Abstract
Patterns of biodiversity are often explained by ecological processes, where traits that promote novel ways of interacting with the environment (key innovations) play a fundamental role in promoting diversification. However, sexual selection and social competition can also promote diversification through rapid evolution of ornamental traits. Because selection can operate only on existing variation, the tendency of ornamental traits to constrain or enable the production of novel phenotypes is a crucial but often overlooked aspect of diversification. Starlings are a speciose group characterized by diverse iridescent colors produced by nanometer-scale arrays of melanin-containing organelles (melanosomes) that play a central role in sexual selection and social competition. We show that evolutionary lability of these colors is associated with both morphological and lineage diversification in African starlings. The solid rod-like melanosome morphology has evolved in a directional manner into three more optically complex forms that can produce a broader range of colors than the ancestral form, resulting in (i) faster color evolution, (ii) the occupation of novel, previously unreachable regions of colorspace, and ultimately (iii) accelerated lineage diversification. As in adaptive radiations, key innovations in ornament production can provide high phenotypic trait variability, leading to dramatic effects on the tempo and mode of diversification.
Keywords: biophotonics, macroevolution, phylogenetics, structural color, Sturnidae
Adaptive radiation—the rapid ecological diversification of related species triggered by ecological opportunity like colonization of novel environments (1)—is one of the central processes responsible for the observed diversity of life, and its study has provided a powerful framework for understanding patterns of animal and plant diversification. Adaptive radiations are characterized by rapid lineage diversification and/or accumulation of morphological disparity resulting from ecological opportunity (2, 3), such as upon the colonization of islands (4). Many classic examples of adaptive radiations, such as the Galapagos finches (5), Hawaiian Drosophilids (6, 7), Caribbean anoles (8), and African cichlid fishes (9, 10), exemplify this pattern. However, ecological opportunity may also emerge due to intrinsic properties of a lineage or clade (3, 11). Traits known as “key innovations” [i.e., features that allow organisms to interact with the environment in novel ways (12)] relax constraints of adaptive evolution and/or enhance competitive ability (13) and can lead to rapid bursts of diversification through occupation of new adaptive zones (2, 14). Innovations such as the pharyngeal jaw of labroid fishes (15, 16), which has evolved independently at least six times in this exceptionally diverse clade (17), and the highly labile beak shape of Madagascar vangas (18) exemplify how such naturally selected novelties can have dramatic effects in both lineage and morphological diversification.
Although key innovations are a major component of adaptive radiations (1, 14), their role in nonecological diversification has rarely been considered. One of the primary nonecological drivers of diversification is sexual selection, where signals used in mate choice or social competition can rapidly diverge, leading to reproductive isolation (19, 20). There is growing evidence that sexual selection is an important but overlooked component in many recognized adaptive [e.g., cichlids (9, 10) and Hawaiian Drosophilids (6, 21)] and nonadaptive radiations [e.g., Laupala crickets (22) and Xerocrassa snails (23)]. Innovations that expand a lineage’s perceptual environment have been suggested to facilitate this process. For example, neuroanatomical features allowing for novel communication modalities (24, 25) or visual receptors adapted to novel lighting environments (26) can affect the channels available for mate recognition and promote diversification. However, ornamental traits themselves could also differ in their ability to produce variation and respond to evolutionary forces, thereby directly influencing patterns of diversification. Ornamental traits can diverge even between allopatric populations living in similar ecological conditions and are crucial for maintaining reproductive isolation when in sympatry (27). Therefore, traits that favor the rapid evolution of novel ornamental phenotypes can provide opportunities for divergent phenotypic evolution and rapid population differentiation.
Avian colors provide a unique opportunity to examine how proximate mechanisms constrain or enable the evolution and diversification of ornaments under sexual selection. For example, divergences in socially selected traits such as plumage color and song are essential components in avian diversification, but many clades are characterized by convergent and parallel evolution of plumage color and patterns (28). Although commonly attributed to similar selection pressures, convergent evolution may actually reflect preexisting developmental bias arising from the mechanisms of color production and body patterning (28, 29). Feather coloration results from two broad categories of mechanisms: pigments and structural colors. Pigments are highly constrained in their expression, with a narrow color palette that can be expanded only by evolving novel metabolic pathways (30, 31). Not surprisingly, lineages with pigment-based colors are characterized by extensive phenotypic convergence in ornaments (32, 33). By contrast, a wide array of structural iridescent colors is produced by nanometer-scale arrangements of keratin, melanosomes, and air within feather barbules. This modular template can produce a broad range of colors through minor alteration of its dimensions and organization (34–37). Birds have evolved several unique melanosome morphologies with additional structural parameters that can be independently modified to further alter color and that are associated with many classic examples of avian radiations [e.g., hummingbirds, birds-of-paradise, sunbirds, and trogons (34)]. This extreme lability, coupled with the direct form-to-function mapping of the nanostructural morphology to the color phenotype, makes iridescent colors an ideal trait with which to investigate whether key morphological innovations in ornamental evolution promote diversification.
We investigated the evolution of color-producing nanostructures and their effect on morphological disparity and lineage diversification in African starlings (Sturnidae), the only monophyletic avian group to display all of the melanosome morphotypes that have been identified in birds (38). Sexual selection and social competition have been shown to drive the evolution of coloration in both males and females in this clade (39). Moreover, iridescent colors may be sexually selected and involved in mutual mate choice in a variety of species in this family (40, 41), suggesting a likely role for iridescent colors in mate choice and social competition (39).
