Significance
This research highlights the discovery and genome reconstruction of a member of the globally distributed yet uncultivated candidate phylum TM6 (designated TM6SC1). In addition to the 16S rRNA gene, no other genomic information is available for this cosmopolitan phylum. This report also introduces a mini-metagenomic approach based on the use of high-throughput single-cell genomics techniques and assembly tools that address a widely recognized issue: how to effectively capture and sequence the currently uncultivated bacterial species that make up the “dark matter of life.” Amplification and sequencing random pools of 100 events enabled an estimated 90% recovery of the TM6SC1 genome.
Keywords: genome assembly, metagenomics, single-cell genomics, MDA, symbiotic bacteria
Abstract
The “dark matter of life” describes microbes and even entire divisions of bacterial phyla that have evaded cultivation and have yet to be sequenced. We present a genome from the globally distributed but elusive candidate phylum TM6 and uncover its metabolic potential. TM6 was detected in a biofilm from a sink drain within a hospital restroom by analyzing cells using a highly automated single-cell genomics platform. We developed an approach for increasing throughput and effectively improving the likelihood of sampling rare events based on forming small random pools of single-flow–sorted cells, amplifying their DNA by multiple displacement amplification and sequencing all cells in the pool, creating a “mini-metagenome.” A recently developed single-cell assembler, SPAdes, in combination with contig binning methods, allowed the reconstruction of genomes from these mini-metagenomes. A total of 1.07 Mb was recovered in seven contigs for this member of TM6 (JCVI TM6SC1), estimated to represent 90% of its genome. High nucleotide identity between a total of three TM6 genome drafts generated from pools that were independently captured, amplified, and assembled provided strong confirmation of a correct genomic sequence. TM6 is likely a Gram-negative organism and possibly a symbiont of an unknown host (nonfree living) in part based on its small genome, low-GC content, and lack of biosynthesis pathways for most amino acids and vitamins. Phylogenomic analysis of conserved single-copy genes confirms that TM6SC1 is a deeply branching phylum.
Bacteria that have not been obtained by conventional culturing techniques are the central target of single-cell sequencing (1), which is accomplished using multiple displacement amplification (MDA) (2–5) of genomic DNA to obtain sufficient template. We applied a high-throughput strategy to capture and sequence genomes of bacteria from a biofilm in a hospital sink including pathogens, such as the oral periodontal pathogen (Porphyromonas gingivalis) (6) and uncultivated members (this study). Despite the fact that a typical person spends ∼90% of their time indoors (7), our knowledge of the microbial diversity of the indoor environment has only recently begun to be explored using culture-independent methods (8, 9). Biofilms within water distribution systems in particular are thought to be diverse microbial communities and potential reservoirs of disease-causing organisms in the indoor environment. Several pathogens including Escherichia coli, Legionella pneumophila (10–13), Vibrio cholerae (14), and Helicobacter pylori (15, 16) have been detected in biofilms within water distribution systems. A recent 16S rRNA gene (abbreviated henceforth as 16S unless otherwise stated) molecular survey also revealed significant loads of Mycobacterium avium in showerhead biofilms (17). Based on these findings, indoor environments can clearly serve as significant reservoirs of pathogenic bacteria, and therefore there is great interest in investigating the rare and abundant bacterial species within biofilms in these environments.
One approach to capture uncultivated bacteria is to isolate single bacterial cells by fluorescent activated cell sorting (FACS). The DNA of the sorted cells can then be amplified by MDA and screened for the presence of amplified bacterial genomes, typically by PCR and sequencing of the 16S (3). However, environmentally derived biofilms pose particularly difficult challenges because they can contain low overall cell numbers, and there are abundant inorganic and organic particulates present, which can contribute fluorescent signals that can be mistaken for bacteria. Less than 1% of the single-cell MDA reactions initially attempted in pilot studies were positive for 16S sequences, and therefore discovery of rare species was not statistically favored. Indeed, if a rare bacterial species “X” represents just 0.1% of cells in a sample, then sequencing 1,000 randomly selected cells would result in only a 37% chance of capturing a single cell from that particular species. However, if one generates pools consisting of 100 randomly selected single cells (mini-metagenome), then 10 pools would be sufficient to capture a cell of interest within one of the pools with the same probability but more economically. Thus this strategy, although attractive, faces two computational challenges: (i) assembling a mini-metagenome consisting of up to 100 genomes with highly nonuniform coverage and (ii) identifying contigs from the species of interest within a metagenome with high confidence. In this paper we focus mainly on the latter challenge as well as on the experimental techniques for generating mini-metagenomes from biofilms. The former computational challenge is addressed with simulated and real single-cell datasets in a separate publication (18). By flow-sorting pools of 100 fluorescent detection events into 384-well plates, ∼19–60% were positive for bacterial DNA based on 16S compared with the 1% success rate previously obtained. Because there is no accurate way to determine the total number of cells that were in the original pool, the number of cells that lysed with the single lysis method used, and the number of resulting genomes successfully amplified, several species are assumed to be pooled together. Shotgun metagenomic sequencing of very highly diverse communities combined with methods for assembly and binning contigs has previously revealed genomes of uncultivated organisms including acidophilic members from a low-complexity biofilm community (19), symbionts (20), rumen host-associated organisms (21), marine group II Euryarchaeota (22, 23), as well as the candidate division (CD) designated WWE1 (24). In addition, approaches leveraging metagenomic and single-cell datasets have enabled reconstruction of genomes from uncultivated marine organisms (25, 26). Although recently developed assemblers (27) enhance sequencing of single cells, assembly of our MDA-obtained metagenomes (of up to 100 different bacterial species in this case) is a more difficult computational problem, particularly because mixed cells feature even more nonuniform read coverage compared with single-cell sequencing, for example, due to differing GC content (percent of bases that are guanine or cytosine) between species (28, 29).
A strategy we refer to as a “mini-metagenomic approach” was therefore used to capture low-abundance bacteria. Partial 16S sequences representing a member of the candidate phylum TM6 were recovered from three different wells, each a MDA-amplified pool of 100 events. The candidate phyla TM6 and TM7 were first identified by Rheims et al. (30) based on culture-independent molecular surveys and appear to include common, low-abundance members of microbial communities in diverse environments including domestic water sources. An assembly tool designed for coping with the wide variations in coverage from MDA samples, SPAdes (31), was used to assemble the mini-metagenomes. Computational strategies were used to reconstruct and bin contigs representing genomes of individual species from the mixed genomes similar to published metagenomic methods, revealing a near complete genome for TM6. Single-cell whole-genome amplification techniques have previously allowed partial recovery of genomes from several elusive CD organisms: TM7 (32, 33), OP11 (34), and Poribacteria symbiotically associated with marine sponges (35). In contrast, complete genomes of the Elusimicrobia (previously named the termite group 1 division) were recovered from amplification of pooled clonal single cells (36). The mini-metagenomic approach in combination with single-cell assembly tools and contig binning methods that we used here resulted in the recovery of 1.07 Mb of a TM6 genome (TM6SC1) within only seven contigs. Analysis of core single-copy marker genes from these contigs resulted in a conservative estimate of 91% recovery for this TM6 genome. High nucleotide identity between a total of three TM6 genome drafts generated from pools that were independently captured, amplified, and assembled provided strong confirmation of a correct genomic sequence. From the genomic information available, this TM6 is likely a Gram-negative and facultatively anaerobic representative. Based on its small genome, adenine–thymine (AT) bias, and apparent lack of biosynthetic capability for most amino acids and vitamins, TM6 may represent an obligate community member or symbiont of an unknown host.
Results
Sampling and Sorting Cells from Biofilm Samples.
