseor2 mutant plants, suggesting that presence of these proteins is beneficial to the insect. Forisome dispersion also was not observed in faba bean (Vicia faba) in response to penetration of sieve elements by stylets of Acyrthosiphon pisum (pea aphid) (Walker and Medina-Ortega, 2012). However, when forisome dispersal was induced in Megoura viciae (vetch aphid)-infested faba bean leaves by burning of leaf-tips, changes were observed in insect feeding behavior. EPG analysis showed that the insect switched from phloem sap ingestion to secretion of water saliva, presumably to reverse phloem occlusion (Will et al., 2007). A transition from phloem-sap consumption to salivation has also been observed in similar experiments conducted with several other plants and aphids (Will et al., 2009), thus suggesting that this behavioral response is likely a general response of aphids to phloem occlusion. Will et al. (2007) showed that watery saliva from vetch aphid was capable of reversing forisome dispersal in vitro, thus suggesting that aphid saliva contains factors that have the ability to reverse phloem occlusion mediated by forisome dispersion.
Antibiotic factors in the phloem. The phloem sap of Arabidopsis contains an antibiotic factor that is detrimental to GPA. Phloem sap-enriched petiole exudates collected from leaves of uninfested Arabidopsis plants when added to synthetic diet lowered GPA fecundity (Louis et al., 2010a,b, 2012a; Singh et al., 2011; Nalam et al., 2012). The Arabidopsis ssi2 mutant, which exhibits heightened resistance to GPA, contains elevated levels of this antibiosis activity (Louis et al., 2010a,b). The Arabidopsis PAD4 and MYZUS PERSICAE-INDUCED LIPASE 1 (MPL1) genes were required for the increased antibiosis observed in the ssi2 mutant (Louis et al., 2010a,b). In agreement with a role for this PAD4- and MPL1-dependent antibiotic activites in controlling overall severity of GPA infestation, the ssi2-depdendent antibiosis activity was lower in ssi2 pad4 and ssi2 mpl1 double mutant plants. Furthermore, petiole exudates from uninfested leaves of pad4 and mpl1 contained lower levels of this antibiotic activity (Louis et al., 2010a,b). The identity of this antibiotic factor that is altered in the ssi2, pad4, and mpl1 mutants is not known. As discussed below, Arabidopsis phloem sap contains proteins and non-protein metabolites that are detrimental to GPA. Accumulation of one or more of these factors could potentially be dependent on PAD4 and/or MPL1 activity. Despite the presence of these detrimental factors in the phloem sap, GPA is capable of colonizing Arabidopsis. This is in part because GPA adapts on Arabidopsis to suppress the accumulation and/or detoxify one or more of these factors. As discussed later, oxylipins produced by the host facilitate adaptation of GPA on the host plant allowing it to suppress accumulation of this antibiosis activity (Nalam et al., 2012).
Plants in the Brassicaceae family, which includes Arabidopsis, contain glucosinolates, which are defensive compounds. Glucosinolate accumulation is under control of JA signaling (Mewis et al., 2005), which as discussed below has been suggested to promote resistance against GPA. The metabolism of glucosinolates and their role in plant defense has been reviewed recently (Hopkins et al., 2009; Sønderby et al., 2010; Wittstock and Burow, 2010; Winde and Wittstock, 2011). When acted upon by myrosinases, glucosinolates can release toxic breakdown products that are detrimental to insects (Chew, 1988; Louda and Mole, 1991; Rask et al., 2000). The accumulation of glucosinolates and myrosinases is compartmentalized, thus preventing their mixing in the absence of physical damage to the plant tissue. Glucosinolates are stored in the sulfur-rich S-cells that are in close proximity to the phloem (Koroleva et al., 2000). Myrosinases on the other hand are stored in the myrosin cells and guard cells (Andréasson and Jørgensen, 2003; Zhao et al., 2008). In Arabidopsis, GPA infestation results in elevated levels of glucosinolates that can also be detected in the phloem sap (Kim and Jander, 2007; Louis et al., 2010a). Expression of genes putatively involved in glucosinolate metabolism were also upregulated in Arabidopsis leaves infested with the GPA and in leaves treated with saliva from the GPA (Mewis et al., 2006; De Vos and Jander, 2009). A defensive role for glucosinolates was suggested by the observation that resistance to the GPA was increased in the Arabidopsis atr1D mutant, which accumulates higher amounts of indole-glucosinolates than the WT plant (Kim et al., 2008). By contrast, in comparison to the WT plant, the cyp79B2 cyp79B3 double mutant that does not accumulate indole-glucosinolates exhibited enhanced susceptibility to the GPA. Mutations in the CYP81F2 gene, which encodes a cytochrome P450 monooxygenae that is required for synthesis of 4-hydroxy-indole-3-yl-methyl glucosinolate also resulted in lowered resistance to GPA (Pfalz et al., 2009). The feeding style of aphids, which causes minimal physical damage to cells surrounding the phloem, limits the mixing of glucosinolates and myrosinases, thus allowing intact glucosinolates to be ingested from the phloem by the aphid. Indeed, glucosinolates have been detected in the honeydew of GPA reared on Arabidopsis (Kim and Jander, 2007). However, the indole class of glucosinolates were metabolized in the insect gut (Kim et al., 2008) and the products were found to have a negative effect of GPA settling and growth (Kim and Jander, 2007; Kim et al., 2008; Pfalz et al., 2009). Glucosinolate level and composition was not significantly impacted in the pad4 and mpl1 mutants, suggesting that their accumulation is not under control of PAD4 and MPL1 (Kim and Jander, 2007; Louis et al., 2010a).
Lectins, also known as agglutinins, are proteins that can reversibly and specifically bind to carbohydrates. Many lectins are toxic to phytophagous insects (Vandenborre et al., 2011). The Arabidopsis Phloem Protein2-A1 (PP2-A1), which is a component of phloem protein bodies, is also a lectin. When added to a synthetic diet, recombinant PP2-A1 affected weight gain in GPA and soybean aphid nymphs (Beneteau et al., 2010). Lectins have an affinity for carbohydrates, which may interfere with physiological processes in the insect gut, thus controlling insect infestation (Carlini and Grossi-de-Sa, 2002; Vasconcelos and Oliveira, 2004). Confirming a role for PP2-A1 in Arabidopsis defense against GPA, constitutive expression of PP2-A1 adversely impacted the ability of GPA to feed from the sieve elements (Zhang et al., 2011). The PP2-A1 gene was also required for harpin to promote resistance against GPA (Zhang et al., 2011). Harpin is a hypersensitive response-inducing protein produced by gram negative pathogenic bacteria that also elicits defenses against pathogens.
Nδ-acetylornithine is a novel class of non-protein amino acid that was identified in the phloem sap of methyl-JA-treated Arabidopsis (Adio et al., 2011). GPA infested plants contained elevated levels of Nδ-acetylornithine. Furthermore, GPA reproduction was significantly reduced when the aphids were fed on a diet containing Nδ-acetylornithine, suggesting that this non-protein amino acid has a defensive role against aphids (Adio et al., 2011). The toxic effect of this compound is specific to phloem-feeding insects, since it did not have any effect on the growth of lepidopteran caterpillars. Expression of the NATA1 gene, which is involved in the biosynthesis of Nδ-acetylornithine in Arabidopsis, is induced upon GPA infestation. Furthermore, its expression is high in vascular tissues, thus bolstering the notion that this aphidicidal compound is synthesized and/or accumulates in the phloem (Adio et al., 2011).
Contribution of arabidopsis lipid metabolism to defense against GPA
Besides functioning as major structural components of cell membranes, plant lipids also function as precursors of antibiotic compounds and signaling molecules (Wang, 2004; Shah, 2005; Wasternack, 2007; Upchurch, 2008; Scherer, 2010; Yan et al., 2013). In addition, lipids have also been implicated in cross-kingdom communication (Christensen and Kolomiets, 2011). Lipid metabolism in Arabidopsis also impacts interaction with the GPA. A loss-of-function mutation in the SSI2 gene, which encodes the major plastidyl stearoyl acyl-carrier protein desaturase that catalyzes the synthesis of oleic acid in Arabidopsis, resulted in enhanced resistance against the GPA (Louis et al., 2010b). As mentioned above, the ssi2 mutant exhibits a hyper-senescence phenotype that is characterized by constitutively elevated expression of the SAG13 gene and spontaneous cell death. Furthermore, petiole exudates from ssi2 accumulate elevated levels of antibiosis activity against the GPA. EPG analysis indicated that the GPA feeding behavior was not adversely impacted on the ssi2 mutant compared to the WT plant. The ssi2 mutant also accumulates elevated levels of SA. Since, SA added to a synthetic diet had an adverse impact on insect fecundity, Louis et al. (2010b) conducted experiments with the ssi2 nahG plants, in which the ssi2-dependent accumulation of SA is attenuated, to determine if SA is indeed required for the ssi2-conferred enhanced resistance against GPA. Although presence of nahG had a weak effect on the strength of antibiosis activity in petiole exudates, this was not sufficient to weaken the ssi2-conferred resistance against GPA, thus indicating that SA is not a major factor contributing to the ssi2-conferred enhanced resistance (Louis et al., 2010b).