Results and Discussion
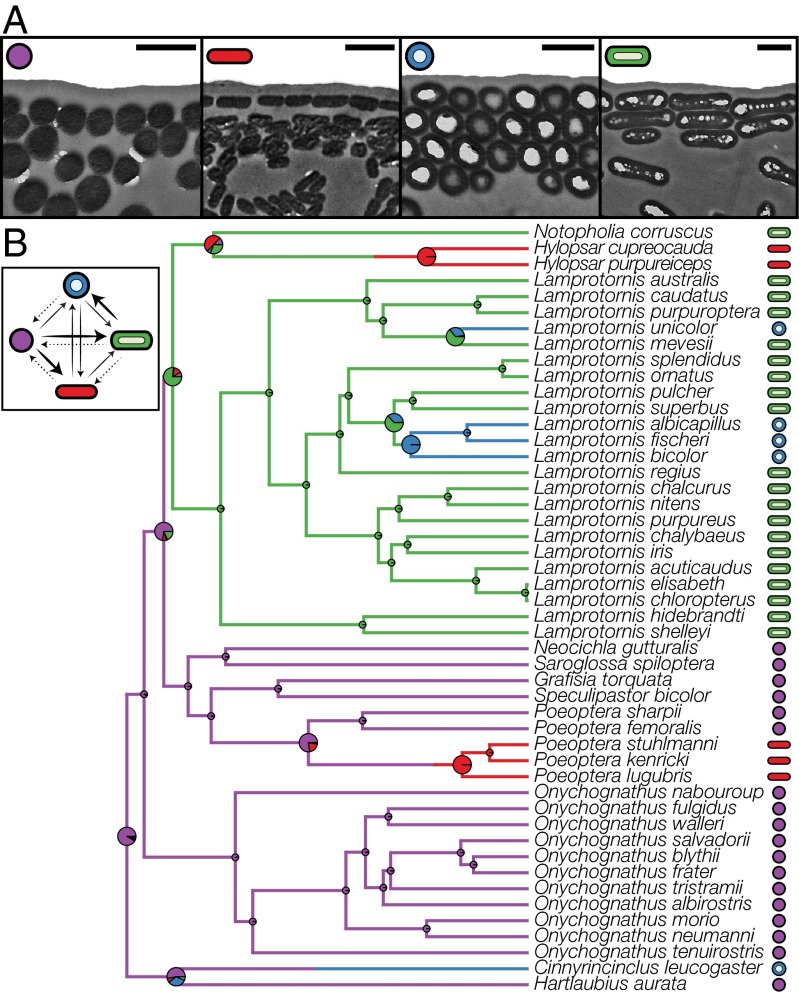
Most iridescent bird species, including non-African starlings, have solid rod-shaped melanosomes arranged in arrays beneath a keratin film (34, 38) and produce iridescence through thin-film interference (Fig. 1A). Using transmission electron microscopy (TEM), we confirmed (34, 38, 42) three innovations in melanosome shape from African starling feathers that provide additional optically relevant parameters: (i) flattened melanosomes that allow melanin layers to be thinner and more densely packed or stacked to form multilayers; (ii) hollow melanosomes that provide sharp optical interfaces between air and melanin, the relative content of which can also be varied to influence color; and (iii) platelet-shaped (hollow and flattened) melanosomes that can form color-producing single layers, multilayers, or alternating platelet–keratin layers (Fig. 1A). Importantly, each starling species has only one of these four morphologies of iridescence-producing melanosome (38).
Fig. 1.
Morphology and evolution of melanosomes responsible for iridescence in African starlings. (A) Melanosome morphologies observed in African starlings. (Scale bars: 500 nm.) (B) Maximum clade credibility tree from the Bayesian phylogenetic analysis. Pie charts represent posterior probabilities for each morphology at the node; large pie charts indicate nodes at which transitions are observed or the ancestral state is uncertain. Branch colors show a representative mapping of character state transition from SIMMAP used in color diversification analyses. (Inset) Transition posterior probabilities between melanosome morphologies. Arrow thickness corresponds to probability of transition being zero, with dashed gray lines indicating transitions with greatest probability of being zero (Fig. S1; for trait-dependent diversification estimates of transition rates see Fig. 3).
We combined phylogenetic comparative approaches across a posterior sample of trees to test the hypothesis that the evolution of these novel traits drives color and lineage diversification in the group. Ancestral state reconstruction identified the typical, solid rod-shaped melanosome as the ancestral morphology of the African starling clade (Fig. 1B). We then compared two models of melanosome evolution: in the first, transitions occurred only from the ancestral to the derived forms, but not vice versa (irreversible model), whereas in the second, modifications in shape and hollowness occurred independently and were reversible (stepwise model). Our results strongly support the irreversible model, indicating that melanosome evolution in this clade occurs in a directional manner from the ancestral to the three derived forms [2ln Bayes factor (BF) irreversible, 6.22; stepwise, 4.07; irreversible vs. stepwise, 2.15; Fig. 1B]. This model is also supported when trait-dependent diversification is taken into account (see below). Thus, we show that an optically simple ancestral melanosome template has repeatedly evolved into more optically complex morphologies at least three times and that these derived forms transition among themselves, but never revert to the ancestral form (Fig. 1B and Fig. S1).
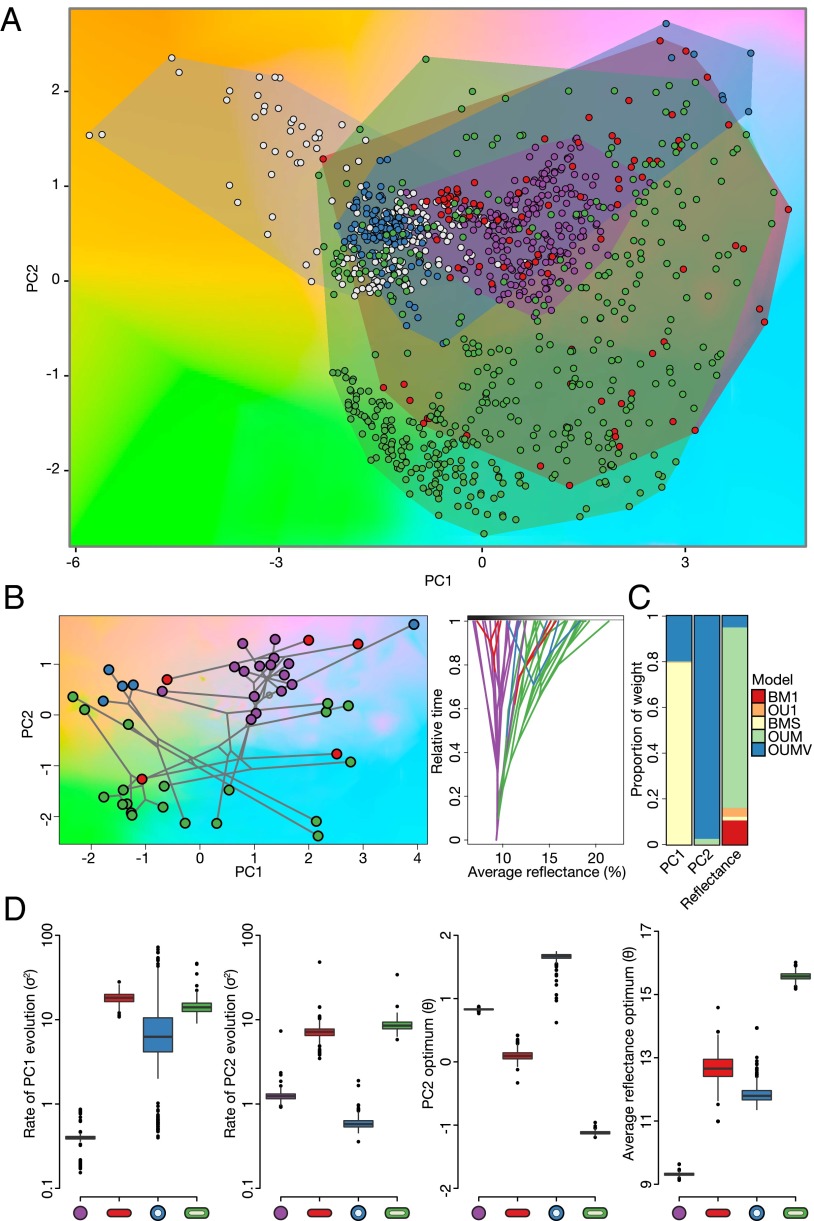
Evolution of these morphological novelties had a marked effect on color diversification (Fig. 2A). We used avian visual models (43) to project the colors of African starling feathers in a colorspace defined by brightness (average reflectance) and two chromatic (wavelength-related) axes of variation (Fig. 2 A and B). We then tested evolutionary models (44) in which color evolves independently of melanosome morphology or not (Fig. 2 C and D). In the latter models, color produced by different melanosomes is allowed to accumulate disparity at different rates and/or evolve toward occupation of different areas of the colorspace (i.e., different optimal values). Brightness evolution showed a melanosome-dependent pattern in which colors of derived morphologies are brighter than those produced by the ancestral morphology [αlog(average reflectance) = 1.81, 95% confidence interval (CI) = 1.18–4.51]. Color disparity in the first principal component (PC1, the short-long wavelength axis; Table S1) accumulated through time with rates 10–40 times higher for the derived morphologies than for the ancestral melanosome morphology. In PC2 (the mid- to extreme-wavelength axis), colors produced by different melanosomes had both differential rates of evolution and different optima in colorspace (αPC2 = 3.76, 95% CI = 2.99–4.47). The ancestral rod-shaped melanosome lineages had an optimum closest to the achromatic center (unsaturated colors), with the derived morphologies, in particular the hollow ones, showing optima in more chromatic areas of the colorspace (Fig. 2 A, B, and D). Additionally, the two flattened melanosome morphologies had the fastest rates of color evolution.
