Abstract
The transcriptional repressor BTB and CNC homology 2 (Bach2) is thought to be mainly expressed in B cells with specific functions such as class switch recombination and somatic hypermutation, but its function in T cells is not known. We found equal Bach2 expression in T cells and analyzed its function using Bach2-deficient (−/−) mice. Although T-cell development was normal, numbers of peripheral naive T cells were decreased, which rapidly produced Th2 cytokines after TCR stimulation. Bach2−/− naive T cells highly expressed genes related to effector-memory T cells such as CCR4, ST-2 and Blimp-1. Enhanced expression of these genes induced Bach2−/− naive T cells to migrate toward CCR4-ligand and respond to IL33. Forced expression of Bach2 restored the expression of these genes. Using Chromatin Immunoprecipitation (ChIP)-seq analysis, we identified S100 calcium binding protein a, Heme oxigenase 1, and prolyl hydroxylase 3 as Bach2 direct target genes, which are highly expressed in effector-memory T cells. These findings indicate that Bach2 suppresses effector memory-related genes to maintain the naive T-cell state and regulates generation of effector-memory T cells.
Keywords: transcription factor, innate-like lymphocytes, CNC family
T cells are selected and differentiate in the thymus. Immature CD4+CD8+ double-positive (DP) thymocytes differentiate into mature CD4 or CD8 single-positive (SP) T cells. Several transcription factors (TFs) are up-regulated during the DP to SP differentiation and contribute to development of T cells (1, 2).
Kruppel-like factor 2 (KLF2) is one of those TF, which regulates the expression of the sphingosine-1-phosphate receptor-1 (S1pr1) in SP cells (2). S1pr1 is required for the egress of SP cells from the thymus, and thus KLF2-deficient (−/−) SP cells accumulate in the thymus. KLF2 positively regulates S1pr1 expression through direct binding to its promoter. KLF2−/− mice also develop an unusual population of innate-like CD8+ T cells (3). This phenotype is not T cell-intrinsic but is caused by IL-4 secreted from bystander promyelocytic leukemia zinc finger protein (PLZF)-positive T cells (3). Forkhead box protein O1 (Foxo1) is another TF up-regulated during thymic differentiation. T cell-specific Foxo1 deficiency led to spontaneous activation, to diminished survival, and to a defect in trafficking of naive T cells (1, 4). The impaired expression of CD62L, CCR7, and IL7R by Foxo1−/− T cells may be partly responsible for these phenotypes (1).
The TF Bach2 also regulates lymphocyte development. It was reported to be expressed mainly in B cells, especially at the stage before plasma cell differentiation (5, 6), and its disruption results in impaired B cell-specific functions (class switch recombination and somatic hypermutation of Ig genes) although the differentiation into plasma cells is intact (6). Bach2 belongs to the Cap’n’collar (CNC) gene family (7), which consists of four transcriptional activators [nuclear factor, erythroid-derived 2 (NF-E2), NF-E2-like 1/2/3 (Nrf1/2/3)] and two repressors (Bach1/2). A basic leucine zipper domain and an adjacent conserved CNC domain are the defining features of this family. The CNC proteins form heterodimers with small maf proteins (MafK/F/G), and those dimers bind to a specific DNA sequence, the antioxidant response element (ARE) (8). The CNC proteins have important roles in stress responses. For example, NF-E2 and Nrf2 are essential for an antioxidant and detoxification system (9), respectively. Bach1 regulates the antioxidant genes through the same cis-elements as Nrf2 (10).
Although Bach2 was reported to be expressed specifically in B cells (11, 12), we found that Bach2 was expressed in normal T cells at levels comparable with B cells. Peripheral T cells consist of mainly two populations: naive and memory T cells. Naive T cells are activated upon antigen recognition and become effector T cells. Thereafter whereas the majority of effector T cells die, a minor population of the effector T cells remain and develop into memory T cells. Memory T cells further consist of two types: central- and effector-memory T cells on the basis of their homing and functional capacity. Central-memory T cells express high levels of CD62L and CCR7 and mainly circulate through lymphoid organs and rapidly proliferate after restimulation whereas effector-memory T cells are preferentially distributed within nonlymphoid tissues and rapidly exhibit effector functions after restimulation (13, 14). In the CD4 T-cell populations, effector and memory T cells with the capacity of different cytokine secretion are developed, such as T helper 1 (Th1) cells producing IFNγ and Th2 cells producing IL-4/IL-5/IL-9/IL-13 (15, 16). Th2 is the subset related to the induction of allergic reaction and is developed in the presence of IL-4.