Oxylipins (oxidized lipids) contribute to Arabidopsis defense to the GPA. Oxidation of polyunsaturated fatty acid (e.g., linoleic and linolenic acids) is the first step in the synthesis of oxylipins (Feussner and Wasternack, 2002; Mosblech et al., 2009). Enzymatic oxidation of polyunsaturated fatty acids is mediated by enzymes like lipoxygenases (LOXs) and α-dioxygenases (α-DOXs). The resultant oxidized fatty acids can be further processed enzymatically or non-enzymatically to yield a variety of oxylipins, several of which have roles in plant stress response. JA is one of the best studied oxylipin that is derived via the LOX pathway. Its role in Arabidopsis-GPA interaction is discussed later. Oxidation of fatty acids by α-DOXs yields 2(R)-hydroperoxides, which can be further processed into other products. In Arabidopsis, expression of the α-DOX1 gene was up-regulated in response to GPA infestation (Avila et al., 2013). Similarly, in tomato expression of the Slα-DOX1 gene was up-regulated in response to potato aphid infestation (Avila et al., 2013). In both, Arabidopsis and tomato, knock-down of α-DOX1 function resulted in increased susceptibility to aphids. In comparison to the WT plant, GPA population size was larger on the Arabidopsis α-dox1 mutant (Avila et al., 2013). Similarly, virus-induced gene silencing of Slα-DOX1 in tomato resulted in an increase in the size of potato aphid population compared to the non-silenced plants (Avila et al., 2013). α-DOX1-derived lipids have known antibiotic and signaling functions, either or both of which could contribute to the α-DOX1-mediated resistance against aphids. As discussed later, recent studies indicate that oxylipins also are susceptibility factors in Arabidopsis interaction with GPA.
MPL1, which is required for the ssi2-dependent enhanced resistance and hyper-accumulation of antibiosis activity in petiole exudates, encodes a protein with homology to α/β-fold acyl hydrolases/lipases (Louis et al., 2010a). A signal peptide at the N-terminus suggests that MPL1 is likely targeted to the endoplasmic reticulum. MPL1 contains a signature GXSXG esterase/lipase catalytic motif and recombinant MPL1 possesses lipase activity. MPL1 expression was induced in response to GPA infestation and petiole exudates from the mpl1 mutant contained lower levels of antibiosis activity compared to WT plant (Louis et al., 2010a). In contrast, MPL1 overexpression resulted in an increase in antibiosis activity, which was paralleled by an increase in resistance against GPA (Louis et al., 2010a). Loss of MPL1 function had no impact on antixenosis against GPA. The feeding behavior of GPA was comparable when feeding on WT and mpl1 mutant (Louis et al., 2010a). Although the exact identity of the MPL1-dependent antibiotic factor is not known, it could possibly be a lipid/lipid-derived product present in the phloem sap. The phloem sap is known to contain a variety of lipids including oxylipins (Benning et al., 2012). One or more products of MPL1 activity could be directly toxic to the insect. Alternatively, an MPL1-derived lipid might indirectly impact GPA infestation. MPL1 also contains a HX4D acyltransferase motif suggesting that it could be involved in lipid modifications. Whether the lipase activity or the acyltransferase motif of MPL1 are required for its involvement in Arabidopsis-GPA interaction remains to be determined.
Regulation of defenses
Role of salicylic acid, jasmonic acid and ethylene in arabidopsis-GPA interaction
SA and JA have important signaling functions in plant defense against pathogens. In some cases of plant-pathogen interaction they function together to promote resistance, while in other cases SA and JA have an antagonistic relationship (Mur et al., 2006; Pieterse et al., 2009; Robert-Seilaniantz et al., 2011). SA and JA signaling also have a role in some cases of plant defense against aphids. In tomato, SA signaling is required for Mi-1.2-mediated resistance against the potato aphid (Li et al., 2006). Furthermore, gene expression studies conducted in Arabidopsis indicate that GPA infestation triggers the induction of the SA and JA pathway. For example, expression of the PATHOGENESIS-RELATED 1 (PR1) gene, which for long has been used as a molecular marker for the activation of SA signaling, and the PDF1.2 gene, which is a marker for the activation of JA and ethylene signaling, were upregulated in GPA-infested leaves (Moran and Thompson, 2001; Moran et al., 2002; De Vos et al., 2005; Pegadaraju, 2005; Mewis et al., 2006). Similarly in tomato, induction of the SA-inducible P4 gene, which is homologous to the Arabidopsis PR1 gene, has also been noted in plants infested with the potato aphid (Li et al., 2006). Expression of the isochorismate synthase encoding ICS1 (SID2) gene, which is involved in the synthesis of SA in Arabidopsis, was also upregulated in response to GPA infestation (Pegadaraju, 2005). Similarly, expression of ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5), which is required for SA synthesis, was also induced in GPA-infested plants (Pegadaraju, 2005). However, De Vos et al. (2005) reported that GPA infestation did not result in any observable increase in SA content in Arabidopsis. As suggested by them, this inability to see an increase in SA could be due to fewer cells responding to GPA infestation as opposed to that seen in pathogen-infected tissues. Genetic studies conducted by several groups have indicated that although GPA infestation activates SA signaling, SA signaling is not important for promoting resistance against GPA. Mutations in the ICS1 and EDS5 genes did not result in increased colonization by GPA on the ics1 and eds5 mutants, compared to WT plants (Moran and Thompson, 2001; Pegadaraju et al., 2005). Furthermore, Pegadaraju et al. (2005) reported that GPA population size was comparable between WT and transgenic Arabidopsis plants expressing the bacterial nahG-encoded salicylate hydroxylase, which converts SA to catechol. Loss of NPR1 (NON-EXPRESSER OF PR GENES1), a key SA signaling regulator and a putative SA receptor (Wu et al., 2012), also did not result in improved performance of GPA on the npr1 mutant than the WT plant (Moran and Thompson, 2001; Mewis et al., 2005; Pegadaraju et al., 2005). Finally, application of (1, 2, 3) thiadiazole-7-carbothioic acid (S) methyl ester (BTH), a synthetic functional analog of SA, also did not curtail GPA colonization on Arabidopsis (Moran and Thompson, 2001). Taken together, these results confirm that SA signaling is not critical for controlling GPA infestation on Arabidopsis. Quite to the contrary, Mewis et al. (2005) reported that GPA population size was smaller on the NahG and npr1 plants than on the WT plant, suggesting that SA signaling in fact might be promoting susceptibility to GPA.
According to a “decoy” hypothesis involving SA, some insects may have evolved to trick the host into activating SA signaling, which in many cases is known to antagonize the activation of JA signaling (Walling, 2008). In support for a potential function of the JA pathway in promoting resistance against GPA, Ellis et al. (2002a) reported that the cev1 (constitutive expression of VSP1) mutant, which contains higher levels of JA than the WT plant, was more resistant to GPA than the WT plant. They further showed that exogenously applied MeJA also promoted resistance to GPA in Arabidopsis (Ellis et al., 2002a). In contrast, GPA numbers were higher on JA-insensitive coi1 (coronatine insensitive1) mutant compared to the WT plant (Ellis et al., 2002a; Mewis et al., 2005, 2006). JA is known to promote the accumulation of metabolites like indole-glucosinolates, camalexin and the non-protein amino acid Nδ-acetylornithine (Zhou et al., 1999; Mikkelsen et al., 2003; Adio et al., 2011) that are detrimental to GPA. However, the simultaneous deficiency of ω3 fatty acid desaturases encoded by the Arabidopsis FAD7 and FAD8 genes, which are required for the synthesis of trienoic fatty acids, the precursors for JA synthesis, resulted in enhanced resistance to GPA (Avila et al., 2012). Similar to the results with the Arabidopsis fad7 fad8 double mutant, in tomato, loss of FAD7 activity in the spr2 mutant resulted in enhanced resistance against the potato aphid (Avila et al., 2012). This increase in resistance in spr2 was not associated with JA signaling, but instead resulted from elevated content of SA and increased Slα-DOX1 gene activity in the spr2 mutant (Avila et al., 2012, 2013). Mutations in other genes involved in JA synthesis also did not influence potato aphid colonization on tomato (Avila et al., 2012), although previous studies indicated that MeJA application enhanced resistance against potato aphid in tomato (Cooper et al., 2004). Considering that COI1 is a component of the proteasomal protein turnover mechanism that likely also impacts processes other than JA (He et al., 2012; Ralhan et al., 2012), and Arabidopsis CEV1 is a subunit of cellulose synthase, the lack of which has pleiotropic effects in the cev1 mutant that include the simultaneous hyperactivation of ethylene and JA signaling (Ellis et al., 2002b), the results with the fad7 fad8 mutant (Avila et al., 2012), in conjunction with a previous report by De Vos et al. (2005), who failed to see any significant increase in JA in GPA-infested Arabidopsis, call for a careful reevaluation of JA's role in Arabidopsis defense against GPA.
Ethylene production is elevated in aphid infested plants. For example, increases in ethylene production was observed in tomato infested with potato aphid, alfalfa infested with Therioaphis maculate (spotted alfalfa aphid), wheat infested with Schizaphis graminum (greenbug), and barley infested with Rhopalosiphum padi (oat aphid) and Russian wheat aphid (Dillwith et al., 1991; Anderson and Peters, 1994; Miller et al., 1994; Argandoña et al., 2001; Mantelin et al., 2009). Changes in expression of genes putatively involved in ethylene metabolism/signaling also have been observed in melon plants responding to the melon and cotton aphid infestation, in tomato infested with the potato aphid, and Arabidopsis infested with the GPA (Moran et al., 2002; Anstead et al., 2010; Mantelin et al., 2009). In melon, the expression of ethylene pathway and response genes was reported to be highly upregulated during the resistant interaction mediated by the Vat resistance gene, thus leading the authors to suggest that ethylene may have a role in Vat-mediated resistance (Anstead et al., 2010). Quite to the contrary, genetic studies in tomato indicated that ethylene signaling contributes to susceptibility to the potato aphid in the absence of the Mi-1.2 resistance gene function (Mantelin et al., 2009). In choice test assays, the potato aphid was observed to prefer the WT plant to the ethylene-insensitive Never ripe mutant. Similarly, in Arabidopsis, Mewis et al. (2006) reported that GPA and cabbage aphid populations were significantly smaller on the ethylene-insensitive etr1 mutant than the corresponding WT plant, thus suggesting that like in the tomato-potato aphid interaction, ethylene likely contributes to Arabidopsis susceptibility to the GPA and the cabbage aphid. However, in another study the same group did not observe significant differences in GPA population size on the etr1 mutant compared to WT plants (Mewis et al., 2005). Similarly, Kettles et al. (2013) did not observe significant differences in GPA fecundity on insects reared on WT and etr1 mutant. However, a higher fecundity was observed for GPA reared on the ethylene-insensitive ein2 mutant than the WT plant (Kettles et al., 2013).