Fig. 2.
Models of color evolution in African starlings. (A) Colorspace with all 10 color patches measured from male and female African starlings overlaid. Noniridescent colors (white) occupy a different area of colorspace than iridescent colors, and the ancestral rod-shaped melanosome (purple) occupies a smaller area close to the achromatic center than the derived morphologies (red, flattened; blue, hollow rods; green, platelets). Convex polygons represent the area of colorspace occupied by each morphology. (B) Chromatic (Left) and brightness (Right) phylocolorspace of the iridescent mantle coloration of African starlings. (C) Mean Akaike weights of models from the posterior distribution of 500 trees and melanosome regime mappings used in the analysis. BM1, Brownian motion with a single rate; OU1, Ornstein–Uhlenbeck process with a single optimum; BMS, Brownian motion with melanosome-dependent rates; OUM, Ornstein–Uhlenbeck process with melanosome-dependent optima; OUMV, Ornstein–Uhlenbeck process with melanosome-dependent rates and optima (Figs. S2–S5 and Tables S2 and S3). (D) Maximum-likelihood parameter estimates for models across the posterior distribution of trees and melanosome regime mappings for the best models for each color trait, back-transformed to the original trait scale.
These results demonstrate that, as hypothesized, the origin of novel melanosomes promotes both (i) occupation of previously unreachable areas of colorspace and (ii) accelerated accumulation of color disparity, thus underlying macroevolutionary patterns of ornamental trait evolution. Taken together with the directional mode of melanosome evolution, these results suggest that derived melanosomes act as an ornamental release of constraint, favoring signal elaboration and diversification (45). Further, lineages with derived morphologies occupy a broader area of the colorspace as a combined consequence of different rates of hue disparity accumulation and higher optimal values of both saturation and brightness (Fig. 2 A and B and Fig. S2). Therefore, irreversible changes to derived melanosomes favor the evolution of brighter and more saturated colors, consistent with predictions of a functional role in intraspecific communication through sexual or social selection that are otherwise constrained in expression due to a limitation in available forms. This melanosome-dependent color evolution was observed across body patches and in both males and females (Tables S2 and S3), consistent with the idea that social competition has driven plumage evolution in both sexes in African starlings (39).
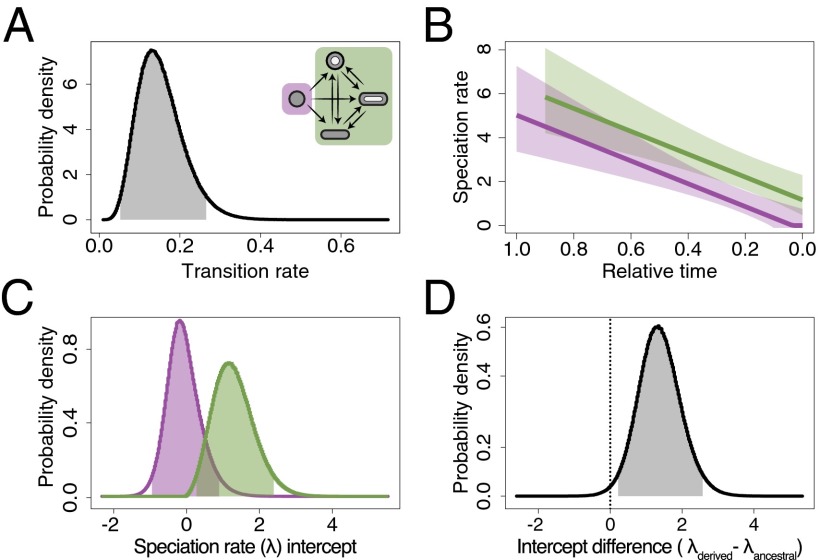
Lineages with derived melanosomes also diversify faster (46) (Fig. 3 and Table S4). An irreversible model with a slowdown in speciation rates over time [compared with the best time-independent model: ΔAICc = 13.16 (AICc, Second-Order Akaike Information Criterion); Fig. 3 A and B] and a greater average speciation rate in lineages with derived melanosome forms than in those with the ancestral morphology (compared with the best trait-independent model: ΔAICc = 3.82, Fig. 3 C and D) best explained speciation as a function of melanosome type. Thus, speciation in this clade can be described by two important aspects (Fig. 3): (i) a slowdown in speciation that is general to all African starlings and (ii) greater speciation rates, controlling for time, for derived melanosomes than for ancestral ones. The time-dependent component observed in this radiation (Fig. 3 A and B) is consistent with diversity-dependent dynamics where diversification rates are initially high as lineages colonize a novel environment, but subsequently slow down as ecological opportunities diminish (1). On the other hand, the higher average speciation rate observed for lineages with novel melanosomes (Fig. 3 C and D) likely reflects the heightened opportunity for diversification resulting from the enhanced ornamental variation that these labile traits provide. We also found the evolution of novel melanosome morphologies to be independent of social system evolution [MCMCglmm: estimate = 172.7, 95% CI = −223.3–705.06, Markov Chain Monte Carlo probability (pMCMC) = 0.36], suggesting that the opportunity for accelerated diversification was a consequence of ornamental evolvability and not due to the potentially confounding effects of the complex cooperatively breeding systems found in this clade (39).