In this study, we found that Bach2 deficiency results in the reduction of naive T cells and enhances effector-memory T cells, particularly Th2 cells. We identified Bach2 target genes related to effector-memory cells. These analyses reveal that Bach2 has a critical function to maintain the naive state of T cells by suppressing several effector memory-related genes in naive T cells.
Results
Expression of Bach2 in T Cells.
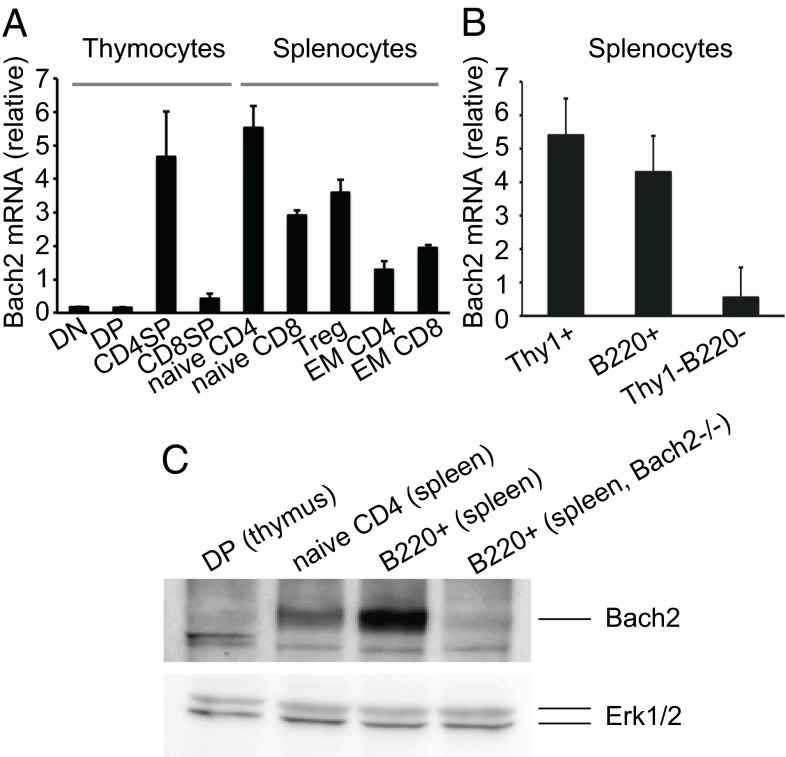
To uncover the molecular mechanism of T-cell differentiation, we used microarray data (RefDIC; http://refdic.rcai.riken.jp) to search for genes whose expression is altered during T-cell differentiation. We found that Bach2 transcripts are up-regulated during this differentiation process. Quantitative RT-PCR (qPCR) confirmed that Bach2 expression is very low in DN and DP, and higher in SP thymocytes. Whereas CD4 SP thymocytes and CD4+ and CD8+ splenic T cells express a high level of Bach2, there is relatively low expression in CD8SP thymocytes (Fig. 1A). These results indicate that Bach2 is highly expressed in T cells and up-regulated during their maturation.
Fig. 1.
The expression of Bach2 mRNA in T cells. (A) qPCR for Bach2 in thymocytes and spleen T cells. The mRNA expression was normalized to cyclophilin A. (B) qPCR for Bach2 in spleen cells. Data are expressed as mean ± SD, n = 3 in A and B. (C) Western blot detection of Bach2 in DP thymocytes, naive CD4 T cells, and B220+ cells from WT and Bach2−/− mice. Erk1/2 was used as a loading control. DN, double negative; EM, effector-memory; Treg, regulatory T cells.
Although we initially reported that Bach2 was mainly expressed in B cells (17), we found now that it is also expressed in T cells and at levels almost compatible to B cells whereas non-T/B cells express a much lower level (Fig.1B). Among T-cell subpopulations, naive T cells have the highest level, and effector-memory T cells show a much reduced level of Bach2 expression (Fig.1A). The high expression in naive T cells was confirmed at the protein level although it was lower than that in B cells (Fig.1C), indicating that Bach2 expression is differentially regulated posttranscriptionally in T and B cells.
Effects of Bach2 Deficiency on T-Cell Populations.
To investigate the role of Bach2 in T cells, we analyzed T cells from Bach2−/− mice. Although Bach2 mRNA is up-regulated during thymocyte differentiation, the cell number and frequency of the CD4/CD8 subpopulation were normal in Bach2−/− thymocytes (Fig. S1 A and B). The differentiation markers CD5, TCRβ, CD69, and heat stable antigen (HSA) in each subpopulation were expressed normaly (Fig. S1C), indicating that Bach2 does not play a critical role in thymocyte differentiation.