Ethylene signaling was also required in Arabidopsis for harpin-induced resistance against GPA (Dong et al., 2004). Harpin treatment limited insect settlement, feeding and fecundity on Arabidopsis (Dong et al., 2004; Liu et al., 2010; Zhang et al., 2011). Unlike the WT plants, harpin was unable to limit insect fecundity on the etr1 and ein2 mutants (Dong et al., 2004). Liu et al. (2010) further demonstrated that the ethylene and harpin up-regulated the AtMYB44 gene, which encodes a Myb-family transcription factor, was required for harpin-induced resistance to GPA. AtMYB44 expression is also induced in response to GPA infestation and application of GPA saliva (De Vos and Jander, 2009). However, whether AtMYB44 is required for basal resistance against GPA was not reported. AtMYB44 was required for the harpin-induced up-regulation of EIN2 expression. Also required for harpin-induced resistance against GPA, but not for EIN2 induction, were AtMYB15, AtMYB51, and AtMYB73, three other Myb-family transcription factor encoding genes, which are also ethylene inducible (Liu et al., 2010). Ethylene and harpin also induced expression of AtMYB30, AtMYB108, AtZFP6, and AtRAP2.6L. However, mutations in these genes enhanced harpin-induced resistance to GPA, suggesting that these genes might be involved in dampening of harpin- and likely ethylene-induced defense signaling. Ethylene-signaling thus may be involved in promoting expression of both, genes that promote resistance as well as those that promote susceptibility to GPA. This might explain the discrepancies observed between various studies (Dong et al., 2004; Mewis et al., 2005, 2006; Kettles et al., 2013) on the impact of ethylene-insensitive mutants on Arabidopsis-GPA interaction.
PAD4, a regulator of antixenosis, antibiosis and premature leaf senescence in GPA-infested arabidopsis
The Arabidopsis PAD4 gene, which encodes a nucleocytoplasmic protein, is required for defense against a variety of pathogens and the GPA (Weirmer et al., 2005; Louis et al., 2012a). PAD4 was found to be required for modulating SA accumulation in pathogen-infected plants (Zhou et al., 1998; Jirage et al., 1999; Rietz et al., 2011), and for systemic acquired resistance, an inducible defense mechanism that increases resistance against subsequent infections in plants that previously experienced a localized infection (Shah and Zeier, 2013). As mentioned above, in Arabidopsis-GPA interaction, PAD4 function is required for antibiosis and antixenosis. When given a choice between the WT and the pad4 mutant, GPA preferred to settle on the pad4 mutant (Pegadaraju et al., 2007). In contrast, when given a choice between the WT and plants constitutively expressing PAD4 from the CaMV 35S promoter, GPA preferred the WT plant, thus suggesting that PAD4 is required for the accumulation of a factor that deters insect settling on Arabidopsis. EPG analysis indicated that compared to GPA feeding on the WT plant, insects on the pad4 mutant spent more time in the sieve element phase (Pegadaraju et al., 2007), suggesting that the pad4 mutant lacked a factor that controls insect feeding from sieve elements. In contrast, on plants constitutively expressing PAD4, GPA spent less time in the sieve element phase and the plants exhibited enhanced resistance to GPA (Pegadaraju et al., 2007). Petiole exudates collected from the pad4 mutant lacked the antibiosis activity that is present in petiole exudates collected from WT plants (Louis et al., 2010b). Analysis of plants expressing the coding sequence of the GUS reporter from the PAD4 promoter indicated that the PAD4 promoter directs GUS expression in cells at the feeding site (Louis et al., 2012b). Thus, the feeding deterrence function of PAD4 is likely due to its function in the phloem or cells surrounding the phloem. Glucosinolate content was not affected in the pad4 mutant (Kim and Jander, 2007), thus suggesting that the PAD4-mediated resistance likely does not involve glucosinolates.
PAD4 is also required for the timely activation of leaf senescence in GPA-infested Arabidopsis. Senescence associated chlorophyll loss was slower in the pad4 mutant than the WT plant (Pegadaraju et al., 2005). Expression of the SAG genes was induced slower in the pad4 mutant (Pegadaraju et al., 2005; Louis et al., 2010b, 2012a). Although overexpression of PAD4 did not constitutively activate SAG gene expression, GPA infestation resulted in the faster activation of SAG genes (Pegadaraju et al., 2007). The fact that mere expression of PAD4 was not sufficient for the induction of leaf senescence, suggests that additional factors are required for promoting leaf senescence in response to aphid infestation.
PAD4 exhibits homology to acyl hydrolases/esterases/lipases. The N-terminal half of PAD4 contains a triad of amino acids (S118, D178, and H229) (Jirage et al., 1999; Feys et al., 2005), which in other acyl hydrolases/lipases are part of the catalytic triad (Blow, 1990). However, no hydrolytic activity has been demonstrated for PAD4. On plants expressing missense versions of PAD4 (PAD4[S118A] and PAD4[D178A]) GPA population size was larger than on plants expressing WT PAD4, thus suggesting that S118 and D178 are critical for PAD4's involvement in defense against GPA (Louis et al., 2012a). However, the PAD4[S118A] allele did not impact plant choice by GPA. Furthermore, the PAD4-modulated expression of SAGs in response to GPA infestation also was not adversely impacted by this mutant alleles. Feeding behavior analysis with the EPG technique revealed that insects spent more time feeding from the sieve elements of plants expressing the PAD4[S118A] allele (Louis et al., 2012a). In addition, petiole exudates from plants expressing PAD4[S118A] were also deficient in the PAD4-dependent antibiosis activity. These results suggest that S118 in PAD4 is required for its antibiosis function and for deterring feeding from sieve elements. Thus, two distinct PAD4 activities contribute to defenses against GPA. The first activity, which limits insect feeding and promotes antibiosis, is dependent on S118. The second activity, which deters insect settling and promotes PAD4 expression, does not require S118.
In pathogen-infected Arabidopsis, PAD4 functioning along with its interacting protein EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1), regulates its own expression and functions in a feed-positive SA amplification loop with EDS1 to regulate expression of defense genes (Weirmer et al., 2005). EDS1 expression is induced in GPA-infested plants (Pegadaraju et al., 2007). However, EDS1 was not required for controlling GPA infestation (Pegadaraju et al., 2007; Louis et al., 2012a,b). GPA population size was comparable between the WT and the eds1 mutant. As mentioned above, SA is also not critical for controlling GPA infestation. Taken together, these studies indicate that PAD4-mediated defense against GPA is unique in that it does not invoke EDS1 and SA. It is plausible that the two different PAD4 molecular activities that are required for controlling GPA infestation involve PAD4 interaction with separate protein(s).
PAD4 was identified by Glazebrook et al. (1997) in a screen for Arabidopsis mutants that were deficient in the accumulation of the phytoalexin, camalexin. PAD4 regulates expression of PAD3, which encodes a P450 monooxygenase involved in camalexin biosynthesis (Zhou et al., 1999). Expression of PAD3 was up-regulated in GPA-infested Arabidopsis leaves as well as in response to GPA saliva (Pegadaraju et al., 2005; De Vos and Jander, 2009). Furthermore, camalexin levels were found to increase in response to GPA infestation (Louis, 2011; Kettles et al., 2013). However, PAD4 function was not required for this increase in camalexin in GPA-infested Arabidopsis (Louis, 2011). Furthermore, Pegadaraju et al. (2005) observed that GPA population size was comparable between the WT and the pad3 mutant, thus leading them to conclude that camalexin is not important for the PAD4-mediated resistance against GPA. In their bioassays with GPA on Arabidopsis, Pegadaraju et al. (2005) used insects that were reared on a mix of mustard and radish plants. Furthermore, the bioassays were conducted over a 2 day period. In contrast to the observations of Pegadaraju et al. (2005), Kettles et al. (2013) observed that in a different experimental set-up involving 2 week-long fecundity assays, GPA nymphs that were born on the pad3 mutant and reared to adulthood on the pad3 mutant exhibited higher fecundity than nymphs born and raised on WT plants. Furthermore, they observed that camalexin provided in an artificial diet had a detrimental effect on GPA fecundity. Taken together, these two studies (Pegadaraju et al., 2005; Kettles et al., 2013) suggest that the role of PAD3 and camalexin in defense against GPA might be influenced by the length of time the insects are reared on Arabidopsis. PAD3 and camalexin synthesis are also required for controlling cabbage aphid infestation on Arabidopsis (Kuśnierczyk et al., 2008).
TPS11-dependent trehalose metabolism regulates defense against GPA
The non-reducing α,α-1,1-linked glucose disaccharide trehalose, which serves as an energy source and an osmoprotectant in lower organisms, has a role in the protection of plants from stress (Schluepmann et al., 2003; Paul et al., 2008; Fernandez et al., 2010). Trehalose-6-phosphate (T6P) and trehalose are suggested to have signaling function in plants (Paul et al., 2008; Fernandez et al., 2010). In plants, trehalose is synthesized in two steps. The first step involves the synthesis of T6P by T6P synthase. In the subsequent step, T6P is dephosphorylated by T6P phosphatase to yield trehalose. Recently, it was suggested that trehalose or a derivative has a novel signaling function in plant defense against GPA (Singh et al., 2011; Hodge et al., 2012). Trehalose levels increased in response to GPA infestation. This increase in trehalose accumulation in GPA-infested plants was paralleled by a transient elevation of TPS11 expression (Singh et al., 2011). TPS11 encodes a protein with T6P synthase and T6P phosphatase activities. The GPA infestation-induced accumulation of trehalose was attenuated in the tps11 mutant, allowing increased colonization of the mutant by GPA. In contrast, resistance was enhanced in tre1 (trehalase) mutant, which accumulates elevated levels of trehalose due to deficiency in the ability to degrade trehalose (Singh et al., 2011). Similarly, trehalose content and resistance to GPA were higher in transgenic otsB plants, which overexpress the T6P phosphatase-encoding bacterial otsB coding sequence. Like the pad4 mutant, the tps11 mutant was preferred by GPA over WT plants and petiole exudates of the tps11 plants contained reduced antibiosis activity. EPG characterization of GPA feeding behavior indicated that GPA spent more time in the sieve element phase on the tps11 mutant than the WT plant. Trehalose application restored resistance to the tps11 mutant, thus confirming that trehalose deficiency was cause of the tps11 phenotypes.