Fig. 3.
Speciation rates for African starlings estimated from MuSSE models. (A) Bayesian posterior probabilities for the transition rate in the best-supported model (irreversible evolution with equal transition rates from the ancestral to the derived and among the derived melanosome morphologies; Inset and Table S4) across the posterior sample of trees. Shaded area represents the 95% highest probability density region for the parameter estimate. (B) Estimates for the time-dependent speciation rates for the ancestral (violet) and derived (green) melanosome morphologies. Lines represent the posterior mode estimates and shaded areas the 95% CI (pMCMC(λslope < 0) < 0.001). (C) Posterior probabilities for the speciation rate intercepts for ancestral and derived melanosome morphologies. Although estimates can be negative to provide the overall time-dependent estimate, they are treated as zero when the linear estimate reaches that value (violet line in B). (D) Difference between the estimates from C. The 95% highest posterior density does not overlap and is therefore greater than zero (pMCMC(λancestral > λderived) = 0.009).
Divergence in social traits is ubiquitous in avian diversification, particularly in geographically isolated allospecies, where new and different phenotypes can accumulate in different populations (28, 47). Neural network models suggest that, when new stimulatory signals arise in such populations, they can be favored and fixated even in the absence of initial divergence in perceptual systems or mating preferences, by exploring intrinsic biases in perception and recognition (48, 49). This condition may be particularly important for birds, where sexual imprinting, learning flexibility, and generalization play a major role in species recognition and mate selection (50, 51) and where innate preferences for novel traits may be ubiquitous (28, 52). This process is analogous to a mutation-order speciation process, where geographically isolated populations experiencing similar selection pressures can diverge by fixating different advantageous mutations (53, 54), with the caveat that the social selective pressures themselves can evolve and diverge in response to these novel phenotypes (20, 55). Stochasticity plays an important role in building up divergence in this process, but the selective advantages of these novel phenotypes in social and sexual selection distinguish it from drift (53, 54).
Together, these findings highlight that constraints on trait expression can limit diversity and disparity of ornamental traits. Although nonecological divergence between allopatric populations over time is expected, it will do so only as a function of the rate at which new phenotypes can originate in different populations (28). In the African starlings, where sexual selection and social competition have been hypothesized to drive color evolution (39), we find that the influence of ornamental key innovations is essential to producing patterns of morphological and lineage diversification. Conserved or parallel mechanisms of iridescent color production in this clade can produce widely divergent phenotypes, and diverse innovations in melanosome morphology further enhance the rates of plumage color evolution and the breadth of achievable colors. Moreover, the stability of honest signals has been suggested to limit the role of sexual selection in diversification, because the preference for signals of individual quality would favor the evolution of a few convergent solutions (55, 56). Because iridescent colors may be costly to produce (57-59) and maintain (60), the modular shared template of iridescent colors, which allows for the evolution of disparate phenotypes that share honest signaling properties, may be a unique example of a trait that can maintain honesty without hampering diversification.
If these allopatric populations establish sympatry, accumulated differentiation during allopatry may be crucial for the stability of new species. Intrinsic reproductive isolation in birds can take millions of years, and therefore prezygotic isolation mediated by sexual signals, which can occur over a much shorter timescale, is critical for rapid avian divergence (27, 47). Such social signal-mediated prezygotic isolation can occur even in the absence of ecological divergence (23) or change in sexual selection regime if ornamental traits are sufficiently labile (61). Thus, the high evolvability of ornaments such as iridescent feather colors may facilitate the early divergence of ornamental traits crucial for rapid diversification in allopatry or the stability of recent species in sympatry, both in adaptive and in nonadaptive radiations (19). In fact, in African starlings, sympatry seems to be more common in sister species characterized by derived melanosomes (e.g., Lamprotornis superbus, Lamprotornis albicapillus, and Lamprotornis unicolor are sympatric, as are Poeoptera femoralis and Poeoptera stuhlmanni) than in lineages with the ancestral morphology (such as the Onychognathus clade). Although the extremely dynamic African vegetation and climatic cycling that shaped African avifauna macroevolution (62) complicate inference of past distribution and the geography during speciation (63), these suggest a possible role for labile ornamental traits in maintaining stable, currently sympatric species after initial allopatric divergence. Phylogeographic studies in clades of both constrained and labile ornaments should test whether populations indiscernible by coloration have accumulated lower levels of genetic differentiation than color-divergent, derived melanosome sister species.
The influence of novelty in communication systems on diversification has been considered only in terms of innovations that expand a lineage’s perceptual environment (24, 25). Our results show that the evolvability of ornamental traits themselves, under preexisting communication channels, can play a major and largely unappreciated role in this process. Tests of key innovations (whether ornamental or not) do not preclude the influence of ecological or geographic factors driving diversification (12, 13, 64, 65). Other effects, such as those encompassed by the time-dependent [and potentially diversity-dependent (11, 66)] component of speciation, are probably acting in concert with sexual selection to produce the observed diversity of African starlings. Nonetheless, the origin and irreversible evolution of optically complex melanosomes show a remarkable influence on the accumulation of morphological disparity and lineage diversification. Thus, considering the proximate basis of ornament expression provides important insights into the role played by ornamental traits in adaptive and nonadaptive radiations. In the same way that key innovations and ecological opportunity have been pivotal for understanding why diversification rates vary under ecological speciation (1, 14), key ornamental innovations provide a unique framework for how and when sexual selection should be expected to be an engine of diversification.