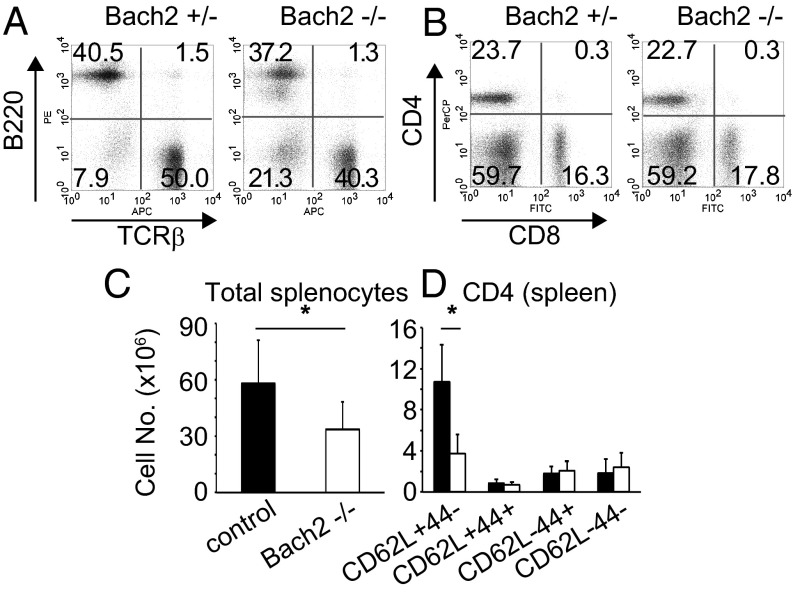
As for mature peripheral T cells, the cell number in the spleen was slightly decreased, but the ratio of T/B cells and CD4/CD8 expression were unaffected (Fig. 2 A and B). On the other hand, the percentages and the numbers of naive (CD62LhiCD44lo) T cells in spleen (Fig. 2C) and lymph node of Bach2−/− mice were decreased compared with other populations (Fig. 2 C and D, Fig. S2 A and B). These results suggest that Bach2 is required for the homeostasis of peripheral naive T cells.
Fig. 2.
Analysis of spleen subpopulations in Bach2−/− and Bach2+/− mice. (A and B) FACS profiles of T (TCRβ+) and B cells (B220+) (A) and CD4/CD8 expression (B) are shown. (C and D) The cell numbers of total splenocytes (C) and each T-cell subpopulation (D). The filled and open bars indicate control and Bach2−/− cells, respectively. Data are expressed as mean ± SD, n = 7, *P < 0.05.
Up-Regulated Expression of Effector Memory-Related Genes in Bach2−/− Naive T Cells.
We then examined the effects of Bach2 deficiency on gene expression and functions of naive T cells. Splenic naive CD4 T cells were stimulated with anti-CD3/CD28 antibodies (Abs). Whereas weak stimulation (anti-CD3/28 = 1/0.1 μg/mL) resulted in moderate reduction of Bach2−/− cells, there was no difference in proliferation with strong stimulation (anti-CD3/28 = 1/1 μg/mL), indicating a limited effect on proliferation (Fig. S2C). Under these stimulatory conditions, we analyzed mRNA expression using a cDNA microarray. We selected genes that were differentially expressed between WT and Bach2−/− cells in at least two of the three stimulatory conditions (Dataset S1). Gene ontology enrichment analysis [FuncAssociate 2.0 (18)] showed that the genes up-regulated in Bach2−/− T cells were enriched in those related to biodefense and immune response, including the stress response (GO:0006950, P = 9.83 × 10−11) and innate responses (GO:0045087, P = 5.71 × 10−7) whereas the down-regulated genes did not show a remarkable enrichment for any particular function.