Trehalose application induces PAD4 expression, which was constitutively higher in the trehalose hyper-accumulating otsB plant as well as TPS11 overexpressing plants (Singh et al., 2011). GPA infestation-induced expression of PAD4 was delayed in the tps11 mutant compared to the WT plant, thus indicating that TPS11-dependent trehalose increases are required for the timing and strength of PAD4 expression in GPA-infested Arabidopsis (Singh et al., 2011). TPS11 also influences starch accumulation in GPA-infested plants. Starch content was lower in the GPA-infested tps11 mutant compared to the WT plant (Singh et al., 2011). In comparison, sucrose content was higher on the tps11 mutant. These results suggest that TPS11-dependent trehalose, promotes the flux of C from sucrose into starch, thereby contributing to overall defense against GPA. As mentioned above, starch accumulation is a likely physiological adaptation that helps the plant to counter the GPA.
The phloem sap of GPA-infested Arabidopsis contains elevated levels of trehalose (Hodge et al., 2012), thus suggesting that trehalose can potentially act as a phloem-mobile signal. Hodge et al. (2012) further observed that trehalose content in GPA was higher when cultivated on Arabidopsis than on synthetic medium or on other plants that did not accumulate trehalose in response to GPA infestation, leading them to suggest that when on Arabidopsis the insect assimilates trehalose from phloem sap. Although trehalose is a major sugar in the insect hemolymph, when added to a synthetic diet trehalose was detrimental to GPA (Singh et al., 2011), thus suggesting that de novo synthesized trehalose in the hemolymph might have a different physiological effect than trehalose in the gut. Thus, trehalose consumed by the insect when feeding on Arabidopsis could potentially be detrimental to the insect. The mid-gut of pea aphid and soybean aphid contain trehalase (Cristofoletti et al., 2003; Bansal et al., 2013), suggesting that some aphids likely utilize trehalase in the gut to counter the presence of trehalose consumed from plants. Thus, trehalose might have a dual role in plant-aphid interaction, functioning as a signaling molecule to promote plant defenses and simultaneously having a detrimental effect on insect physiology.
GPA infestation also promoted trehalose accumulation in leaves of tomato plants (Singh and Shah, 2012). Expression of the TPS11 and PAD4 homologs were up-regulated in GPA-infested tomato, which also accumulated elevated levels of sucrose and starch. Furthermore, trehalose application induced expression of the tomato PAD4 homolog, promoted starch accumulation and limited insect colonization, thus suggesting that the role of trehalose in defense against GPA is likely conserved in other species, as well. Hodge et al. (2012) however, did not detect trehalose accumulation in GPA infested spring cabbage and black mustard as compared to Arabidopsis, suggesting that the interaction between the GPA and additional hosts needs to be studies in order to determine the extent to which the engagement of trehalose metabolism in plant defense against aphids has been conserved.
Arabidopsis contains more than 20 genes putatively involved in trehalose metabolism (Rolland et al., 2006; Paul et al., 2008). Whether the trehalose that accumulates in response to GPA infestation in Arabidopsis leaves is a product of TPS11 activity, or is synthesized by one or more of the other enzymes, is not known. It is plausible that TPS11-synthesized T6P and/or trehalose functions as a signal to activate expression and/or activity of one or more of the other trehalose biosynthesis genes/enzymes, which contribute to the overall increase in trehalose in GPA-infested plants. TPS11 expression is also altered in plants exposed to other abiotic and biotic stressors (Golem and Culver, 2003; Suzuki et al., 2005; Fujita et al., 2007; Iordachescu and Imai, 2008), suggesting that the TPS11-regulated mechanism(s) has a wider role in plant stress response.
Role of small RNAs in arabidopsis-GPA interaction
Small RNAs, which are 20–40 nucleotide-long non-coding RNAs, regulate gene expression at the transcriptional and post-transcriptional level. In plants, small RNAs are known to regulate JA and ethylene signaling, and influence defense against microbes and herbivores (Pandey and Baldwin, 2007; Pandey et al., 2008; Katiyar-Agarwal and Jin, 2010). Genetic studies in Arabidopsis indicated that mutations in plant microRNA (miRNA) pathway significantly impacted GPA fecundity (Kettles et al., 2013). GPA fecundity was lower on plants that contained mutations in the DCL1 (DICER-LIKE1), HYL1 (HYPONASTIC LEAVES 1), HEN1 (HUA ENHANCER1), SE (SERRATE), and HST (HASTY) genes, which are involved in miRNA generation, processing or transport, and in the AGO1 (ARGONAUTE1) gene, which is one of the slicer in the miRNA pathway. Expression of the CYP83B1 and CYP81F2 genes, which are involved in the synthesis of indolic glucosinolates, and the CYP79B2 gene, which is involved in the synthesis of indole-3-acetaldoxime, a precursor for indolic glucosinolates as well as auxin and camalexins, was constitutively higher in the dcl1 mutant. In addition, PAD3 induction and camalexin accumulation in response to GPA infestation were higher in the dcl1 mutant than the WT plant. GPA fecundity was significantly higher on the dcl1 pad3 double mutant compared to the dcl1 mutant plant, thus suggesting that increased PAD3 activity contributes to the dcl1-conferred reduction in GPA fecundity.
As mentioned above, GPA as well as several other aphids also vector viral diseases. Genomes of many viruses encode suppressors of RNA silencing that impact plant defense signaling. For example, the 2b protein of cucumber mosaic virus (CMV) inhibits anti-viral miRNA silencing mechanisms, as well as interferes with JA and SA signaling (Ji and Ding, 2001; Lewsey et al., 2010). Since CMV is transmitted by GPA, CMV infection could potentially influence GPA performance on plants. Indeed, GPA survival was higher on CMV-infected tobacco, as compared to mock-infected plants (Ziebell et al., 2011). By comparison, to the mock- or CMV-infected tobacco, GPA survival was lower on tobacco plants infected with a 2b gene-deletion mutant CMVΔ2b virus (Ziebell et al., 2011). EPG monitoring of GPA behavior confirmed that in comparison to insects feeding on plants infected with the WT CMV, GPA feeding on plants infected with the CMVΔ2b virus exhibited reduced feeding from sieve elements. Whether this effect of 2b on promoting GPA infection is due to its impact on RNA-silencing is not known. The impact of viral infection on aphid performance is not limited to GPA and CMV. By suppressing JA signaling in the host plant, the begomovirus Tomato yellow leaf curl China virus is beneficial to the insect vector, the whitefly (Zhang et al., 2012). Suppression of transcriptional silencing by the viral beta-satellite DNA-encoded factor βC1 was responsible for suppression of JA signaling during this tritrophic interaction. The interaction between Arabidopsis, the GPA and viruses vectored by GPA is also being utilized for high-throughput screens for Arabidopsis genes involved in this interaction (Chen et al., 2012).
Roots as a source of a signal influencing arabidopsis-GPA interaction
In recent years, studies on the role of roots in plant responses to above-ground herbivory have gained momentum (Kaplan et al., 2008; Rasmann and Agrawal, 2008; Erb et al., 2009; Erb, 2012; Nalam et al., 2013b). For example, the secondary metabolite and insecticide nicotine is synthesized in the roots from where it is transported to the shoots to be stored in vacuoles and provide defense against herbivores (Morita et al., 2009). The maize Mir1-CP cysteine protease, which accumulates at elevated levels in leaves in response to fall armyworm (Spodoptera frugiperda) infestation, is also suggested to be synthesized in the roots from where it is transported to the shoots (Lopez et al., 2007). Very recently, Nalam et al. (2012) demonstrated that communication between the roots and shoots also has a critical role in the interaction between Arabidopsis and GPA. GPA feeding on Arabidopsis leaves resulted in the up-regulation of LOX5 (LIPOXYGENASE 5) expression in roots. LOX5 encodes a 9-LOX, which is involved in the oxidation of fatty acids to yield oxylipins that can be further modified to yield a variety of biologically active lipids. Levels of 9-LOX-derived oxylipins also increased in roots, shoots and petiole exudates of plants that had GPA on their shoots. Loss of LOX5 function resulted in reduced colonization of the lox5 mutant by GPA. Insects reared on the lox5 mutant had significantly lower water content compared to insects reared on the WT plant. Irrigation with 9-LOX products restored water content and restored insect colonization on the lox5 mutant, suggesting that a 9-LOX lipid or product thereof is required for facilitating GPA colonization on WT plants. EPG monitoring of insect feeding behavior indicated that on the lox5 mutant, GPA spent less time feeding from sieve elements and drinking from the xylem, thus accounting for the reduced water content in the insects. Grafting experiments in which the WT shoots were grafted on lox5 roots and vice-versa indicated that LOX5 function is required in roots to promote GPA colonization on Arabidopsis shoots (Nalam et al., 2012).