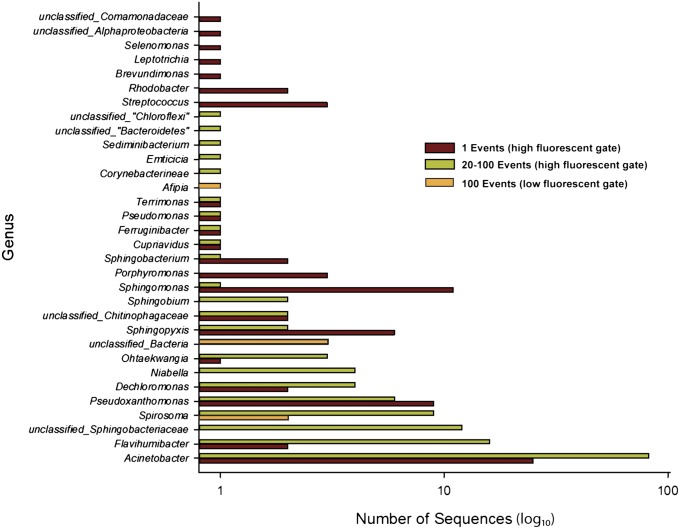
We modified our single-cell genomic methods developed for marine samples (25, 27) and healthy human microbiome samples (gastrointestinal, oral, and skin) (1, 37) to acquire microbial genomes from biofilms in the indoor environment. The marine-derived samples contained relatively high bacterial content and were a rich source of single cells for FACS isolation and genomic sequencing, with about 20–30% of single-cell amplifications yielding a positive 16S (25, 27). In contrast, FACS analysis of the untreated biofilm samples from this environment (indoor surface) was more challenging to analyze due to (i) the presence of autofluorescent nonbacterial particles that produced elevated background signals and (ii) difficulties in disrupting the intact biofilm to access individual cells. The typical success rate for capturing single cells from these difficult indoor environmental samples was roughly 1% (wells yielding a positive 16S). To address these issues, the biofilm sample was vortexed, filtered through a 5-µm filter, and concentrated to purify the bacterial fraction within a Nycodenz gradient (Methods) before FACS and DNA amplification by MDA. This processing raised the number of FACS-positive DNA-stained events within the bacterial size range to roughly 20% and the overall success rate of bacterial cell sorting to 18% based on sequencing of 16S PCR products derived from the MDA reactions. From the high-fluorescence gate, we sorted single events into a total of 416 wells, and to more fully capture the bacterial diversity as well as increase the odds of capturing low-abundance species in the sample, we also sorted multiple events into wells. A total of 128 wells received 20 events and another 128 wells received 100 events. In addition, 32 wells from a low-fluorescence gate in a defined forward scatter size range received 100 events (SI Appendix, Fig. S1). The overall success rate for a positive 16S sequence increased to 60% in these high-fluorescent multievent wells and 19% for the wells that received low-fluorescence events.
Plates containing the sorted events were processed on an automated high-throughput single-cell platform (Methods and SI Appendix, Fig. S2) to amplify genomes by MDA and screen the amplified DNAs for 16S sequences. The relative abundance of various genera found in the 100-cell low-fluorescent sort and the parallel high-throughput single-cell and multiple-cell sort from the high-fluorescence population from this same sink biofilm sample are shown in Fig. 1. Across all gated events, 232 total 16S sequences were within the domain Bacteria. Some of the most highly detected genera such as Acinetobacter and Sphingomonas are consistent with those found in microbial communities associated with drinking water distribution systems (17, 38, 39). From the 32 wells with 100 low-fluorescent events in each, 6 wells produced a 16S sequence. Two wells contained an unclassified member of the genus Spirosoma at 91% sequence identity and 1 well a member of genus Afipia (97% identity). The final three wells contained nearly identical sequences (>99.5%), which had a maximum level of sequence identity to a previously deposited clone (GenBank accession no. GU368367 belonging to the CD TM6 at ∼94% identity). The three amplified DNAs containing the partial TM6 16S sequences were sequenced on the Roche 454 and Illumina GAIIx platforms.
Fig. 1.
Summary of genera found in the biofilm sample from single and multievent sorts. The total number of 16S rRNA gene sequences for each observed bacterial genera recovered in individual MDA-amplified wells. Data are presented for wells in which 1, 20, or 100 events were sorted from either a high- or a low-fluorescence event population. Data from the 20- and 100-event wells that were sorted from the high fluorescence population are grouped together.
Genome Assembly and Contig Identification.
Three different assembly approaches were used to obtain the assemblies for these genomes. Due to the fact that there was no close reference genome for distantly related CD TM6, both unsupervised and supervised contig classification and binning approaches were then needed to confidently identify those contigs belonging to this organism. For assembly, we used one assembler designed for sequences of cultured cells (CLC) (www.clcbio.com) and two assembly tools specifically designed for data generated from single-cell MDA reactions: Velvet-SC (27) and SPAdes (31) (Table 1). Previous studies (27, 31) demonstrated that the Velvet-SC and SPAdes assemblers are significantly better than Velvet (40) and SoapDenovo (41) in assembly of single-cell datasets because they are able to cope with the wide variations in coverage characteristic of MDA samples. SPAdes was further designed to cope with the elevated number of chimeric reads and read pairs characteristic of single-cell assemblies. For this TM6 study, more complete assemblies were obtained with SPAdes than with Velvet-SC or CLC by most assembly metrics (Table 1). The SPAdes TM6 assembly showed remarkably superior results with respect to N50 and longest contig size (Table 1), and, because this software has a low rate of assembly errors (31), it was chosen for this study.
Table 1.
Assembly statistics
| MDA1 |
MDA2 |
MDA3 |
|||||||
| No. of input reads: 15M |
No. of input reads: 26M |
No. of input reads: 24M |
|||||||
| Contigs (bp) | SPAdes | Velvet-SC | CLC | SPAdes | Velvet-SC | CLC | SPAdes | Velvet-SC | CLC |
| ≥110 | 1,518 | 319 | 4,768 | 1,111 | 372 | 3,744 | 1,369 | 291 | 5,475 |
| ≥201 | 1,496 | 288 | 2,126 | 1,094 | 338 | 1,635 | 1,357 | 260 | 2,225 |
| ≥501 | 636 | 267 | 741 | 479 | 305 | 564 | 526 | 220 | 657 |
| Total no. | 1,537 | 319 | 5,972 | 1,150 | 373 | 5,196 | 1,386 | 298 | 7,280 |
| N50 | 45,160 | 25,874 | 29,529 | 42,853 | 28,061 | 27,458 | 36,106 | 6,872 | 8,601 |
| N75 | 10,907 | 9,399 | 2,510 | 10,356 | 8,909 | 2,896 | 2,844 | 2,263 | 377 |
| Largest | 329,507 | 96,603 | 161,072 | 464,047 | 139,431 | 229,829 | 489,351 | 51,642 | 246,467 |
The number of contigs filtered by minimum sizes 110, 201, and 501 bp and the total number of contigs are shown. N50 (respectively, N75) is the largest contig size, L, such that at least 50% (respectively, 75%) of all bases in the assembly are contained in contigs of size at least L. Boldfaced values indicate the best of the three assemblers on that dataset in that metric, although metrics should not be considered in isolation.
A 273-kb contig in TM6 MDA2 with an average GC content of 36% contained a 16S rRNA gene with a flanking 23S. Taxonomic affiliations of the predicted protein sequences derived from this contig were assigned using the Automated Phylogenetic Inference System software (APIS) (42), which generates a phylogenetic tree for each ORF in a genomic or metagenomic sample using homologous proteins from complete genomes. APIS classifies each ORF taxonomically and functionally based on phylogenetic position. APIS trees showed that the majority of ORFs of this 273-kb contig were very distantly related to any sequenced genome, consistent with the contig belonging to an uncharacterized organism such as CD TM6.