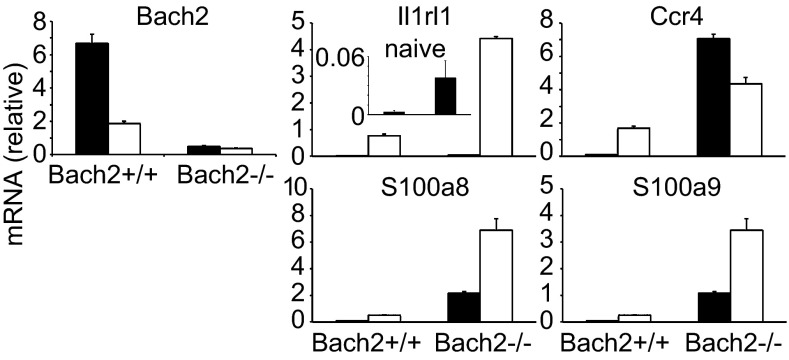
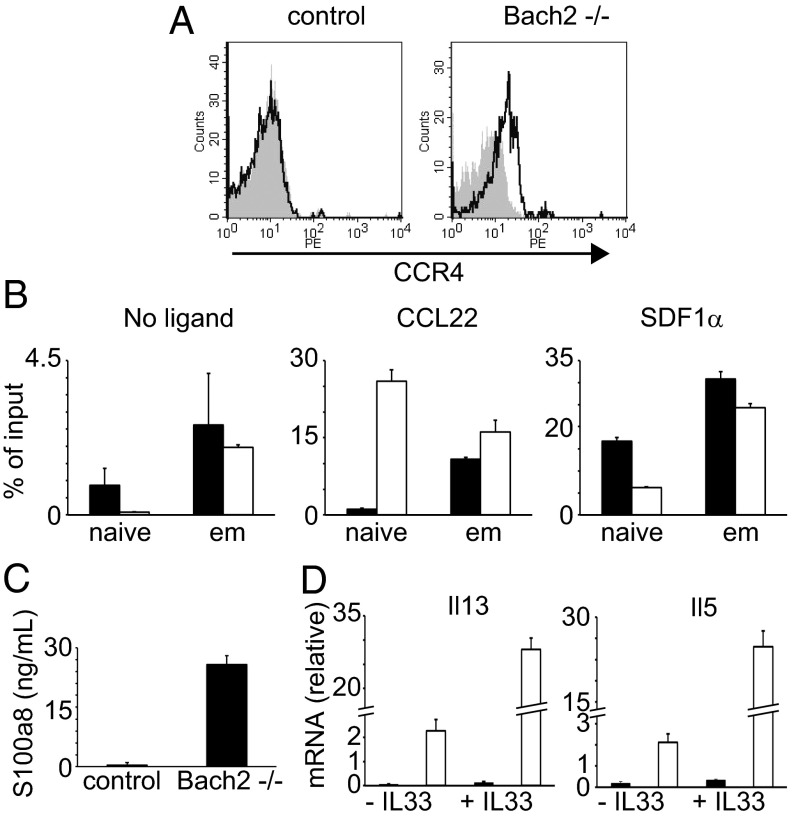
Interestingly, we found that these affected genes partially overlapped with those of IL2-inducible T-cell kinase (Itk)−/− T cells (Dataset S1) (19). Itk−/− T cells have been shown to possess memory- and innate cell-like properties. In fact, many of the overlapping genes are known to be related to innate immunity (Dataset S1). Because we observed that Bach2 expression was lower in effector-memory T cells than naive cells (Fig. 1A) and it has been shown that memory T cells and innate cells have common gene expression features (20), we speculated that Bach2 might suppress effector memory-related genes in naive T cells. To this end, we examined whether effector-memory T cells expressed the same genes that were up-regulated in Bach2−/− naive T cells. Indeed, those genes such as CCR4, S100 calcium binding protein a (S100a), and ST-2 were expressed in normal effector-memory T cells at higher levels than in naive cells (Fig. 3 and Fig. S3A). We confirmed up-regulation at the protein and/or cell function levels (Fig. 4): the cell surface CCR4 expression, the serum level of S100a, and the migration toward CCL22 were greatly enhanced in effector-memory T cells. These results suggest that Bach2 is required for the suppression of effector memory-related genes and functions in naive T cells.
Fig. 3.
Bach2 regulates effector-memory T cell-related genes. The expression of genes that are up-regulated in Bach2-deficient unstimulated and stimulated naive CD4 T cells in the microarray were examined by qPCR in naive and effector-memory CD4 T cells. The filled and open bars indicate naive and effector-memory CD4 T cells, respectively. Data are expressed as mean ± SD, n = 3. Il1rl1, Il1 receptor-like 1.
Fig. 4.