A variety of LOX-derived oxylipins are known to function as defense modulating signaling molecules in plants. Similarly, a 9-LOX product could potentially be involved in suppressing Arabidopsis defense against GPA. Alternatively, GPA might have evolved to target the LOX5 pathway to produce oxylipins that suppress and/or counter Arabidopsis defenses against GPA. When added to synthetic diet, 9-LOX products enhanced insect fecundity, suggesting that these lipids might be directly acting on the insect to stimulate feeding and simultaneously promote fecundity. Previously it was shown that GPA reared on plants contained 9-LOX-derived lipids, while insects reared on synthetic diet lacked these lipids (Gosset et al., 2009), suggesting that GPA derives these lipids from the host. Indeed, in comparison to GPA reared on WT Arabidopsis, which contained 9-LOX synthesized oxylipins, GPA reared on the lox5 mutant lacked these lipids (Nalam et al., 2013a), supporting the notion that the GPA derives these 9-LOX synthesized lipids from Arabidopsis. The presence of 9-LOX products in the phloem sap-enriched petiole exudates of Arabidopsis supports this suggestion (Nalam et al., 2012). However, one cannot rule out the possibility that GPA synthesizes these oxylipins only in response to a plant-derived stimuli that is missing in the lox5 mutant.
If 9-LOX lipids are susceptibility factors, then why should plants increase synthesis of 9-LOX products when colonized by GPA? Recently, Nalam et al. (2013a) suggested that root-synthesized 9-LOX lipids are engaged by the plant to activate defenses in the shoots. The GPA infestation-induced up-regulation of PAD4 expression was attenuated in the lox5 mutant (Nalam et al., 2013a). Furthermore, 9-LOX products when applied to WT Arabidopsis induced PAD4 expression (Nalam et al., 2013a). Thus, as indicated in Figure 2, while Arabidopsis utilizes LOX5-derived lipids to activate defenses, the GPA has likely evolved to utilize 9-LOX-derived oxylipins, or products thereof, consumed from the host plant as a cue to facilitate feeding and promoting fecundity.
Concluding remarks
Although for quite some time Arabidopsis has been utilized as a model plant to understand the molecular basis of plant growth, development and response to pathogens, it is only in the last 10 years that significant progress has been made on utilizing Arabidopsis to study plant interaction with phloem-sap consuming insects. The compatible interaction between Arabidopsis and GPA has provided new insights on the physiological and molecular adaptations that contribute to controlling the severity of infestation. In addition, this interaction has also begun to provide information on how the insect targets host mechanisms to facilitate colonization. The near completion of the GPA genome sequence (AphidBase; www.aphidbase.org) and the recent development of plant delivered RNAi approaches to silence GPA gene expression should now facilitate understanding the function of GPA genes in this interaction, as well.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Nick Sloff and John Diaz-Montano are acknowledged for providing images of aphids. This work was made possible by grants from the National Science Foundation (MCB-0920600 and IOS-0919192) to Jyoti Shah.
References
- Adio A. M., Casteel C. L., De Vos M., Kim J. H., Joshi V., Li B., et al. (2011). Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 23, 3303–3318 10.1105/tpc.111.088989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J., Peters D. C. (1994). Ethylene production from wheat seedlings infested with biotypes of Schizaphis graminum (Homoptera: Aphidae). Environ. Entomol. 23, 992–998 [Google Scholar]
- Andréasson E., Jørgensen L. B. (2003). Localization of plant myrosinases and glucosinolates, in Integrative Phytochemistry: From Ethnobotany to Molecular Ecology, Vol. 37 ed Romeo J. T. (Amsterdam: Elsevier; ), 79–99 10.1016/S0079-9920(03)80019-9 [DOI] [Google Scholar]
- Anstead J. A., Froelich D. R., Knoblauch M., Thompson G. A. (2012). Arabidopsis P-protein filament formation requires both AtSEOR1 and AtSEOR2. Plant Cell Physiol. 53, 1033–1042 10.1093/pcp/pcs046 [DOI] [PubMed] [Google Scholar]
- Anstead J. A., Samuel P., Song N., Wu C., Thompson G. A., Goggin F. (2010). Activation of ethylene-related genes in response to aphid feeding on resistant and susceptible melon and tomato plants. Entomol. Exp. Appl. 134, 170–181 10.1111/j.1570-7458.2009.00945.x [DOI] [Google Scholar]
- Argandoña V. H., Chaman M., Cardemil L., Muñoz O., Zúñiga G. E., Corcuera L. J. (2001). Ethylene production and peroxidase activity in aphid-infested barley. J. Chem. Ecol. 27, 53–68 10.1023/A:1005615932694 [DOI] [PubMed] [Google Scholar]
- Ashford D. A., Smith W. A., Douglas A. E. (2000). Living on a high sugar diet: the fate of sucrose ingested by a phloem-feeding insect, the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 46, 335–341 10.1016/S0022-1910(99)00186-9 [DOI] [PubMed] [Google Scholar]
- Avila C. A., Arevalo-Soliz L. M., Jia L., Navarre D. A., Chen Z., Howe G. A., et al. (2012). Loss of function of FATTY ACID DESATURASE 7 in tomato enha-nces basal aphid resistance in a salicylate-dependent manner. Plant Physiol. 158, 2028–2041 10.1104/pp.111.191262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila C. A., Arevalo-Soliz L. M., Lorence A., Goggin F. (2013). Expression of α-DIOXYGENASE 1 in tomato and Arabidopsis contributes to plant defenses against aphids. Mol. Plant Microbe. Interact. [Epub ahead of print] 10.1094/MPMI-01-13-0031-R [DOI] [PubMed] [Google Scholar]
- Bansal R., Rouf Mian M. A., Mittapalli O., Michel A. P. (2013). Molecular characterization and expression analysis of soluble trehalase gene in Aphis glycines, a migratory pest of soybean. Bull. Entmol. Res. 10.1017/S0007485312000697 [DOI] [PubMed] [Google Scholar]
- Beneteau J., Renard D., Marché L., Douville E., Lavenant L., Rahbé Y., et al. (2010). Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 153, 1345–1361 10.1104/pp.110.153882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning U. F., Tamot B., Guelette B. S., Hoffmann-Benning S. (2012). New aspects of phloem-mediated long-distance lipid signaling in plants. Front. Plant Sci. 3:53 10.3389/fpls.2012.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R. L., Eastop V. F. (2000). Aphids on the World's Crops. Chichester: John Wiley and Sons [Google Scholar]
- Blow D. (1990). More of the catalytic triad. Nature 343, 694–695 10.1038/343694a0 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Bos J. I. B., Prince D., Pitino M., Maffei M. E., Win J., Hogenhout S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (Green Peach Aphid). PLoS Genet. 6:e1001216 10.1371/journal.pgen.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhssini M. E., Ogbonnaya F. C., Ketata H., Mosaad M. M., Street K., Amri A., et al. (2011). Progress in host plant resistance in wheat to Russian wheat aphid (Hemiptera: Aphididae) in North Africa and West Asia. Aus. J. Crop Sci. 5, 1108–1113 [Google Scholar]
- Cabrera y Poch H. L., Ponz F., Fereres A. (1998). Searching for resistance in Arabidopsis thaliana to the green peach aphid Myzus persicae. Plant Sci. 138, 209–216 10.1016/S0168-9452(98)00144-7 [DOI] [Google Scholar]
- Campbell B. C., Jones K. C., Dreyer D. L. (1986). Discriminative behavioral responses by aphids to various plant matrix polysaccharides. Entomol. Exp. Appl. 41, 17–24 10.1111/j.1570-7458.1986.tb02166.x [DOI] [Google Scholar]
- Carlini C. R., Grossi-de-Sa M. F. (2002). Plant toxic proteins with insecticidal properties: a review on their potentialities as bioinsecticides. Toxicon 40, 1515–1539 10.1016/S0041-0101(02)00240-4 [DOI] [PubMed] [Google Scholar]
- Chen X., Vosman B., Visser R. G. F., van der Vlugt R. A. A., Broekgaarden C. (2012). High throughput phenotyping for aphid resistance in large plant collections. Plant Methods 8, 33 Available online at: http://www.plantmethods.com/content/8/1/33 10.1186/1746-4811-8-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew F. S. (1988). Biological effects of glucosionlates, in Biologically Active Natural Products. 1. Vol. 8. ed Cutler H. G. (Washington, DC: American Chemical Society; ), 155–181 10.1021/bk-1988-0380.ch012 [DOI] [Google Scholar]
- Christensen S. A., Kolomiets M. V. (2011). The lipid language of plant-fungal interactions. Fungal Genet. Biol. 48, 4–14 10.1016/j.fgb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Cooper W. C., Jia L., Goggin F. L. (2004). Acquired and R-gene mediated resistance against the potato aphid in tomato. J. Chem. Ecol. 30, 2527–2542 10.1007/s10886-004-7948-9 [DOI] [PubMed] [Google Scholar]
- Cristofoletti P. T., Riberio A. F., Deraison C., Rahbé Y., Terra W. R. (2003). Midgut adaptation and digestive enzyme distribution in a phloem feeding insect, the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 49, 11–24 10.1016/S0022-1910(02)00222-6 [DOI] [PubMed] [Google Scholar]
- Devonshire A. L., Field L. M., Foster S. P., Moores G. D., Williamson M. S., Blackman R. L. (1998). The evolution of insecticide resistance in the peach-potato aphid, Myzus persicae. Philos. Trans. Royal Soc. Lond. B Biol. Sci. 353, 1677–1684 [Google Scholar]