Materials and Methods
Phylogeny.
We reconstructed phylogenetic relationships for all 113 starling species and five outgroups, using sequences from up to five mitochondrial coding genes and four nuclear introns (67, 68). We followed the taxonomic suggestions of Lovette and Rubenstein (67), with the exception of Lamprotornis elisabeth, which the International Ornithological Congress considers a species and whose markedly different coloration is informative to our analysis. We also conducted analyses excluding L. elisabeth, yielding qualitatively identical results (Figs. S3 and S4 and Tables S2–S4). We inferred phylogenetic relationships and branch lengths in a Bayesian framework in BEAST (69), using a relaxed molecular clock for mtDNA sequences with Passeriformes rates (70) as a normally distributed prior. Four independent MCMC chains were run for 3 × 107 generations, sampling every 3,000 after 20% burn-in. We verified stationarity and convergence of independent chains, using split frequency distributions and correlations between chains, using the AWTY software (71), and parameter effective sample sizes were checked to be greater than 200 with TRACER (69). We estimated the maximum clade credibility (MCC) (Fig. 1B and Fig. S1B) tree and randomly subsampled 500 trees from the combined posterior distribution, pruned to the 48 African subclade species and standardized to a total clade root-to-tip distance of 1 for comparative analyses.
Melanosome Evolution.
Although the morphology of African starling melanosomes has been described previously (34, 38, 42), we confirmed these by examining transmission electron micrographs of at least one species per genus, as well as species from genera or clades reported not to share the morphology of their closest relatives. Feathers were collected from specimens from The American Museum of Natural History and prepared following standard procedures (72).
We estimated melanosome ancestral states and transition rates between morphologies, using reversible-jump Markov chain Monte Carlo (RJMCMC) in BayesTraits (73). We ran RJMCMC analyses on the sample of 500 trees, with exponential priors for transition rates with mean drawn from a uniform hyperprior (range 0−0.5), after tuning for adequate sampling and acceptance rates (73). We ran five 108-generation chains, sampling every 1,000 after 106 burn-ins, estimating ancestral states for all nodes in the MCC tree, accounting for phylogenetic uncertainty by using the most recent common ancestor approach (74) across the 500 posterior trees.
We tested the stepwise and irreversible models of melanosome evolution by calculating the BF, using the ratio of posterior to prior odds (73), derived by the frequency in which model variants were sampled in the posterior relative to their prior expectation as calculated from the expanded Stirling number. Bayes factors were compared in a 2ln scale, in which values between 2 and 6 are considered positive evidence against the null model, 6 and 10 are strong evidence, and values over 10 are very strong (75). The RJMCMC approach has the advantage of thoroughly exploring both model and parameter space, but asymmetry in transition rates can be confounded by differences in trait-dependent cladogenesis (76). Therefore, tests of irreversibility were confirmed under trait-dependent diversification models (Diversification Rates section).
Color Evolution.
We measured reflectance from 747 museum skins of 47 of 48 African starling species (specimens of Onychognathus neumanni were not available, but its melanosome morphology has been described previously (42) and is similar to that of its congeners). Up to 10 male and 10 female (median 8.4) specimens in full adult plumage showing no signs of molt were measured from each species. Reflectance spectra were taken at coincident normal geometry from 10 plumage patches (Fig. S1B), and additional color patches were measured when distinctly identified.
We represented colors using the avian colorspace model (43) in which reflectance spectra are converted to relative cone stimuli and projected in a tetrahedron, where each vertex represents one of the four cones characterizing avian color vision. The common starling (Sturnus vulgaris) visual system (77) was used to calculate stimuli from reflectance spectra under idealized illumination (43). This is the sensory equivalent of a morphospace where similar colors fall in close proximity in the colorspace and disparate colors are far apart. Given that relative cone stimuli are often correlated, we conducted a principal component analysis to represent the colorspace in fewer orthogonal variables. This approach was preferred over analyzing commonly used chromatic values of hue and chroma (which are included in Fig. S2D) for two reasons. First, hue measurements in tetrachromatic space are represented by angular variables, which cannot be adequately used in generalized Hansen models, and the variability of the species studied spanned the whole range of angular hueθ (Fig. S2 B and C). Direct measurement of hue as the wavelength of peak reflectance of spectral curves was also complicated due to nonspectral colors (characterized by multiple peaks). Second, disparity in color phenotypes scales as a function of both hue and saturation, such that using these variables independently would not adequately capture phenotypic differences (Fig. S2A). We used the first two components (PCs, accounting for 95.65% of original variation; Table S1), which were equivalent to a chromaticity diagram, but without assumptions of opponency mechanisms (78). To represent plumage brilliance, we used the log-transformed average reflectance value of spectral curves. Color analyses were conducted using the R package pavo (79).
To determine evolutionary regimes governing color diversification, we generated stochastic maps of melanosome evolution, using SIMMAP (80) for each of the 500 trees. We used these to test five evolutionary models of Brownian motion (BM) and Ornstein–Uhlenbeck (OU) processes in a generalized Hansen framework (44): single-rate BM (BM1), single-optimum OU (OU1), melanosome-dependent BM rates (BMS), OU optima (OUS), and OU with both melanosome-dependent rates and optima (OUMV). BM models are characterized by a rate (σ2) indicating how fast disparity accumulates over time, and OU models have additional optima (the phenotypic value θ to which lineages are being attracted) and attraction rate (α) parameters.
We included only species that had iridescent or eumelanic colors (that is, those expressing the material components necessary to produce iridescent color) for each patch in the color evolution analyses (Fig. S1B). We chose a representative patch (male mantle) that was iridescent for the greatest number of species (46) and included most species with each melanosome morphology, obtaining maximum-likelihood (ML) parameter estimates for the five models for each PC1, PC2, and brightness across the 500 trees and stochastic maps. Fit of competing models was accessed using second-order AIC (AICc) (Fig. S3). Finally, to assess the power of our analyses and identify the best, most parsimonious models, we conducted pairwise phylogenetic Monte Carlo comparisons (81) (Figs. S4 and S5). For the remaining body patches and female colors, we conducted model fitting and AICc selection, using the MCC tree (Fig. S1B and Tables S2 and S3). Analyses were conducted using R packages OUwie (44) and phytools (82).