An independent metagenomic binning approach using an autonomous method, principal coordinate analyses (PCA) of the penta-nucleotide frequency, followed by k-means clustering, revealed a small grouping of contigs clustered near the putative TM6 contig. In a second independent approach for taxonomic classification of the contigs, MGTAXA, a software that performs taxonomic classification of metagenomic sequences with machine-learning techniques (http://andreyto.github.com/mgtaxa and http://mgtaxa.jcvi.org), was used to classify the contigs. MGTAXA, which is also fundamentally based on the frequency of kmers, can allow users to input sequences as training sets to then further classify their own metagenomic sequences. Using the putative TM6 16S and 23S rRNA gene-containing contig as a training sequence, MGTAXA identified contigs (contigs of length >300 bp) with similar taxonomic classification (SI Appendix, Fig. S3). Taxonomic affiliations of the non-TM6 contigs were dominated by Bacteriodetes (Sphingobacteriales) and Flavobacteriaceae (Chryseobacterium) (SI Appendix, Fig. S3). Contigs identified as TM6 from the intersection of these independent approaches sharing a GC content of 36 ± 2% were chosen as the final set. Each approach was in general agreement for the final contig set that was originally identified using MGTAXA, providing confidence in the final contigs chosen. The nucleotide frequency approach identified eight additional contigs compared with MGTAXA, but these either deviated slightly from the expected GC (within ±5%) or were classified as belonging to Bacteriodetes by MGTAXA and/or APIS and were therefore excluded from the set. In the case of uncultivated genomes where there is no closely related reference genome, the identification and use of contigs containing a marker gene for the genome of interest (such as the 16S rRNA gene) is helpful to guide nucleotide frequency binning and critical for the effective grouping of contigs.
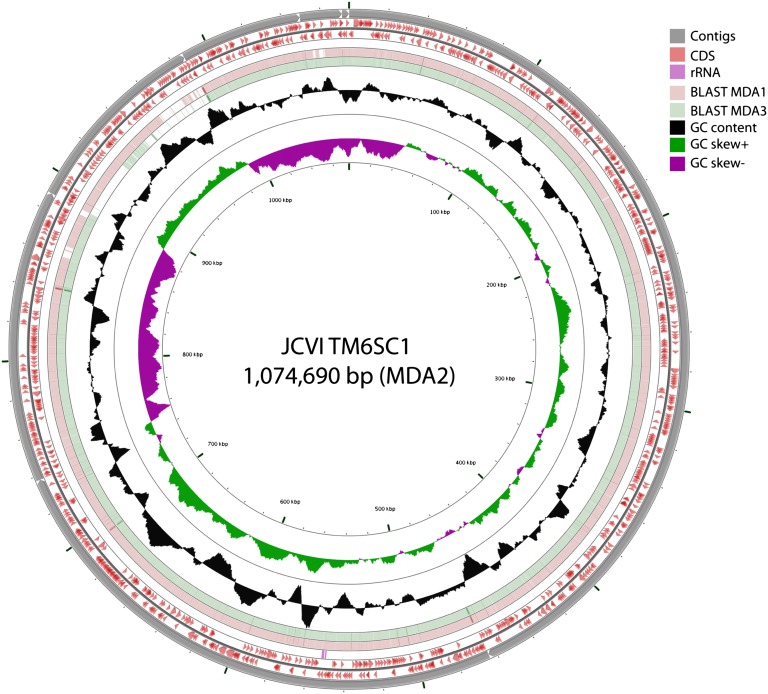
All three amplified TM6 SPAdes assemblies were processed as described above to yield draft TM6 genomes. MDA2 contained the largest TM6 assembly with 1,074,690 bp contained in seven contigs (Fig. 2). Comparative genomic analyses on the three sets of contigs using ProgressiveMauve (43) and LAST alignments (44) confirmed that the assembled contigs of MDA1 and MDA3 were contained within MDA2 with highly conserved synteny (SI Appendix, Fig. S4). At the nucleotide level, BLASTN comparisons on the concatenated contigs representing genome MDA2 (Fig. 2) indicate nearly identical assemblies. To further confirm the agreement between the MDA1, -2, and -3 TM6 contigs, all reads for each MDA were mapped to the MDA2 genome and SNP analyses were performed. The MDA2 TM6 genome recruited 33% (5.8 of 15 M), 64% (16.8 of 26 M) and 70% (16.9 of 24 M) of the reads for MDA1, MDA2, and MDA3, respectively (SI Appendix, Fig. S5). At a minimum, 10× coverage and a cutoff at 50% frequency, there were fewer than 20 SNPs identified (SI Appendix, Fig. S5). Unless stated, further analyses focus on only the assembled, binned, and annotated TM6 contigs from MDA2 (designated as TM6SC1).
Fig. 2.
Circular representation of the TM6SC1 genome as a pseudomolecule derived from the concatenated contigs for MDA2. From the inner to the outer ring: GCskew−, GCskew+, G+C content, BLASTN alignment against MDA1 contigs, BLASTN alignment against MDA3 contigs, predicted CDS, rRNA, and MDA2 contigs (contigs were ordered by length and then concatenated).
Genome General Features.
The coding density of the TM6SC1 assembly is relatively high at 89%, which includes the coding sequence (CDS) and RNA genes, and there is excellent fit to the expected number of CDS per genome size (based on plots of predicted CDS per genome size) (SI Appendix, Fig. S6). Analysis of conserved single-copy marker genes using a set of 111 genes (25) revealed that the assembly includes 101 of 111 genes (SI Appendix, Table S2), and thus a conservative estimate of genome completeness is 91% for this TM6 genome. In the recent single-cell genomic study describing a partial genome of 270 kb for CD OP11 (designated ZG1) (34), roughly 45% of the 423 protein-coding genes had no function prediction (27% of those with no prediction were conserved hypothetical proteins with similarities in the databases, and 72% were hypothetical proteins unique to the ZG1 genome). The authors noted that several candidate division genomes shared a similar percentage of protein-coding annotated genes, e.g., 54% for CD TM7 (32) and 48% for CD WWE1 (24) compared with Escherichia coli K-12 (14%) and Bacillus licheniformis American Type Culture Collection 14580 (27%) (34). ZG1 also has the lowest percentage (41%) of protein-coding genes assigned to clusters of orthologous groups (COGs) relative to genomes of other CDs (e.g., 53% for TM7, 63% for WWE1, and 80% for Elusimicrobia). The TM6 genome (Table 2) also has a low percentage of functionally annotated genes (43%) with 34% of these assigned to COGs. Based on these studies, it is clear that single-cell sequencing techniques can tap into diverse genomes with few similar ORF matches in existing databases, greatly expanding the known diversity.
Table 2.
Statistics characterizing the assembled and annotated TM6SC1 genome
| Genome features | Value |
| Assembly size (bp) | 1,074,690 |
| % G+C content | 36 |
| No. of ORFs | 1,056 |
| No. of tRNA genes | 29 |
| No. of rRNA genes | 2 |
| Protein-coding genes (CDS) | 993 |
| No. conserved single-copy genes | 101/111 (91%) |
| No. ORFs with functional annotation | 428 |
| No. ORFs without function prediction | 565 |
| Average CDS length | 952 |
| No. ORFs connected to KEGG pathways | 322 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Phylogenetic and Phylogenomic Analyses of Candidate Phylum TM6.