- De Vos M., Jander G. (2009). Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant Cell. Environ. 32, 1548–1560 10.1111/j.1365-3040.2009.02019.x [DOI] [PubMed] [Google Scholar]
- De Vos M., Van Oosten V. R., Van Poecke R. M., Van Pelt J. A., Pozo M. J., Mueller M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18, 923–937 10.1094/MPMI-18-0923 [DOI] [PubMed] [Google Scholar]
- Dillwith J. W., Berberet R. C., Bergman D. K., Neese P. A., Edwards R. M., McNew R. W. (1991). Plant biochemistry and aphid populations: studies on the spotted alfalfa aphid, Therioaphis maculata. Arch. Insect Biochem. Physiol. 17, 235–251 10.1002/arch.940170407 [DOI] [Google Scholar]
- Divol F., Vilaine F., Thibivilliers S., Kusiak C., Sauge M. H., Dinant S. (2007). Involvement of the xyloglucan endotransglycosylase/ hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ. 30, 187–201 10.1111/j.1365-3040.2006.01618.x [DOI] [PubMed] [Google Scholar]
- Dixon A. F. G. (1998). Aphid Ecology: An Optimization Approach, 2nd Edn. New York, NY: Chapman and Hall [Google Scholar]
- Dong H.-P., Peng J., Bao Z., Meng X., Bonasera J. M., Chen G., et al. (2004). Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol. 136, 3628–3638 10.1104/pp.104.048900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. (2006). Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot. 57, 747–754 10.1093/jxb/erj067 [DOI] [PubMed] [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J. G. (2002a). Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae and Myzus persicae. Mol. Plant Microbe Interact. 15, 1025–1030 10.1094/MPMI.2002.15.10.1025 [DOI] [PubMed] [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J. G. (2002b). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566 10.1105/tpc.002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M. (2012). The role of roots in plant defence, in Plant Defence: Biological Control, eds Mérillon J. M. M., Ramawats K. G. G. (Netherlands: Springer; ), 291–309 10.1007/978-94-007-1933-0_12 [DOI] [Google Scholar]
- Erb M., Lenk C., Degenhardt J., Turlings T. C. J. (2009). The underestimated role of roots in defense against leaf attackers. Trends Plant Sci. 14, 653–659 10.1016/j.tplants.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Fernandez O., Bethencourt L., Quero A., Sangwan R. S., Clement C. (2010). Trehalose and plant stress response: friend or foe. Trends Plant Sci. 15, 409–417 10.1016/j.tplants.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Feys B. J., Wiermer M., Bhat R. A., Moisan L. J., Medina-Escobar N., Neu C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613 10.1105/tpc.105.033910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297 10.1146/annurev.arplant.53.100301.135248 [DOI] [PubMed] [Google Scholar]
- Fujita M., Mizukado S., Fujita Y., Ichikawa T., Nakazawa M., Seki M., et al. (2007). Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 364, 250–257 10.1016/j.bbrc.2007.09.124 [DOI] [PubMed] [Google Scholar]
- Gassmann W., Bhattacharjee S. (2012). Effector-triggered immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol. Plant Microbe Interact. 2, 862–868 10.1094/MPMI-01-12-0024-IA [DOI] [PubMed] [Google Scholar]
- Gaupels F., Knauer T., van Bel A. J. E. (2008). A combinatory approach for analysis of protein sets in barley sieve-tube samples using EDTA facilitated exudation and aphid stylectomy. J. Plant Physiol. 165, 95–103 10.1016/j.jplph.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2011). Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 155, 1566–1577 10.1104/pp.110.170399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georghiou G. P., Lagunes-Tejada A. (1991). The Occurrence of Resistance to Pesticides in Arthropods. An Index of Cases Reported Through 1989. Rome: FAO [Google Scholar]
- Girousse C., Moulia B., Silk W., Bonnemain J. L. (2005). Aphid infestation causes different changes in carbon and nitrogen allocation in alfalfa stems as well as different inhibitions of longitudinal and radial expansion. Plant Physiol. 137, 1474–1484 10.1104/pp.104.057430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E. E., Crute I. R., et al. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golem S., Culver J. (2003). Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis. Mol. Plant Microbe Interact. 16, 681–688 10.1094/MPMI.2003.16.8.681 [DOI] [PubMed] [Google Scholar]
- Gosset V., Harmel N., Göbel C., Francis F., Haubruge E., Wathelet J.-P., et al. (2009). Attacks by a piercing-sucking insect (Myzus persicae Sulzer) or a chewing insect (Leptinotarsa decemlineata Say) on potato plants (Solanum tuberosum L.) induce differential changes in volatile compound release and oxylipin synthesis. J. Exp. Bot. 60, 1231–1240 10.1093/jxb/erp015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez S., Michalakis Y., Van Munster M., Blanc S. (2013). Plant feeding by insect vectors can affect life cycle, population genetics and evolution of plant viruses. Funct. Ecol. 10.1111/1365-2435.12070. [DOI] [Google Scholar]
- Hao P., Liu C., Wang Y., Chen R., Tang M., Du B., et al. (2008). Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 146, 1810–1820 10.1104/pp.107.111484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chung E.-H., Hubert D. A., Tornero P., Dangl J. L. (2012). Specific missense alleles of the Arabidopsis jasmonic acid co-receptor COI1 regulate innate immune receptor accumulation and function. PLoS Genet. 8:e1003018 10.1371/journal.pgen.1003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helden M. C., Tjallingii W. F., Dieleman F. L. (1993). The resistance of lettuce (Lactuca sativa L.) to Nasonovia ribisnigri: bionomics of Nasonavia ribisnigri on near isogenic lettuce lines. Entomol. Exp. Appl. 66, 653–658 10.1111/j.1570-7458.1993.tb00692.x [DOI] [Google Scholar]
- Hill C. B., Li Y., Hartman G. L. (2006). A single dominant gene for resistance to the soybean aphid in the soybean cultivar Dowling. Crop Sci. 46, 1601–1605 10.2135/cropsci2005.11-0421 [DOI] [Google Scholar]
- Hodge S., Ward J. L., Beale M. H., Bennett M., Mansfield J. W., Powell G. (2012). Aphid-induced accumulation of trehalose in Arabidopsis thaliana is systemic and dependent upon aphid density. Planta 237, 1057–1064 10.1007/s00425-012-1826-1824 [DOI] [PubMed] [Google Scholar]
- Hopkins R. J., van Dam N. M., van Loon J. J. A. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54, 57–83 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Iordachescu M., Imai R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50, 1223–1229 10.1111/j.1744-7909.2008.00736.x [DOI] [PubMed] [Google Scholar]
- Ji L. H., Ding S. W. (2001). The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant Microbe Interact. 14, 715–724 10.1094/MPMI.2001.14.6.715 [DOI] [PubMed] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan I., Halitschke R., Kessler A., Sardanelli S., Denno R. F. (2008). Constitutive and Induced defenses to herbivory in above- and belowground plant tissues. Ecology 89, 392–406 10.1890/07-0471.1 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Jin H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246 10.1146/annurev-phyto-073009-114457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempema L. A., Cui X., Holzer F. M., Walling L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs: similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865 10.1104/pp.106.090662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J. S., Day M. F., Eastop V. F. (1962). A Conspectus of Aphids as Vectors of Plant Viruses. London: Commonwealth Institute of Entomology, 114 [Google Scholar]
- Kerchev P. I., Fenton B., Foyer C. H., Hancock R. D. (2012). Infestation of potato (Solanum tuberosum L.) by the peach-potato aphid (Myzus persicae Sulzer) alters cellular redox status and is influenced by ascorbate. Plant Cell Environ. 35, 430–440 10.1111/j.1365-3040.2011.02395.x [DOI] [PubMed] [Google Scholar]
- Kettles G. J., Drurey C., Schoonbeek H., Maule A. J., Hogenhaut S. A. (2013). Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 198, 1178–1190 10.1111/nph.12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R. (2012). Leaf senescence and abiotic stress share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481 10.1007/s00709-011-0308-z [DOI] [PubMed] [Google Scholar]
- Kim J. H., Jander G. (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49, 1008–1019 10.1111/j.1365-313X.2006.03019.x [DOI] [PubMed] [Google Scholar]
- Kim J. H., Lee B. W., Schroeder F. C., Jander G. (2008). Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J. 54, 1015–1026 10.1111/j.1365-313X.2008.03476.x [DOI] [PubMed] [Google Scholar]
- Kim K., Hill C. B., Hartman G. L., Hyten D. L., Hudson M. E., Diers B. W. (2010). Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor. Appl. Genet. 121, 599–610 10.1007/s00122-010-1333-6 [DOI] [PubMed] [Google Scholar]
- Klein C., Waterhouse D. F. (2000). Distribution and Importance of Arthropods Associated with Agriculture and Forestry in Chile. Canberra: ACIAR Monograph N 68 [Google Scholar]
- Kogan M., Ortman E. F. (1978). Antixenosis-a new term proposed to define Painter's “nonpreference” modality of resistance. Entomol. Soc. Am. Bull. 24, 175–176 [Google Scholar]