Diversification Rates.
To verify the robustness of asymmetrical models of melanosome evolution and test for melanosome-dependent diversification rates, we used multistate speciation and extinction (MuSSE) models implemented in the R package diversitree (46). We considered model combinations based on the model of melanosome evolution (all transition rates allowed, irreversible, stepwise), transition rates (all rates different, all rates equal), trait-dependent diversification (melanosome independent, different speciation rates for ancestral and derived melanosomes, and different rates for each melanosome type), and time-dependent and time-independent speciation (Table S4). We focused on speciation rates, because initial extinction rate (μ) estimates were virtually zero, resulting in net diversification (speciation minus extinction rates: r = λ − μ) approximately equal to speciation rates (λ), a common feature given the difficulty in estimating extinction rates from molecular phylogenies (66, 83, 84).
We performed 5,000 independent ML estimations for each model, using random combinations of starting parameters (from a 0–5 uniform distribution) on the MCC tree (66, 85). Models were compared using AICc (Table S4). We then ran a MCMC analysis with 104 generations after 1,000 burn-ins with a uniform prior for all parameters (range −10–10) for the best model across all 500 posterior trees, combined for a total posterior distribution of 5 × 106 samples. We recorded the proportion of samples (pMCMC) where the slope of the time-dependent speciation rate was zero and where the speciation rate for derived morphologies was higher than the ancestral one (84). Finally, to tease apart confounding effects of other aspects of social competition such as social system variation, we tested for the correlated evolution of melanosome morphology and cooperative breeding behavior (39) through a Bayesian phylogenetic mixed-effects generalized linear model implemented in the MCMCglmm package (86). Our model included cooperative breeding as a binomial response variable (sensu ref. 39) and melanosome morphology as a binomial predictor (rod-shaped ancestral/derived, given that the best diversification model inferred no difference between the derived morphologies in speciation rates), with species identity weighted by the phylogenetic covariance from the MCC tree as the random effect (86).
Supplementary Material
Acknowledgments
We thank Irby Lovette for providing sequence data for phylogenetic analyses and The American Museum of Natural History (AMNH) for access to specimens. We thank Luke Harmon, Matt Pennell, Joseph Brown, Carlos Botero, Rebecca Safran, Jeremy Beaulieu, Richard FitzJohn, Carl Boettiger, members of the Shawkey laboratory, and two anonymous reviewers for invaluable comments and methodological guidance. Computational resources were supported by the National Science Foundation (NSF) through the Extreme Science and Engineering Discovery Environment (XSEDE) Science Gateways program. This work was supported by NSF Grant DEB-1210630, AMNH Chapman research grant, and Sigma XI Grants-in-Aid of Research (to R.M.); Human Frontier Science Program Grant RGY0083, Air Force Office of Scientific Research Grant FA9550-09-1-0159, and NSF Grant EAR-1251895 (to M.D.S.); and NSF Grants IOS-1121435 and IOS-1252946 (to D.R.R.) and Columbia University (D.R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Dryad Repository (http://dx.doi.org/10.5061/dryad.5sr48).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220784110/-/DCSupplemental.
References
- 1. Schluter D (2000) The Ecology of Adaptive Radiation (Oxford Univ Press, New York)
- 2. Simpson GG (1953) The Major Features of Evolution (Columbia Univ Press, New York)
- 3.Yoder JB, et al. Ecological opportunity and the origin of adaptive radiations. J Evol Biol. 2010;23(8):1581–1596. doi: 10.1111/j.1420-9101.2010.02029.x. [DOI] [PubMed] [Google Scholar]
- 4.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457(7231):830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 5. Grant PR, Grant BR (2008) How & Why Species Multiply: The Radiation of Darwin’s Finches (Princeton Univ Press, Princeton)
- 6.Kambysellis MP, et al. Pattern of ecological shifts in the diversification of Hawaiian Drosophila inferred from a molecular phylogeny. Curr Biol. 1995;5(10):1129–1139. doi: 10.1016/s0960-9822(95)00229-6. [DOI] [PubMed] [Google Scholar]
- 7.Morales-Hojas R, Vieira J. Phylogenetic patterns of geographical and ecological diversification in the subgenus Drosophila. PLoS ONE. 2012;7(11):e49552. doi: 10.1371/journal.pone.0049552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Losos JB (2009) Lizards in an Evolutionary Tree: The Ecology of Adaptive Radiation in Anoles (Univ of California Press, Berkeley, California)
- 9.Wagner CE, Harmon LJ, Seehausen O. Ecological opportunity and sexual selection together predict adaptive radiation. Nature. 2012;487(7407):366–369. doi: 10.1038/nature11144. [DOI] [PubMed] [Google Scholar]
- 10.Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18(2):169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- 11.Etienne RS, Haegeman B. A conceptual and statistical framework for adaptive radiations with a key role for diversity dependence. Am Nat. 2012;180(4):E75–E89. doi: 10.1086/667574. [DOI] [PubMed] [Google Scholar]
- 12. Losos JB, Mahler DL (2010) Evolution Since Darwin: The First 150 Years, eds Bell MA, Futuyma DJ, Eanes WF, Levinton JS (Sinauer, Sunderland, MA), pp 381–420.