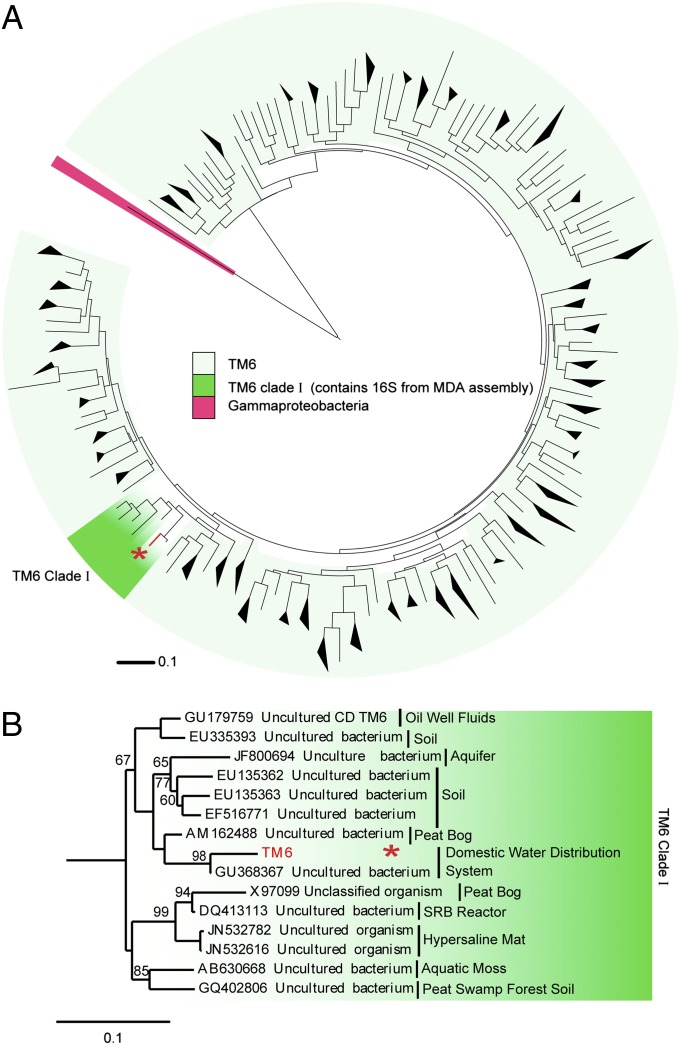
A large number of TM6-related 16S rRNA gene sequences have been identified from geographically varied sampling sites (Fig. 3A and high resolution in SI Appendix, Fig. S7), which suggests that this phylum has a cosmopolitan distribution, although typically found at low relative abundance. Its ecological distribution (derived from published and unpublished studies that have deposited related sequences in GenBank) includes domestic water sources (17, 39, 45), acidic cave biofilms, acid mine drainage biofilms (46), wastewater biofilms (47), soil, contaminated groundwater and subsurface sites (48, 49), aquatic moss, hypersaline mats, peat bogs, and peat swamps (30, 50). These and additional environments where TM6 was detected in 16S rRNA gene clone libraries, including a number of biofilm-related samples, are highlighted in SI Appendix, Table S1. Notably, only a few TM6 sequence signatures have so far been identified as associated with a human host (51). We designate the clade that our TM6 16S fell within as TM6 clade I (Fig. 3 A and B) because it also includes the 16S from a peat bog clone library that led to the designation of TM6 (30). The name is derived from “Torf, Mittlere Schicht” (“peat, middle layer”) (30) (Fig. 3B). Candidate division TM7 was also first designated based on a sequence from that clone library. Interestingly, at the time of this study, the closest sequence in the National Center for Biotechnology Information nr database to our assembled TM6 genome is from a biofilm in a corroded copper water pipe (GenBank accession no. GU368367) (39) (Fig. 3B). Several studies indicate that TM6 16S sequences are commonly detected in biofilms from domestic water systems (SI Appendix, Table S1). Five such sequences were discovered along with potential opportunistic pathogens in showerhead biofilms (17). A recent study of a more than 20-y-old drinking water network that compared bacterial core communities in bulk water and associated biofilms revealed that the biofilm samples contained a unique community with no overlapping phylotypes with the bulk water samples (45). TM6 represented 11% of the clones observed in these biofilms. Given the occurrence of TM6 organisms in biofilm communities and their apparent enrichment in biofilm samples, it is interesting to speculate that they may play a role in biofilm development or be dependent upon communal living.
Fig. 3.
Evolutionary relationships of candidate division TM6. (A) Phylogenetic relationship of 16S rRNA gene sequences designated as members of TM6 in public databases reveal the global distribution and sequence diversity within this group. An asterisk indicates the TM6 sequences from this study. (B) Unrooted 16S rRNA gene tree based on maximum-likelihood analysis of representative candidate division TM6 and Proteobacteria sequences. One thousand bootstrapped replicate resampled datasets were analyzed. Bootstrap values are indicated as percentages and not shown if below 50%.
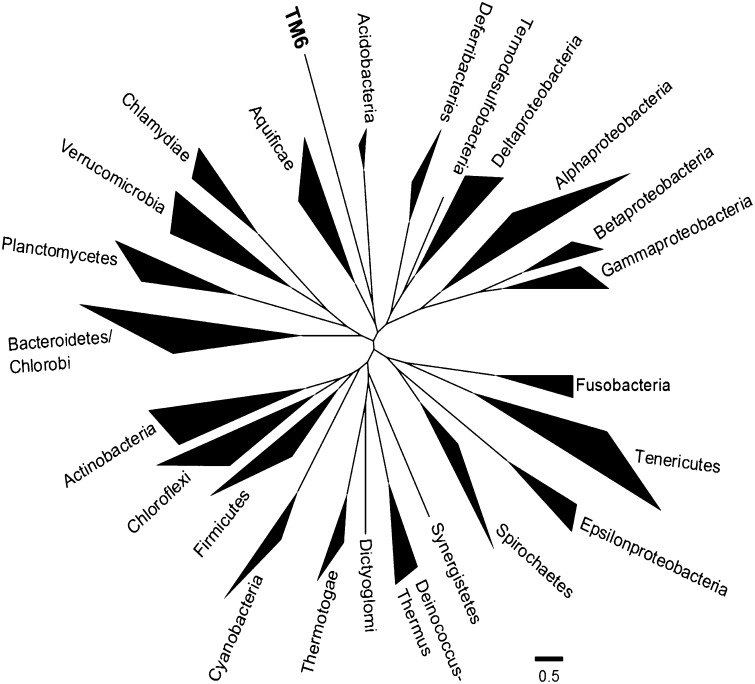
The number of identified candidate phyla within the domain Bacteria has grown from the 11 that were recognized in 1987 (52), to 26 in 1998 (53), to the most recent list of around 30 (www.arb-silva.de) (54). Early phylogenetic identification of the bacterial candidate phyla, including TM6, was accomplished using 16S rRNA gene phylogeny (53, 55, 56). In these studies, the TM6 sequences available did not find phylogenetic congruence with existing divisions and was designated as a phylum-level candidate division. Our goal was to resolve its phylogenetic position with the genome information now available. For this purpose we used the automated pipeline for phylogenomic analyses (AMPHORA2) that uses multiple marker gene analysis (57, 58). We began with a set of 29 genes (of 31 supported by AMPHORA2) that could be identified in the TM6 genome and that were previously chosen based on their universality, low copy number, phylogenetic signal, and low rates of horizontal gene transfer (57). These sequences were aligned and compared by using the AMPHORA2 seed alignment through a hidden Markov model (HMM). We masked the resulting alignments to remove poorly conserved regions using the AMPHORA2-supplied masks and concatenated them together to serve as input to Phyml. The resulting phylogeny showed that the TM6 sequences that were obtained in this study were representative of a deep-branching phylum that claded closest to the Acidobacteria and Aquificae phyla (Fig. 4 and SI Appendix, Fig. S9). A similar topology, where 16S rRNA genes representative of TM6 were clading closest to the Acidobacteria phylum, was also observed when using the SSU-Align program (SI Appendix, Fig. S8). Its other closest 16S rRNA gene-neighbor represent the Elusimicriobim Phylum (SI Appendix, Fig. S8) which is in line with previous 16S rRNA gene analyses (55, 56). However, due to the more robust AMPHORA2 analyses where multiple marker genes with phylogentic signals were used, we propose that the TM6 phylum is most closely related to the Acidobacteria and Aquificeae phyla. Additional genomes will be needed to further refine this phylogenetic position.
Fig. 4.
Phylogenetic tree illustrating the major lineages (phyla) of the domain Bacteria analyzed with AMPHORA2 and 29 protein phylogenetic markers. The TM6 gene sequences were aligned against the AMPHORA2 seed alignment consisting of sequences from over 1,000 genomes through HMM. Tree branch lengths ≤0.4 were collapsed. For the original tree, see SI Appendix, Fig. S9.
Pathways and Processes.