- Koornneef M., Meinke D. W. (2010). The development of Arabidopsis as a model plant. Plant J. 61, 909–921 10.1111/j.1365-313X.2009.04086.x [DOI] [PubMed] [Google Scholar]
- Koroleva O. A., Davies A., Deeken R., Thorpe M. R., Tomos A. D., Hedrich R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124, 599–608 10.1104/pp.124.2.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuśnierczyk A., Winge P., JØrstad T. S., Troczyñska J., Rossiter J. T., Bones A. M. (2008). Towards global understanding of plant defence against aphids – timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115 10.1111/j.1365-3040.2008.01823.x [DOI] [PubMed] [Google Scholar]
- Lankau R. A. (2007). Specialist and generalist herbivores exert opposing selection on a chemical defense. New Phytol. 175, 176–184 10.1111/j.1469-8137.2007.02090.x [DOI] [PubMed] [Google Scholar]
- Larson K. C., Whitham T. G. (1991). Manipulation of food resources by a gall-forming aphid: the physiology of sink-source interactions. Oecologia 88, 15–21 10.1007/BF00328398 [DOI] [PubMed] [Google Scholar]
- Levy M., Wang Q., Kaspi R., Parrella M. P., Abel S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96 10.1111/j.1365-313X.2005.02435.x [DOI] [PubMed] [Google Scholar]
- Lewsey M. G., Murphy A. M., MacLean D., Dalchau N., Westwood J. H., Macaulay K., et al. (2010). Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol. Plant Microbe Interact. 23, 835–845 10.1094/MPMI-23-7-0835 [DOI] [PubMed] [Google Scholar]
- Li Q., Xie Q., Smith-Becker J., Navarre D., Kaloshian I. (2006). Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol. Plant Microbe Interact. 19, 655–664 10.1094/MPMI-19-0655 [DOI] [PubMed] [Google Scholar]
- Liu R., Lü B., Wang X., Zhang C., Zhang S., Qian J., et al. (2010). Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. J. Biosci. 35, 435–450 10.1007/s12038-010-0049-8 [DOI] [PubMed] [Google Scholar]
- Lopez L., Camas A., Shivaji R., Ankala A., Williams P., Luthe D. (2007). Mir1-CP, a novel defense cysteine protease accumulates in maize vascular tissues in response to herbivory. Planta 226, 517–527 10.1007/s00425-007-0501-7 [DOI] [PubMed] [Google Scholar]
- Louda S., Mole S. (1991). Glucosinolates: chemistry and ecology, in Herbivores: Their Interactions with Secondary Plant Metabolites, eds Rosenthal G. A., Berenbaum M. R. (San Diego, CA: Academic Press; ), 123–164 10.1016/B978-0-12-597183-6.50009-7 [DOI] [Google Scholar]
- Louis J. (2011). Molecular basis of plant defense against aphids: Role of the Arabidopsis thaliana PAD4 and MPL1 genes. Ph.D. dissertation, Denton, TX: University of North Texas. Available online at: http://digital.library.unt.edu/ark:/67531/metadc84242/
- Louis J., Gobbato E., Mondal H. A., Feys B. J., Parker J. E., Shah J. (2012a). Discrimination of Arabidopsis PAD4 activities in defense against green peach aphid and pathogens. Plant Physiol. 158, 1860–1872 10.1104/pp.112.193417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J., Mondal H. A., Shah J. (2012b). Green peach aphid infestation induces Arabidopsis PHYTOALEXIN-DEFICIENT4 expression at site of insect feeding. Plant Signal. Behav. 7, 1–3 10.4161/psb.22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J., Singh V., Shah J. (2012c). Arabidopsis thaliana - Aphid Interaction. Arabidopsis Book 10:e0159 10.1199/tab.0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J., Lorenc-Kukula K., Singh V., Reese J. C., Jander G., Shah J. (2010a). Antibiosis against the green peach aphid requires the Arabidopsis thaliana MYZUS PERSICAE-INDUCED LIPASE1 gene. Plant J. 64, 800–811 10.1111/j.1365-313X.2010.04378.x [DOI] [PubMed] [Google Scholar]
- Louis J., Leung Q., Pegadaraju V., Reese J. C., Shah J. (2010b). PAD4-dependent antibiosis contributes to the ssi2-conferred hyper-resistance to the green peach aphid. Mol. Plant Microbe Interact. 23, 618–627 10.1094/MPMI-23-5-0618 [DOI] [PubMed] [Google Scholar]
- Mantelin S., Bhattarai K. K., Kaloshian I. (2009). Ethylene contributes to potato aphid susceptibility in a compatible tomato host. New Phytol. 183, 444–456 10.1111/j.1469-8137.2009.02870.x [DOI] [PubMed] [Google Scholar]
- Matthews R. E. F. (1991). Relationships between plant viruses and invertebrates, in Plant Virology, 3rd Edn., ed Matthews R. E. F. (San Diego, CA: Academic Press; ), 520–561 10.1016/B978-0-12-480553-8.50019-5 [DOI] [Google Scholar]
- Meinke D. W., Cherry J. M., Dean C., Rounsley S. D., Koornneef M. (1998). Arabidopsis thaliana: a model plant for genome analysis. Science 282, 662–682 10.1126/science.282.5389.662 [DOI] [PubMed] [Google Scholar]
- Mewis I., Appel H. M., Hom A., Raina R., Schultz J. C. (2005). Major signaling pathways modulate Arabidopsis thaliana (L.) glucosinolate accumulation and response to both phloem feeding and chewing insects. Plant Physiol. 138, 1149–1162 10.1104/pp.104.053389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis I., Tokuhisa J. G., Schultz J. C., Appel H. M., Ulrichs C., Gershenzon J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67, 2450–2462 10.1016/j.phytochem.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Petersen B. L., Glawischnig E., Jensen A. B., Andreasson E., Halkier B. A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131, 298–308 10.1104/pp.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles P. W. (1999). Aphid saliva. Biol. Rev. 74, 41–85 10.1017/S0006323198005271 [DOI] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M. A., Shulaev V., et al. (2009). The plant NADPH oxidase RbohD mediates rapid, systemic signaling in response to diverse stimuli. Sci. Signal. 2, ra45 10.1126/scisignal.2000448 [DOI] [PubMed] [Google Scholar]
- Miller H., Neese P. A., Ketring D., Dillwith J. W. (1994). Involvement of ethylene in aphid infestation of barley. J. Plant Growth Regul. 13, 167–171 10.1007/BF00226033 [DOI] [Google Scholar]
- Milligan S. B., Bodeau J., Yaqhoobi J., Kaloshian I., Zabel P., Williamson V. M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler T. E., Sylvester E. S. (1961). A comparison of the injury of alfalfa by the aphids Therioaphis maculata and Macrosiphum pisi. J. Econ. Entomol. 54, 615–622 [Google Scholar]
- Moran P. J., Thompson G. A. (2001). Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 125, 1074–1085 10.1104/pp.125.2.1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P. J., Cheng Y. F., Cassell J. L., Thompson G. A. (2002). Gene expression profiling of Arabidopsis thaliana in compatible plant aphid interactions. Arch. Insect Biochem. Physiol. 51, 182–203 10.1002/arch.10064 [DOI] [PubMed] [Google Scholar]
- Morita M., Shitan N., Sawada K., Van Montagu M. C. E., Inzé D., Rischer H., et al. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. U.S.A. 106, 2447–2452 10.1073/pnas.0812512106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosblech A., Feussner I., Heilmann I. (2009). Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47, 511–517 10.1016/j.plaphy.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Mur L. A. J., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcome of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262 10.1104/pp.105.072348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N. S., Louis J., Pappan L. K., Pappan K., Begum K., Chen M. S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U.S.A. 105, 9965–9969 10.1073/pnas.0708958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam V. J., Keeretaweep J., Sarowar S., Shah J. (2012). Root-derived oxylipins promote green peach aphid performance on Arabidopsis thaliana foliage. Plant Cell 24, 1643–1653 10.1105/tpc.111.094110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam V. J., Keereetaweep J., Shah J. (2013a). The green peach aphid, Myzus persicae, acquires a LIPOXYGENASE5-derived oxylipin from Arabidopsis thaliana, which promotes colonization of the host plant. Plant Signal. Behav. 8:e22735 10.4161/psb.22735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam V. J., Shah J., Nachappa P. (2013b). Emerging role of roots in plant responses to aboveground insect herbivory. Insect Sci. 20, 286–296 10.1111/1744-7917 [DOI] [PubMed] [Google Scholar]
- Painter R. H. (1951). Insect resistance in Crop Plants. New York, NY: The Macmillan Co, 520 [Google Scholar]
- Pandey S. P., Baldwin I. T. (2007). RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J. 50, 40–53 10.1111/j.1365-313X.2007.03030.x [DOI] [PubMed] [Google Scholar]
- Pandey S. P., Shahi P., Gase K., Baldwin I. T. (2008). Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc. Natl. Acad. Sci. U.S.A. 105, 4559–4564 10.1073/pnas.0711363105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59, 417–441 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Pauquet J., Burget E., Hagen L., Chovelon V., Le Menn A., Valot N., et al. (2004). Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses, in Proceedings of Cucurbitaceae 2004, the 8th EUCARPIA Meeting on Cucurbit Genetics and Breeding, eds Lebeda A., Paris H. (Olomouc: Palaky University; ), 325–329 [Google Scholar]
- Pedigo L. P. (1999). In Entomology and Pest Management 3rd Edn. Englewood Cliffs, NJ: Prentice-Hall [Google Scholar]
- Pegadaraju V. (2005). Molecular Insights into Arabidopsis Response to Myzus Persicae Sülzer (Green Peach Aphid). Ph.D. thesis, Kansas State University, Manhattan, KS.