- 13.Hunter JP. Key innovations and the ecology of macroevolution. Trends Ecol Evol. 1998;13(1):31–36. doi: 10.1016/s0169-5347(97)01273-1. [DOI] [PubMed] [Google Scholar]
- 14.Heard SB, Hauser DL. Key evolutionary innovations and their ecological mechanisms. Hist Biol. 1995;10(2):151–173. [Google Scholar]
- 15.Liem K. Evolutionary strategies and morphological innovations: Cichlid pharyngeal jaws. Syst Biol. 1973;22(4):425–441. [Google Scholar]
- 16.Price SA, et al. Functional innovations and morphological diversification in parrotfish. Evolution. 2010;64(10):3057–3068. doi: 10.1111/j.1558-5646.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 17.Wainwright PC, et al. The evolution of pharyngognathy: A phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Syst Biol. 2012;61(6):1001–1027. doi: 10.1093/sysbio/sys060. [DOI] [PubMed] [Google Scholar]
- 18.Jønsson KA, et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc Natl Acad Sci USA. 2012;109(17):6620–6625. doi: 10.1073/pnas.1115835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West-Eberhard MJ. Sexual selection, social competition, and speciation. Q Rev Biol. 1983;58(2):155–183. [Google Scholar]
- 20.Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci USA. 1981;78(6):3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carson HL. Sexual selection: A driver of genetic change in Hawaiian Drosophila. J Hered. 1997;88(5):343–352. doi: 10.1093/oxfordjournals.jhered.a023115. [DOI] [PubMed] [Google Scholar]
- 22.Mendelson TC, Shaw KL. Sexual behaviour: Rapid speciation in an arthropod. Nature. 2005;433(7024):375–376. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- 23.Sauer J, Hausdorf B. Sexual selection is involved in speciation in a land snail radiation on Crete. Evolution. 2009;63(10):2535–2546. doi: 10.1111/j.1558-5646.2009.00751.x. [DOI] [PubMed] [Google Scholar]
- 24.Carlson BA, et al. Brain evolution triggers increased diversification of electric fishes. Science. 2011;332(6029):583–586. doi: 10.1126/science.1201524. [DOI] [PubMed] [Google Scholar]
- 25.Ryan MJ. Neuroanatomy influences speciation rates among anurans. Proc Natl Acad Sci USA. 1986;83(5):1379–1382. doi: 10.1073/pnas.83.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455(7213):620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 27.Edwards SV, et al. Speciation in birds: Genes, geography, and sexual selection. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6550–6557. doi: 10.1073/pnas.0501846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price T. Speciation in Birds. Greenwood Village, CO: Roberts and Co.; 2008. [Google Scholar]
- 29.Price T, Pavelka M. Evolution of a colour pattern: History, development, and selection. J Evol Biol. 1996;9(4):451–470. [Google Scholar]
- 30.Prum RO, LaFountain AM, Berro J, Stoddard MC, Frank HA. Molecular diversity, metabolic transformation, and evolution of carotenoid feather pigments in cotingas (Aves: Cotingidae) J Comp Physiol B. 2012;182(8):1095–1116. doi: 10.1007/s00360-012-0677-4. [DOI] [PubMed] [Google Scholar]
- 31.Stoddard MC, Prum RO. How colorful are birds? Evolution of the avian plumage color gamut. Behav Ecol. 2011;22(5):1042–1052. [Google Scholar]
- 32.Price JJ, Friedman NR, Omland KE. Song and plumage evolution in the New World orioles (Icterus) show similar lability and convergence in patterns. Evolution. 2007;61(4):850–863. doi: 10.1111/j.1558-5646.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 33.Prager M, Andersson S. Convergent evolution of red carotenoid coloration in widowbirds and bishops (Euplectes spp.) Evolution. 2010;64(12):3609–3619. doi: 10.1111/j.1558-5646.2010.01081.x. [DOI] [PubMed] [Google Scholar]
- 34.Durrer H. Schillerfarben der vogelfeder als evolutionsproblem [Iridescent colors of bird feathers as an evolutionary question] Denkschr Schweiz Naturforsch Ges. 1977;91:1–127. [Google Scholar]
- 35.Greenewalt C, Brandt W, Friel D. Iridescent colors of hummingbird feathers. J Opt Soc Am. 1960;50(10):1005–1013. [Google Scholar]
- 36.Eliason CM, Shawkey MD. A photonic heterostructure produces diverse iridescent colours in duck wing patches. J R Soc Interface. 2012;9(74):2279–2289. doi: 10.1098/rsif.2012.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shawkey MD, Hauber ME, Estep LK, Hill GE. Evolutionary transitions and mechanisms of matte and iridescent plumage coloration in grackles and allies (Icteridae) J R Soc Interface. 2006;3(11):777–786. doi: 10.1098/rsif.2006.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durrer H. Schillerfarben der stare (Sturnidae) [Iridescent colors of the starlings (Sturnidae)] J Ornithol. 1970;111(2):133–153. [Google Scholar]
- 39.Rubenstein DR, Lovette IJ. Reproductive skew and selection on female ornamentation in social species. Nature. 2009;462(7274):786–789. doi: 10.1038/nature08614. [DOI] [PubMed] [Google Scholar]
- 40.Bennett ATD, Cuthill IC, Partridge JC, Lunau K. Ultraviolet plumage colors predict mate preferences in starlings. Proc Natl Acad Sci USA. 1997;94(16):8618–8621. doi: 10.1073/pnas.94.16.8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komdeur J, Oorebeek M, van Overveld T, Cuthill IC. Mutual ornamentation, age, and reproductive performance in the European starling. Behav Ecol. 2005;16:805–817. [Google Scholar]
- 42.Craig AJFK, Hartley AH. The arrangement and structure of feather melanin granules as a taxonomic character in African Starlings (Sturnidae) Auk. 1985;102(3):629–632. [Google Scholar]
- 43.Stoddard MC, Prum RO. Evolution of avian plumage color in a tetrahedral color space: A phylogenetic analysis of new world buntings. Am Nat. 2008;171(6):755–776. doi: 10.1086/587526. [DOI] [PubMed] [Google Scholar]
- 44.Beaulieu JM, Jhwueng D-C, Boettiger C, O’Meara BC. Modeling stabilizing selection: Expanding the Ornstein-Uhlenbeck model of adaptive evolution. Evolution. 2012;66(8):2369–2383. doi: 10.1111/j.1558-5646.2012.01619.x. [DOI] [PubMed] [Google Scholar]
- 45.Endler JA, Westcott DA, Madden JR, Robson T. Animal visual systems and the evolution of color patterns: Sensory processing illuminates signal evolution. Evolution. 2005;59(8):1795–1818. doi: 10.1554/04-669.1. [DOI] [PubMed] [Google Scholar]
- 46. FitzJohn RG (2012) Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol 3(6):1084–1092.