Due to the distant homology of TM6 proteins with existing genomes, only 43% of the protein-coding regions were functionally annotated (428 genes). As with many genomes of uncultivated species using single-cell genomic techniques, it is still possible to gain insight into the predicted metabolic abilities of TM6SC1 using the captured genomic information. In our case, having an estimated 91% of a genome, we are still cautious in our interpretation. The fact that we generated three separate sets of contigs that are nearly identical at the nucleotide level provides additional confidence in the functional interpretations based on presence or absence of key genes and operons but does not rule out the possibility that we are missing these from the three assemblies.
Cell-Wall Biogenesis and Pili.
TM6SC1 contains evidence for a Gram-negative envelope including outer-membrane proteins Omp18, YaeT, RomA, and OmpH and outer-membrane-related genes homologous to the type II general secretion pathway (gspD, gspE, and gspG) as well as murJ (peptidoglycan lipid II flippase). There were very few genes that gave an indication of a possible phenotype, but there is some evidence that this organism may form a type of spore as it contains genes with homology to sporulation, the SpoIID/lytB domain, and SpoVG (SI Appendix, Table S3). An ability to form a spore-like feature (such as an endospore) is consistent with our enrichment of these organisms in the small, weakly fluorescent population (spores are typically of low-fluorescence profile). Only one gene with homology to flagellar genes was found (fliC), indicating that this organism may not be motile via flagella, although it is possible that it could be motile via a gliding motility using the type IV pili-related genes (pilA and pilB). The genome also encodes a sigma factor 54, which is a central transcriptional regulator in many bacteria and has been linked to a multitude of processes like nitrogen assimilation, motility, virulence (host colonization), and biofilm formation (59).
Energy Production and Conservation.
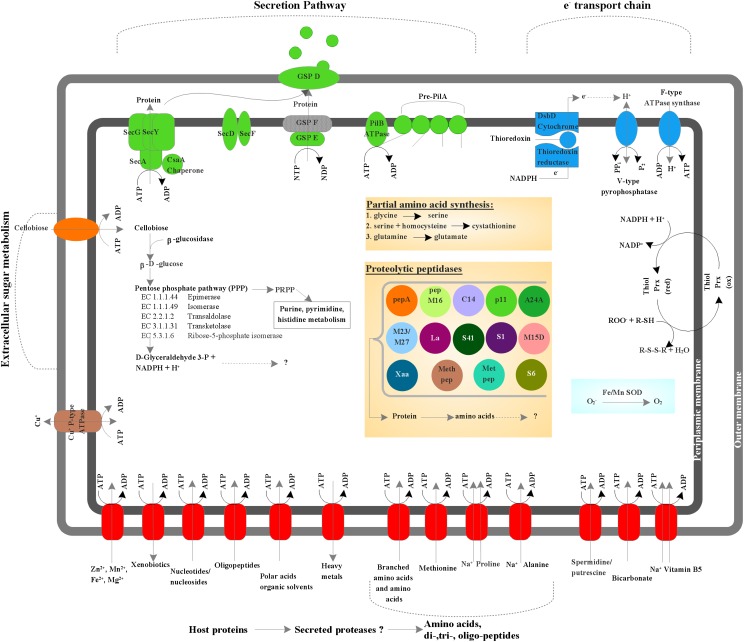
The predicted metabolic pathways of TM6SC1 are shown in Fig. 5. The genome retains a noncatalytic glycoside-binding protein, a lectin B chain, and a β-glucosidase, suggesting that TM6 can bind complex carbohydrates and perform extracellular hydrolysis of the cellulose-derived disaccharide cellobiose. Cellobiose can enter the bacterial cell either via a phosphoenolpyruvate-dependent phosphotransferase system or via protein-dependent ATP-binding-cassette (ABC) transporters; the latter were identified at several locations in the genome. β-Glucosidase acts on the glucose-β(1,4) linkage in cellobiose, which results in the production of β–d-glucose, the unphosphorylated substrate needed for the pentose phosphate pathway for which the genome harbors all enzymes. A putative sodium-dependent bicarbonate transporter and a carbonic anhydrase (EC 4.2.1.1), responsible for inorganic carbon (CO2) uptake (60) and rapid interconversion of carbon dioxide and water to bicarbonate and protons, respectively, were also identified. Potentially, the latter two are used to remove CO2 produced by the oxidative arm of the pentose phosphate pathway and to prevent acidification of the cytoplasm. Alternatively, a pathway for autotrophic carbon fixation unique to TM6 is possibly present. These features reflect an unexpected mixotrophic lifestyle that is unusual for bacteria with small genome sizes (61). The genome contains only one enzyme from the glycolysis pathway, a phosphoglycerate mutase that catalyzes the conversion of 3-phosphoglycerate to 2-phosphoglycerate. It lacks any identifiable tricaboxylic-acid-cycle enzymes but has a modified electron transport chain consisting of an F-type ATPase synthase, a protein disulfide reductase (DsbD), and five thioredoxin reductases that previously were identified as essential for aerobic growth of facultative anaerobic bacteria (62). Thioredoxin reductases are known for acting on sulfur groups of electron donors with NAD+ or NADP+ as acceptors and for shuffling electrons from cytoplasm to periplasm without involvement of additional cofactors such as quinone. A demethylmenaquinone—which functions as a reversible redox component of the electron transfer chain, mediating electron transfer between hydrogenases and cytochromes in anaerobic conditions—was identified; however, both of these proteins are absent. Another gene that indicates TM6’s potential to grow anaerobically is the nitrogen fixation protein (nifU) gene that is responsible for Fe-S cluster assembly and functions as an electron transfer component. No other nif genes were identified, and hence nitrogenase activity, which requires additional genes (e.g., NifH, NifS, NifV), is unlikely. Instead, it seems likely that Fe-S clusters are synthesized for a coproporphyrinogen III oxidase (HemN), the only protein that appears to use this cofactor. The remainder of the heme synthesis pathway appears to be missing; thus hemN may be used to scavenge porphyrins. Also, a V-type pyrophosphate-energized proton pump that can generate a proton motive force (PMF) or use an existing PMF to drive pyrophosphate synthesis such as in acidic environments is present. The genome harbors manganese and iron superoxide dismutases but no catalase for H2O2 degradation. However, it has two peroxidase enzymes, which derive electrons from NADH2 to reduce peroxide to H2O. The genome also encodes a putative copper ion transporter ATPase for copper efflux, which also can prevent oxidative stress in aerobic conditions. Together, these features suggest that TM6SC1 is a facultative anaerobic bacterium able to generate energy from organic carbon sources with a modified electron transport chain adapted for both aerobic and anaerobic conditions.
Fig. 5.
Predicted metabolic pathways of phylotype TM6SC1. Predicted ABC transporters (e.g., amino acid importers, nucleotide/nucleoside importers, divalent ion importers) (red) as well as a cellobiose importer (orange). ATP-driven transporters are indicated by the ATP hydrolysis reaction. The copper ion transporting P-type ATPase is proposed to serve as both uptake and efflux systems, which is shown by a bidirectional arrow. Several protein-secretion components belonging to the type II secretion pathway; GSP and Sec proteins were identified as well as five prepilin related domains and a type IV pilB ATPase (green). A modified electron transport chain was also identified consisting of seven thioredoxins and five thioredoxin reductases, a V-type pyrophosphatase, an F-type ATPase synthase, and a protein disulfide reductase (DsbD) (blue). Thiolperoxidases (Bcp and bacterioferretine-like), superoxide dismutases (manganese and iron), were identified as cytosolic and periplasmic enzymes protecting against oxidative stress. A β-glucosidase that catalyzes the formation of β-d-glucose from disaccharides (e.g., cellobiose) was identified as well as all enzymes involved in the pentose phosphate pathway. However, other enzymes (e.g., enzymes from the Calvin cycle or the citric acid cycle) involved in the conversion of 3-glyceraldehyde-3-P could not be identified. Enzymes for de novo synthesis of amino acids were absent, however several amino acid importing membrane proteins (ABC transporters), and 14 cytosolic and proteolytic peptidases were identified, indicating that TM6 has a high capacity for amino acid scavenging.