- Pegadaraju V., Knepper C., Reese J. C., Shah J. (2005). Premature leaf senescence modulated by the Arabidopsis PAD4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol. 139, 1927–1934 10.1104/pp.105.070433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V., Louis J., Singh V., Reese J. C., Bautor J., Feys B., et al. (2007). Phloem-based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. Plant J. 52, 332–341 10.1111/j.1365-313X.2007.03241.x [DOI] [PubMed] [Google Scholar]
- Pescod K., Quick W., Douglas A. (2007). Aphid responses to plants with genetically manipulated phloem nutrient levels. Physiol. Entomol. 32, 253–258 10.1111/j.1365-3032.2007.00577.x [DOI] [Google Scholar]
- Pfalz M., Vogel H., Kroymann J. (2009). The gene controlling the Indole Glucosinolate Modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21, 985–999 10.1105/tpc.108.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pitino M., Coleman A. D., Maffei M. E., Ridout C. J., Hogenhout S. A. (2011). Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE 6:e25709 10.1371/journal.pone.0025709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M., Hogenhout S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26, 130–139 10.1094/MPMI-07-12-0172-FI [DOI] [PubMed] [Google Scholar]
- Pollard D. G. (1973). Plant penetration by feeding aphids (Hemiptera: Aphidoidea): a review. Bull. Entomol. Res. 62, 631–714 10.1017/S0007485300005526 [DOI] [Google Scholar]
- Pompon J., Quiring D., Giordanengo P., Pelletier Y. (2010). Role of xylem consumption on osmoregulation in Macrosiphum euphorbiae (Thomas). J. Insect Physiol. 56, 610–615 10.1016/j.jinsphys.2009.12.009 [DOI] [PubMed] [Google Scholar]
- Ralhan A., Schöttle S., Thurow C., Iven T., Feussner I., Polle A., et al. (2012). The vascular pathogen Verticillium longisporum requires a jasmonic acid-independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots. Plant Physiol. 159, 1192–1203 10.1104/pp.112.198598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J. S., Wilson A. C., de Vos M., Sun Q., Tamborindeguy C., Winfield A., et al. (2007). Genomic resources for Myzus persicae: EST sequencing, SNP identification, and microarray design. BMC Genomics 8:423 10.1186/1471-2164-8-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113 10.1023/A:1006380021658 [DOI] [PubMed] [Google Scholar]
- Rasmann S., Agrawal A. A. (2008). In defense of roots: a research agenda for studying plant resistance to belowground herbivory. Plant Physiol. 146, 875–880 10.1104/pp.107.112045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J. C., Tjallingii W. F., van Helden M., Prado E. (2000). Waveform comparisons among AC and DC electronic monitoring systems for aphid (Homoptera: Aphididae) feeding behavior, in Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior, eds Walker G. P., Backus E. A. (Lanham, MD: Thomas Say Publications in Entomology, Entomological Society of America; ), 70–101 [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N., et al. (2011). Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119 10.1111/j.1469-8137.2011.03675.x [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. G. (2011). Disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Roche P., Alston F. H., Maliepaard C., Evans K., Vrielink R., Dunemann F., et al. (1997). RFLP and RAPD markers linked to the rosy leaf curling aphid resistance gene (Sd1) in apple. Theor. Appl. Gen. 94, 528–533 10.1007/s00122005044712582537 [DOI] [Google Scholar]
- Rodriguez P. A., Bos J. I. B. (2013). Toward understanding the role of aphid effectors in plant infestation. Mol. Plant-Microbe Interact. 26, 25–30 10.1094/MPMI-05-12-0119-FI [DOI] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709 10.1146/annurev.arplant.57.032905.105441 [DOI] [PubMed] [Google Scholar]
- Rossi M., Goggin F. L., Milligan S. B., Kaloshian I., Ullman D. E., Williamson V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U.S.A. 95, 9750–9754 10.1073/pnas.95.17.9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G. F. E. (2010). Phospholipase A in plant signal transduction, in Plant Cell Monographs-Lipid Signaling in Plants, Vol. 16, ed Munnik T. (Berlin, Heidelberg: Springer-Verlag; ), 3–22 [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 6849–6854 10.1073/pnas.1132018100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. (2005). Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 43, 229–260 10.1146/annurev.phyto.43.040204.135951 [DOI] [PubMed] [Google Scholar]
- Shah J., Zeier J. (2013). Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 4:30 10.3389/fpls.2013.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. X., Bacigalupe L. D., Luna-Rudloff M., Figueroa C. C. (2012a). Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: Aphididae) II: Costs and benefits. PLoS ONE 7:e36810 10.1371/journal.pone.0036810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. X., Jander G., Samaniego H., Ramsey J. S., Figueroa C. C. (2012b). Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: Aphididae) I: a transcriptomic survey. PLoS ONE 7:e36366 10.1371/journal.pone.0036366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V. (2012). Studies on Plant-Aphid Interactions: a Novel Role for Trehalose Metabolism in Arabidopsis Defense Against Green Peach Aphid, Ph.D. dissertation, Denton, TX: University of North Texas. UNT Digital Library [Google Scholar]
- Singh V., Louis J., Ayre B., Reese J. C., Shah J. (2011). TREHALOSE PHOSPHATE SYNTHASE11-dependent trehalose metabolism regulates Arabidopsis thaliana defense against the phloem-feeding insect, Myzus persicae. Plant. J. 67, 94–104 10.1111/j.1365-313X.2011.04583.x [DOI] [PubMed] [Google Scholar]
- Singh V., Shah J. (2012). Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signal. Behav. 7, 1–3 10.4161/psb.20066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M. (2005). Plant Resistance to Arthropods: Molecular and Conventional Approaches. Netherlands: Springer; 10.1007/1-4020-3702-3 [DOI] [Google Scholar]
- Smith C. M., Clement S. L. (2012). Molecular bases of plant resistance to arthropods. Annu. Rev. Entomol. 57, 309–328 10.1146/annurev-ento-120710-100642 [DOI] [PubMed] [Google Scholar]
- Sønderby I. E., Geu-Flores F., Halkier B. A. (2010). Biosynthesis of glucosinolates – gene discovery and beyond. Trends Plant Sci. 15, 283–290 10.1016/j.tplants.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Spiller N. J., Koenders L., Tjallingii W. F. (1990). Xylem ingestion by aphids–a strategy for maintaining water balance. Entomol. Exp. Appl. 55, 101–104 10.1111/j.1570-7458.1990.tb01352.x [DOI] [Google Scholar]
- Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V., Mittler R. (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 139, 1313–1322 10.1104/pp.105.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B. P. H. J., Nürnberg T., Jossten M. H. A. J. (2011). On PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15 10.1105/tpc.110.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii W. F. (1990). Continuous recording of stylet penetration activities by aphids, in Aphid–plant genotype interactions, eds Campbell R. K., Eikenbary R. D. (Amsterdam: Elsevier; ), 89–99 [Google Scholar]
- Tjallingii W. F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57, 739–745 10.1093/jxb/erj088 [DOI] [PubMed] [Google Scholar]
- Tjallingii W. F., Esch T. H. (1993). Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol. Entomol. 18, 313–328 10.1111/j.1365-3032.1993.tb00604.x [DOI] [Google Scholar]
- Upchurch R. G. (2008). Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 30, 967–977 10.1007/s10529-008-9639-z [DOI] [PubMed] [Google Scholar]
- Vandenborre G., Smagghe G., Van Damme E. J. M. (2011). Plant lectins as defense proteins against polyphagous insects. Phytochemistry 72, 1538–1550 10.1016/j.phytochem.2011.02.024 [DOI] [PubMed] [Google Scholar]
- Vasconcelos I. M., Oliveira J. T. (2004). Antinutritional properties of plant lectins. Toxicon 44, 385–403 10.1016/j.toxicon.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Vasquez B. L. (1995). Resistant to most insecticides, Chapter 15, in Book of Insects Records, eds Walker T. J. (Gainesville, FL: University of Florida; ), 34–36 [Google Scholar]
- Vos P., Simons G., Jesse T., Wijbrandi J., Heinen L., Hogers R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotech. 16, 1365–1369 10.1038/4350 [DOI] [PubMed] [Google Scholar]
- Walker G. P. (2000). A beginner's guide to electronic monitoring of homopteran probing behavior, in Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior, eds Walker G. P., Backus E. A. (Lanham, MD: Thomas Say Publications in Entomology, Entomological Society of America; ), 14–40 [Google Scholar]
- Walker G. P., Medina-Ortega K. J. (2012). Penetration of faba bean sieve elements by pea aphid does not trigger forisome dispersal. Entomol. Exp. Appl. 144, 326–335 10.1111/j.1570-7458.2012.01297.x18621720 [DOI] [Google Scholar]
- Walling L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216 [DOI] [PubMed] [Google Scholar]
- Walling L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866 10.1104/pp.107.113142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz C., Giavalisco P., Schad M., Juenger M., Klose J., Kehr J. (2004). Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry 65, 1795–1804 10.1016/j.phytochem.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Wang X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7, 329–336 10.1016/j.pbi.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697 10.1093/aob/mcm079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirmer M., Feys B. J., Parker J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389 10.1016/j.pbi.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Wilkinson T. L., Ashford D. A., Pritchard J., Douglas A. E. (1997). Honeydew sugars and osmoregulation in the pea aphid Acyrthosiphon pisum. J. Exp. Biol. 200, 2137–2143 [DOI] [PubMed] [Google Scholar]
- Will T., Kornemann S. R., Furch A. C. U., Tjallingii W. F., van Bel A. J. E. (2009). Aphid watery saliva counteracts sieve-tube occlusion: a universal phenomenon. J. Exp. Biol. 212, 3305–3312 10.1242/jeb.028514 [DOI] [PubMed] [Google Scholar]
- Will T., Tjallingii W. F., Thonnessen A., van Bel A. J. E. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U.S.A. 104, 10536–10541 10.1073/pnas.0703535104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T., van Bel A. J. E. (2006). Physical and chemical interactions between aphids and plants. J. Exp. Bot. 57, 729–737 10.1093/jxb/erj089 [DOI] [PubMed] [Google Scholar]
- Winde I., Wittstock U. (2011). Insect herbivore counteradaptations to the plant glucosinolate-myrosinase system. Phytochemistry 72, 1566–1575 10.1016/j.phytochem.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Wittstock U., Burow M. (2010). Glucosinolate breakdown in Arabidopsis: mechanisms, regulation and biological significance. Arabidopsis Book 8, e0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yan Y., Borrego E., Kolomiets M. V. (2013). Jasmonate biosynthesis, perception and function in plant development and stress response, in Lipid Metabolism, Chapter 16, ed Baez R. V. (InTech; ), 393–442 Available online at: http://www.intechopen.com/books/lipid-metabolism/jasmonate-biosynthesis-perception-and-function-in-plant-development-and-stress-responses 10.5772/52675 [DOI] [Google Scholar]
- Yu T. S., Lue W. L., Wang S. M., Chen J. (2000). Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol. 123, 319–326 10.1104/pp.123.1.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shi H., Chen L., Wang X., Lü B., Zhang S., et al. (2011). Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 11:11 10.1186/1471-2229-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Luan J. B., Qi J. F., Huang C. J., Li M., Zhou X. P., et al. (2012). Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 21, 1294–1304 10.1111/j.1365-294X.2012.05457.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Myers A. M., James M. G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138, 663–674 10.1104/pp.105.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang W., Stanley B. A., Assmann S. M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20, 3210–3226 10.1105/tpc.108.063263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monoxygenase. Plant Cell 11, 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Tsui F., Klessig D. F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell H., Murphy A. M., Groen S. C., Tungadi T., Westwood J. H., Lewsey M. G., et al. (2011). Cucumber mosaic virus and its 2b RNA silencing suppressor modify plant-aphid interactions in tobacco. Sci. Rep. 1, 187 10.1038/srep00187 [DOI] [PMC free article] [PubMed] [Google Scholar]