- 47.Price TD. The roles of time and ecology in the continental radiation of the Old World leaf warblers (Phylloscopus and Seicercus) Philos Trans R Soc Lond B Biol Sci. 2010;365(1547):1749–1762. doi: 10.1098/rstb.2009.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arak A, Enquist M, Arak A, Enquist M. Hidden preferences and the evolution of signals. Philos Trans R Soc Lond B Biol Sci. 1993;340:207–213. [Google Scholar]
- 49.Enquist M, Arak A. Selection of exaggerated male traits by female aesthetic senses. Nature. 1993;361(6411):446–448. doi: 10.1038/361446a0. [DOI] [PubMed] [Google Scholar]
- 50.Verzijden MN, et al. The impact of learning on sexual selection and speciation. Trends Ecol Evol. 2012;27(9):511–519. doi: 10.1016/j.tree.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Irwin DE, Price T. Sexual imprinting, learning and speciation. Heredity. 1999;82(4):347–354. doi: 10.1038/sj.hdy.6885270. [DOI] [PubMed] [Google Scholar]
- 52.Burley NT, Symanski R. “A taste for the beautiful”: Latent aesthetic mate preferences for white crests in two species of Australian grassfinches. Am Nat. 1998;152(6):792–802. doi: 10.1086/286209. [DOI] [PubMed] [Google Scholar]
- 53.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323(5915):737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 54.Mani GS, Clarke BC. Mutational order: A major stochastic process in evolution. Proc R Soc Lond B Biol Sci. 1990;240(1297):29–37. doi: 10.1098/rspb.1990.0025. [DOI] [PubMed] [Google Scholar]
- 55.Prum RO. The Lande-Kirkpatrick mechanism is the null model of evolution by intersexual selection: Implications for meaning, honesty, and design in intersexual signals. Evolution. 2010;64(11):3085–3100. doi: 10.1111/j.1558-5646.2010.01054.x. [DOI] [PubMed] [Google Scholar]
- 56.Price T. Sexual selection and natural selection in bird speciation. Philos Trans R Soc B. 1998;353:251–260. [Google Scholar]
- 57.Meadows MG, Roudybush TE, McGraw KJ. Dietary protein level affects iridescent coloration in Anna’s hummingbirds, Calypte anna. J Exp Biol. 2012;215(16):2742–2750. doi: 10.1242/jeb.069351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGraw KJ, Mackillop EA, Dale J, Hauber ME. Different colors reveal different information: How nutritional stress affects the expression of melanin- and structurally based ornamental plumage. J Exp Biol. 2002;205(23):3747–3755. doi: 10.1242/jeb.205.23.3747. [DOI] [PubMed] [Google Scholar]
- 59.Hill GE, Doucet SM, Buchholz R. The effect of coccidial infection on iridescent plumage coloration in wild turkeys. Anim Behav. 2005;69(2):387–394. [Google Scholar]
- 60.Eliason CM, Shawkey MD. Decreased hydrophobicity of iridescent feathers: A potential cost of shiny plumage. J Exp Biol. 2011;214(13):2157–2163. doi: 10.1242/jeb.055822. [DOI] [PubMed] [Google Scholar]
- 61.Uyeda JC, Arnold SJ, Hohenlohe PA, Mead LS. Drift promotes speciation by sexual selection. Evolution. 2009;63(3):583–594. doi: 10.1111/j.1558-5646.2008.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fjeldså J, Bowie RC. New perspectives on the origin and diversification of Africa’s forest avifauna. Afr J Ecol. 2008;46(3):235–247. [Google Scholar]
- 63.Jetz W, Rahbek C, Colwell RK. The coincidence of rarity and richness and the potential signature of history in centres of endemism. Ecol Lett. 2004;7(12):1180–1191. [Google Scholar]
- 64. Cracraft J (1990) Evolutionary Innovations, ed Nitecki MH (Univ of Chicago Press, Chicago), pp 21–44.
- 65.Kraaijeveld K, Kraaijeveld-Smit FJL, Maan ME. Sexual selection and speciation: The comparative evidence revisited. Biol Rev Camb Philos Soc. 2011;86(2):367–377. doi: 10.1111/j.1469-185X.2010.00150.x. [DOI] [PubMed] [Google Scholar]
- 66.Rabosky DL, Glor RE. Equilibrium speciation dynamics in a model adaptive radiation of island lizards. Proc Natl Acad Sci USA. 2010;107(51):22178–22183. doi: 10.1073/pnas.1007606107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lovette IJ, Rubenstein DR. A comprehensive molecular phylogeny of the starlings (Aves: Sturnidae) and mockingbirds (Aves: Mimidae): Congruent mtDNA and nuclear trees for a cosmopolitan avian radiation. Mol Phylogenet Evol. 2007;44(3):1031–1056. doi: 10.1016/j.ympev.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 68.Lovette IJ, McCleery BV, Talaba AL, Rubenstein DR. A complete species-level molecular phylogeny for the “Eurasian” starlings (Sturnidae: Sturnus, Acridotheres, and allies): Recent diversification in a highly social and dispersive avian group. Mol Phylogenet Evol. 2008;47(1):251–260. doi: 10.1016/j.ympev.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 69.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17(10):2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- 71.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24(4):581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 72.Maia R, D’Alba L, Shawkey MD. What makes a feather shine? A nanostructural basis for glossy black colours in feathers. Proc Biol Sci. 2011;278(1714):1973–1980. doi: 10.1098/rspb.2010.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167(6):808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 74.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53(5):673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 75.Kass R, Raferty A. Bayes factors. J Am Stat Assoc. 1995;90(430):773–795. [Google Scholar]
- 76.Goldberg EE, Igić B. On phylogenetic tests of irreversible evolution. Evolution. 2008;62(11):2727–2741. doi: 10.1111/j.1558-5646.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 77.Hart NS. The visual ecology of avian photoreceptors. Prog Retin Eye Res. 2001;20(5):675–703. doi: 10.1016/s1350-9462(01)00009-x. [DOI] [PubMed] [Google Scholar]
- 78.Endler JA, Day LB. Ornament colour selection, visual contrast and the shape of colour preference functions in great bowerbirds, Chlamydera nuchalis. Anim Behav. 2006;72:1405–1416. [Google Scholar]
- 79. Maia R, Eliason CM, Bitton P-P, Doucet SM, Shawkey MD (2013) pavo: an R package for the analysis, visualization and organization of spectral data. Methods Ecol Evol, 10.1111/2041-210X.12069.
- 80.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52(2):131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 81.Boettiger C, Coop G, Ralph P. Is your phylogeny informative? Measuring the power of comparative methods. Evolution. 2012;66(7):2240–2251. doi: 10.1111/j.1558-5646.2011.01574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 83.Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64(6):1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 84.Hugall AF, Stuart-Fox D. Accelerated speciation in colour-polymorphic birds. Nature. 2012;485(7400):631–634. doi: 10.1038/nature11050. [DOI] [PubMed] [Google Scholar]
- 85.Price SA, Hopkins SSB, Smith KK, Roth VL. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci USA. 2012;109(18):7008–7012. doi: 10.1073/pnas.1117133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hadfield JD, Nakagawa S. General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J Evol Biol. 2010;23(3):494–508. doi: 10.1111/j.1420-9101.2009.01915.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.