Biosynthesis of Amino Acids, Nucleotides, and Coenzymes.
Evidence for the capacity for de novo synthesis of amino acids is currently absent in the assembled genome because it contains only a few enzymes (e.g., glycine hydroxymethyl transferase) that can be used to synthesize serine, glycine, and cysteine only from intermediate metabolites (Fig. 5). Also, a glutamine-fructose-6-phosphate transaminase is present and has the potential to catalyze the formation of glutamate. Several amino peptidases that catalyze the cleavage of amino acids of proteins or peptide substrates were identified (e.g., pepA, pepM16, methionyl aminopeptidase). In addition, cotransporting proteins such as sodium/proline and sodium/alanine symporters were present, suggesting that proline and alanine can be imported from the environment. Taken together, these results suggest that TM6 relies on energy-saving salvatory pathways including peptidase activity and amino acid import. The same pattern is observed for pyrimidine and purine biosynthesis as the required enzymes for synthesis of the building block inosine 5′-monophosphate from inosine are absent, which excludes the potential for de novo synthesis. However, several enzymes with the capacity to both catabolize and synthesize ADP, GDP, ATP, GTP, dATP, and dGTP are present. The coenzyme vitamin K epoxide reductase (involved in vitamin K recycling), the NADP+ reducer glycerol-3-phosphate-NADP dehydrogenase, and the pyridoxamine 5′-phosphate oxidase (which catalyzes the biologically active form of vitamin B6) were identified as electron carriers. The genome also harbors a symporter for sodium/panthothenate (vitamin B5), showing that this essential vitamin is unlikely to be synthesized de novo as in many other bacterial species (63).
We also applied less stringent comparison rules than were applied by the automated annotation to address the unclassified ORFs. We manually reannotated representative “unclassified” ORFs that were located around interesting gene signatures and that might possibly correspond to horizontal gene transfer events (e.g., phage signatures and short sequence repeats) (SI Appendix, Table S3). The amino acid sequences of these ORFs were manually compared with existing protein sequences by using BLASTP with a low stringency. This resulted in identification of several genes coding for proteins broadly classified as involved in virulence. In addition, a number of potential archaeal genes were also identified (SI Appendix, Table S4).
Discussion
We have demonstrated a mini-metagenomic approach based on single-cell genomic sequencing methodology, to be used with deep sequencing and downstream assembly methods optimized for MDA samples, with the aim of reconstructing genomes from small pools of cells. Using this approach, a near complete genome of a member of the low-abundance yet globally distributed candidate phylum TM6 was recovered in a biofilm from a hospital sink. Previous studies have identified TM6 using a phylogenetic marker (16S rRNA gene) in a number of diverse environments with a global distribution. They appear to be low-abundance members of many microbial communities including those in domestic water sources such as drinking water distribution systems (45) and showerhead biofilms (17). At the time of this study, the closest sequence in the National Center for Biotechnology Information nr database to our assembled TM6 genome is from a biofilm in a corroded copper water pipe (GenBank accession no. GU368367) (39), supporting our discovery of this organism in a sink drain biofilm.
From one of our pooled samples, we reconstructed a draft genome (1.07 Mb in seven contigs) for this species of TM6 that we designated TM6SC1. As with many genomes of currently uncultivated organisms that have been recovered with single-cell or metagenomic approaches, the genome of TM6 presented here is only a portion of the whole, and this makes interpretations difficult to confirm or refute without more genomes or an actual isolate. The near perfect nucleotide identity between a total of three independently amplified and assembled samples, however, provided strong confirmation of a correct genomic sequence for the portion of the genome recovered. An analysis of the conserved single-copy genes using a set of 111 genes (25) revealed that the assembly included 101 of the 111 genes and thus an estimated 91% recovery of the TM6 genome.
From the available genes that were assembled and functionally annotated, TM6 is cautiously a Gram-negative and facultative anaerobe. The predicted genome size of TM6 falls within the range of some of the smallest sequenced bacteria that are predominantly symbionts. This raises the question as to whether TM6 might be a free-living organism or has formed a symbiotic relationship with unknown host. The TM6 genome is in general agreement with some of the characteristics of symbionts reported to date. These features include reduced genome size, AT bias, and loss of biosynthetic pathways (64, 65). In particular, amino acid biosynthetic genes are often lost in obligate symbiotic bacterial genomes (obligate host pathogens and obligate endosymbionts) (64, 66, 67), where amino acids are obtained from the host environment.
Several additional lines of evidence point toward TM6 as possibly having a symbiotic lifestyle. The phylogenetic affiliations of roughly 10% of the CDSs had best hits to known facultative symbionts or obligate symbionts including Parachlamydia acanthamoebae, Candidatus Protochlamydia amoebophilia (Chlamydiae), Candidatus Ameobophilus asiaticus, Legionella, Francisella (Gammaproteobacteria) Rickettsia (Alphaproteobacteria), Borrelia (Spirochaetales), and Planctomyces (Planctomycetaceae). (SI Appendix, Table S5). A recent study reported that an obligate endosymbiont, Candidatus Ameobophilus asiaticus, of a free-living amoeba, also lacking almost all amino acid biosynthesis pathways, contains a large fraction of proteins with eukaryotic domains (67, 68). It was demonstrated that these domains are also significantly enriched in the genomes of other amoeba-associated bacteria (including Legionella pneumophilia, Ricksettsia bellii, Francisella tularensis, and M. avium). TM6 also contains several of these eukaryotic domains within predicted coding regions such as ankyrin repeats (eight identified), an F-box domain protein, tetratricopeptide repeat TPR_1, and WD-40 repeat domains (five identified). Based on the fact that TM6 remains uncultivated to date despite being observed globally and across diverse environments also suggests a host such as a free-living amoeba. Such bacteria would likely only yield to cultivation if it was co-isolated with the host species for which it is symbiotically associated. Amoebas are also well known to be globally distributed across diverse environments (67, 69), which could explain the distribution of TM6. In relation to where we recovered this TM6 genome, often studies of hospital water networks (taps and showerheads) yield many amoeba and their associated bacteria (18, 70). These studies are conducted mainly to evaluate the role of pathogenic amoeba-associated bacteria such as Legionella and Parachlamydia in hospital-acquired infections. A direct report of finding TM6-related 16S rRNA gene signatures associated amoeba hosts was not found in our investigations. So far, in terms of host-related systems, TM6 have been reported as part of the consortia of bacteria intimately associated with marine sponges (35). In the absence of direct evidence, further detailed work is needed to determine the association, if any exists, between members of TM6 and eukaryotic hosts.
Overall, the genomic information presented here may help guide cultivation efforts and efforts to further elucidate the function and ecology for this organism. Further application of this approach in other environments may greatly increase the likelihood of capturing and assembling genomes of elusive, low-abundance microorganisms that continue to remain unyielding to culturing approaches.
Methods
Isolation of Bacterial Cells from Sink Material.
Sink drain samples were collected with sterile cotton-tipped swabs from a publicly accessible restroom adjacent to an emergency waiting room. The initial sample was fixed with ethanol and vortexed briefly for 20 s (SI Appendix, Fig. S1). The sample was filtered through a 35-µm filter. A 2-mL cushion of prechilled Nycodenz gradient solution (Nycoprep Universal, Axis Shield) was placed in a 17-mL ultracentrifuge tube, and 6 mL of supernatant was placed gently over the Nycodenz cushion. The pair of balanced tubes was centrifuged at 9,000 × g for 20 min at 4 °C in an ultracentrifuge SW32.1 rotor. The visible cloudy interface containing the bacterial cells was collected gently and mixed by inversion to create a suspension.
Sorting of Single Cells by Flow Cytometry.
Single-cell sorting was performed on a custom FACS Aria II as described (6). FACS detection was performed on the Nycodenz fractionated bacteria-enriched sample. Filter-sterilized (0.2 μm) PBS (1×) was used as sheath fluid and for sample dilution. Unstained and SYBR Green I (0.5×)-stained material was 35-μm filtered, and a 1:1,000 dilution was assessed for event rate at low flow rate (<2,000 total events/s) and adjusted if necessary. A low flow rate is critical to reduce the likelihood of sorting coincident events. Events were sorted into 4 μL of a low EDTA TE (10 mM Tris, 0.1 mM EDTA, pH 8.0) and immediately frozen on dry ice and held there until transfer to −80 °C for storage before processing.
Multiple Displacement Amplification.
MDA was performed in a 384-well format using a GenomiPhi HY kit (GE Healthcare) using a custom Agilent BioCel robotic system (outlined in SI Appendix, Fig. S2). Briefly, cells were lysed by addition of 2 μL of alkaline lysis solution (645 mM KOH, 265 mM DTT, 2.65 mM EDTA, pH 8.0) and then incubated for 10 min at 4 °C. After lysis, 7 μL of a neutralization solution (2.8 μL of 1,290 mM Tris⋅Cl, pH 4.5, and 4.2 μL of GE Healthcare Sample Buffer) was added, followed by 12 μL of GenomiPhi master mix (10.8 μL of GE Healthcare Reaction Buffer and 1.2 μL GE Healthcare Enzyme Mix) for a reaction volume of 25 μL. Reactions were incubated at 30 °C for 16 h followed by a 10-min inactivation step at 65 °C. MDA yield was determined by Picogreen assay. MDAs with yields ≥50 ng/μL were set aside for the purpose of this study as the relationship between yield and MDA quality is unclear. No-template-control MDA reactions were included to reveal any contaminating sequences and processed in parallel through 16S rRNA gene PCR analysis. These negative controls lacking a sorted cell were run in parallel to determine the relative amount and identity of contaminating bacterial DNA in the MDA reagents, a necessary standard practice in single-cell genomics due to the highly processive strand displacement activity of the phi29 DNA polymerase (71–73). Further amplification of selected MDAs to generate 100–200 μg for sequencing and archival storage was performed as described above with 150–1,500 ng of the original MDA as template.
PCR and Analysis of 16S rRNAs.
Using the Biocel robotics platform processing plates in a 384-well format, 16S rRNA was amplified from diluted MDA product (1:20 into TE) using universal bacterial primers 27f and 1492r (74) as follows: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, and 72 °C for 10 min. PCR products were treated with exonuclease I and shrimp alkaline phosphatase (both from Fermentas) before direct cycle sequencing with 27f and 1492r primers at the Joint Technology Center (J. Craig Venter Institute, Rockville, MD). 16S rRNA gene trace files were analyzed and trimmed with the CLC Workbench software program (CLC Bio). Sanger read lengths of less than 200 bp were discarded. Only a minority of 16S rRNA read pairs could form a contig, and in some cases only the forward or reverse read was used to establish taxonomy. Chromatogram quality was assessed manually, and MDAs with both forward and reverse reads of poor quality were excluded from further analysis. MDAs with 16S taxonomy similar to those in MDA and 16S PCR reactions with no template DNA added were excluded from further analysis.
All full-length 16S rRNA sequences from the three assemblies were 100% identical. A BLASTN analysis against the SILVA SSU Ref NR 102 database (54) was performed to classify the 16S sequences taxonomically and to determine their relationship to TM6. An additional analysis was performed against public databases to retrieve neighboring TM6 and related bacterial sequences for generation of 16S rRNA phylogenetic trees. The 16S rRNA gene phylogenetic tree was created by aligning the related sequences against the SILVA alignment with mothur 1.19.4, triming to eliminate gap-only positions and creating a maximum-likelihood tree with PhyML version 20110919 (70). A phylogenetic-marker gene tree was created by using the AMPHORA2 pipeline (57, 58). AMPHORA2 uses a hidden Markov model trained on a reference database of 571 fully sequenced bacterial genomes to identify and align gene sequences belonging to 31 marker genes. Twenty-nine of these genes could be identified in TM6 and were used in the downstream phylogenic analyses (SI Appendix, Table S2). A single large alignment was generated by concatenating the masked HMM-generated AMPHORA2 alignments from 29 genes. As the AMPHORA2 alignments contained information from hundreds of genomes, a phylogenetically representative subset of the alignment was created for computational feasibility. This alignment was used to create a maximum-likelihood tree with PhyML version 20110919 (70) using the Whelan and Goldman (WAG) amino acid evolutionary model (75).
Library Construction and Sequencing.
Illumina sequencing on the GAII platform was performed on the amplified genomic material using the Genome Analyzer II System according to the manufacturer’s specifications. Three TM6 MDAs were barcoded and pooled for a single-lane generating reads totaling 23 GB of data and 85 million reads that passed a quality score >20.
Single-Cell Assemblies.
Assemblies were produced using Velvet-SC (27) and SPAdes (31) and CLC Bio Version 5.1 (CLC Bio). All three are based on the de Bruijn graph. The first two assemblers have been adapted for uneven coverage found in single-cell MDA datasets. For Velvet-SC, we assembled the data with vertex size k = 55. For SPAdes, we iterated over vertex sizes k = 21, 33, and 55.
Contig Binning Methods and Annotation.
A 273-kb contig in MDA2 with an average GC content of 36% contained a 16S rRNA gene with a flanking 23S. The 16S rRNA gene had a top BLAST hit to a member of TM6. Taxonomic affiliations of the predicted protein sequences derived from this contig were also assigned using APIS (42). APIS generates a phylogenetic tree for each ORF in a genomic or metagenomic sample using homologous proteins from PhyloDB 1.05, a J. Craig Venter Institute database containing proteins from all publically available complete genomes as of August 15, 2012. APIS classifies each ORF taxonomically and functionally based on their phylogenetic position. An independent metagenomic binning approach using an autonomous method, PCA, of the penta-nucleotide frequency, followed by k-means clustering was used. A second independent approach used MGTAXA software (http://mgtaxa.jcvi.org), which performs taxonomic classification of metagenomic sequences with machine-learning techniques. The 273-kb contig containing the TM6 16S rRNA was used as the input sequence in training sets to classify all remaining contigs from the three assemblies using MGTAXA. Identified contigs from the intersection of the separate approaches sharing a GC content of 36 ± 2% were chosen as the final set of contigs. MDA2 had the largest number of base pairs and was concatenated to allow comparisons to the MDA1 and MDA3 contig sets. Whole-contig set comparisons were carried out using ProgressiveMauve (43) and the LAST alignment tool (44). The assembly from MDA2 that represented the largest assembled genome was annotated using the J. Craig Venter Institute metagenomic annotation pipeline (www.jcvi.org/cms/research/projects/annotation-service/overview/), which uses Metagene for gene calling (76). A combination of databases and tools including BLAST, RAST (77), as well as MG-RAST (78) uploaded with nucleotide sequences for the CDS, were used to assess a consensus on the pathways and processes predicted for the TM6 genome.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for their time and guidance to help improve the manuscript. We also thank Pamela Mishra and Mathangi Thiagarajan (J. Craig Venter Institute) for bioinformatics support. This work was supported by the Alfred P. Sloan Foundation (Sloan Foundation-2007-10-19 grants to R.M.F., J.C.V., and R.S.L.); by National Institutes of Health (NIH) Grant 3P41RR024851-02S1 (to P.A.P. and G.T.); by NIH Grant 2R01 HG003647 (to R.S.L.); by Government of the Russian Federation Grant 11.G34.31.0018 (to P.A.P.); by NIH Grant UL1TR000100 (to M.G.Z.); and by NIH National Institute of General Medical Sciences Grant 1R01GM095373 (to J.S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The TM6SC1 final assembly sequence data have been submitted to GenBank under BioProject PRJNA193445 (accession no. ARQD00000000). The associated 16S rRNA sequence from the assembled TM6SC1 genome has been deposited in Genbank (accession no. KF057992).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219809110/-/DCSupplemental.
References
- 1.Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10(9):631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 2.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11(6):1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71(6):3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13(5):954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99(8):5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLean JS, et al. Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res. 2013;23(5):867–877. doi: 10.1101/gr.150433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klepeis NE, et al. The National Human Activity Pattern Survey (NHAPS): A resource for assessing exposure to environmental pollutants. J Expo Anal Environ Epidemiol. 2001;11(3):231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- 8.Tringe SG, et al. 2008. The airborne metagenome in an indoor urban environment. PLoS One 3(4):e1862.
- 9.Hospodsky D, et al. Human occupancy as a source of indoor airborne bacteria. PLoS ONE. 2012;7(4):e34867. doi: 10.1371/journal.pone.0034867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gião MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. Interaction of Legionella pneumophila and Helicobacter pylori with bacterial species isolated from drinking water biofilms. BMC Microbiol. 2011;11:57. doi: 10.1186/1471-2180-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Declerck P. Biofilms: The environmental playground of Legionella pneumophila. Environ Microbiol. 2010;12(3):557–566. doi: 10.1111/j.1462-2920.2009.02025.x. [DOI] [PubMed] [Google Scholar]
- 12.Murga R, et al. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology. 2001;147(Pt 11):3121–3126. doi: 10.1099/00221287-147-11-3121. [DOI] [PubMed] [Google Scholar]
- 13.Walker JT, Sonesson A, Keevil CW, White DC. Detection of Legionella pneumophila in biofilms containing a complex microbial consortium by gas chromatography-mass spectrometry analysis of genus-specific hydroxy fatty acids. FEMS Microbiol Lett. 1993;113(2):139–144. doi: 10.1111/j.1574-6968.1993.tb06504.x. [DOI] [PubMed] [Google Scholar]
- 14.Shikuma NJ, Hadfield MG. Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling. 2010;26(1):39–46. doi: 10.1080/08927010903282814. [DOI] [PubMed] [Google Scholar]
- 15.Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health. 2009;7(3):469–477. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 16.Linke S, Lenz J, Gemein S, Exner M, Gebel J. Detection of Helicobacter pylori in biofilms by real-time PCR. Int J Hyg Environ Health. 2010;213(3):176–182. doi: 10.1016/j.ijheh.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Feazel LM, et al. 2009 Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci USA 106(38):16393–16399. [Google Scholar]
- 18. Nurk S, et al. (2013) Assembling Genomes and mini-metagenomes from highly chimeric reads. Research in Computational Molecular Biology, eds Deng M, Jiang R, Sun F, Zhang X, Lecture Notes in Computer Science (Springer, Berlin), Vol 7821, pp 158–170. [Google Scholar]
- 19.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428(6978):37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 20.Woyke T, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443(7114):950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 21.Hess M, et al. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331(6016):463–467.
- 22.Iverson V, et al. Untangling genomes from metagenomes: Revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 23.Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107(37):16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier E, et al. “Candidatus Cloacamonas acidaminovorans”: Genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol. 2008;190(7):2572–2579. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6(6):1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS ONE. 2009;4(4):e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitsaz H, et al. 2011. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nature Biotechnol 29(10):915–921.
- 28.Abulencia CB, et al. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl Environ Microbiol. 2006;72(5):3291–3301. doi: 10.1128/AEM.72.5.3291-3301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz S, Allgaier M, Hugenholtz P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods. 2010;7(12):943–944. doi: 10.1038/nmeth1210-943. [DOI] [PubMed] [Google Scholar]
- 30.Rheims H,, Rainey FA, Stackebrandt E. 1996. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol 17(3–4):159–169. [DOI] [PubMed]
- 31.Bankevich A, et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477.
- 32.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104(29):11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73(10):3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youssef NH, Blainey PC, Quake SR, Elshahed MS. Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma) Appl Environ Microbiol. 2011;77(21):7804–7814. doi: 10.1128/AEM.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegl A, et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5(1):61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hongoh Y, et al. 2008 Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc Natl Acad Sci USA 105(14):5555–5615. [Google Scholar]
- 37.Fodor AA, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS ONE. 2012;7(7):e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Revetta RP, Pemberton A, Lamendella R, Iker B, Santo Domingo JW. Identification of bacterial populations in drinking water using 16S rRNA-based sequence analyses. Water Res. 2010;44(5):1353–1360. doi: 10.1016/j.watres.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Pavissich J, Vargas I, González B, Pastén P, Pizarro G. 2010. Culture dependent and independent analyses of bacterial communities involved in copper plumbing corrosion. J Appl Microbiol 109(3):771–782. [DOI] [PubMed]
- 40.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Hu Y, Bolund L, Wang J. State of the art de novo assembly of human genomes from massively parallel sequencing data. Hum Genomics. 2010;4(4):271–277. doi: 10.1186/1479-7364-4-4-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badger JH, et al. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J Bacteriol. 2006;188(19):6841–6850. doi: 10.1128/JB.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darling AE, Mau B, Perna NT. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frith MC, Hamada M, Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henne K, Kahlisch L, Brettar I, Hofle MG. Analysis of structure and composition of bacterial core communities in mature drinking water biofilms and bulk water of a citywide network in Germany. Appl Environ Microbiol. 2012;78(10):3530–3538. doi: 10.1128/AEM.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lear G, Niyogi D, Harding J, Dong Y, Lewis G. 2009. Biofilm bacterial community structure in streams affected by acid mine drainage. Appl Environ Microbiol 75(11):3455–3460. [DOI] [PMC free article] [PubMed]
- 47.Kwon S, Kim T-S, Yu G, Jung J-H, Park H-D. 2010. Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J Microbiol Biotechnol 20(12):1717–1723.
- 48.Fields M, et al. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol Ecol 53(3):417–428.
- 49.Lin X, Kennedy D, Fredrickson J, Bjornstad B, Konopka A. 2012. Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ Microbiol 14(2)414–425.
- 50.Dedysh S, Pankratov T, Belova S, Kulichevskaya I, Liesack W. 2006. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol 72(3):2110–2117.
- 51.Maldonado-Contreras A, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5(4):574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hugenholtz P, Goebel B, Pace N. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180(18):4765–4774. [DOI] [PMC free article] [PubMed]
- 54.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 56.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67(1):411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9(10):R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28(7):1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 59.Francke C, et al. Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics. 2011;12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dagnall BH, Saier MH., Jr HatA and HatR, implicated in the uptake of inorganic carbon in Synechocystis PCC6803, contain WD40 domains. Mol Microbiol. 1997;24(1):229–230. doi: 10.1046/j.1365-2958.1997.3231687.x. [DOI] [PubMed] [Google Scholar]
- 61.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468(7320):60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 62.Serata M, Iino T, Yasuda E, Sako T. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology. 2012;158(Pt 4):953–962. doi: 10.1099/mic.0.053942-0. [DOI] [PubMed] [Google Scholar]
- 63.Genschel U. Coenzyme A biosynthesis: Reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol Biol Evol. 2004;21(7):1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 64.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6(7):263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 65.Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: Insights from genomics. Trends Ecol Evol. 2000;15(8):321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 66.Yu XJ, Walker DH, Liu Y, Zhang L. 2009. Amino acid biosynthesis deficiency in bacteria associated with human and animal hosts. Infect Genet Evol 9(4):514–517.
- 67.Schmitz-Esser S, et al. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 2010;192(4):1045–1057. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collingro A, et al. Unity in variety: The pan-genome of the Chlamydiae. Mol Biol Evol. 2011;28(12):3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol. 2004;51(5):509–514. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 70.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 71.Woyke T, et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS ONE. 2011;6(10):e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39(4):e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen LZ, et al. Single virus genomics: A new tool for virus discovery. PLoS ONE. 2011;6(3):e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lane DJ. 1991. 16S/23S rRNA sequencing. Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E, Goodfellow M (John Wiley & Sons, New York), pp 115–175. [Google Scholar]
- 75.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18(5):691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 76.Noguchi H, Park J, Takagi T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34(19):5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyer F, et al. The metagenomics RAST server: A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.