Functional characteristics of Bach2−/− T cells. (A) The expression of CCR4 on naive CD4 T cells. The solid and filled gray lines illustrate the staining with anti-CCR4 and a control mAb, respectively. (B) Migration assay in vitro. Total splenocytes were used for a transwell assay with CCL22 (CCR4 ligand), SDF1α (unrelated ligand), or no ligand. After incubation, cells were stained with anti-CD4, CD62L, and CD44 mAbs. The number of migrating cells was determined by flow cytometry and expressed as % of the input cell numbers. The filled and open bars indicate control and Bach2−/− cells, respectively. (C) Serum levels of S100a protein. The serum S100a level from WT and Bach2−/− mice was analyzed by ELISA. (D) ST-2 function in Bach2−/− naive CD4 T cells. WT or Bach2−/− naive CD4 T cells were stimulated with anti-CD3 and CD28 Abs with or without IL33 for 1 d. The mRNA expression of Il13 and Il5 cytokines was assessed by qPCR The filled and open bars indicate Bach2+/+ and Bach2−/− cells, respectively. Data are expressed as mean ± SD, n = 3.
The innate-like characteristics of Itk−/− T cells are also observed in T cells deficient in KLF2 and cAMP response element binding protein-binding protein (CBP). The mechanism to induce this phenotype was reported to involve TF PLZF (3). Accordingly, we analyzed the expression of Itk, KLF2, CBP, and PLZF genes in Bach2−/− T cells but found no significant change in their expression (Fig. S3B). Intracellular staining confirmed that PLZF expression was not altered in Bach2−/− T cells (Fig. S3C). Thus, the characteristics of Bach2−/− T cells are apparently independent of these factors.
Rapid Production of Th2 Cytokines by Bach2−/− Naive CD4 T Cells.
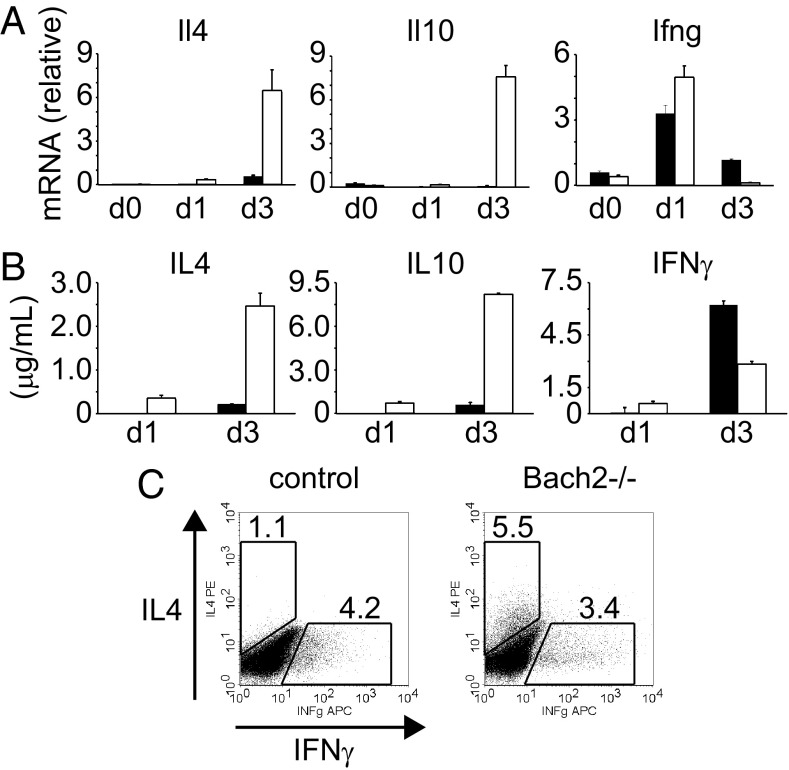
In addition to the enhanced expression of innate/effector memory-related genes, Bach2−/− T cells highly expressed Th2-related genes, such as IL-4, Blimp-1 (Prdm1) and E4BP4 (Nfil3) after TCR stimulation (Dataset S1) (21, 22). We confirmed these data by qPCR, ELISA, and intracellular staining. Bach2−/− T cells had increased expression of IL-4 and IL-10, but not IFNγ, as well as Blimp-1 and Gata3 (Fig. 5, Fig. S4A). Furthermore, the up-regulation of IL-4 was observed even in the presence of IL-12, suggesting that Bach2−/− naive T cells are predisposed to differentiate into Th2 cells (Fig. S4B). Notably, the up-regulation of IL-4 and IL-10 were already observed at day 1, demonstrating that the Bach2−/− naive T cells rapidly produce these effector cytokines (Fig. 5 and Fig. S4A). On the other hand, IFNγ was up-regulated at day 1, but down-regulated at day 3 (Fig. 5). Intracellular staining confirmed that Bach2−/− naive T cells rapidly produce effector cytokines and preferentially differentiate into Th2 cells.
Fig. 5.
Up-regulation of Th2-related genes in Bach2−/− CD4 T cells. (A) mRNA expression of each gene in naive CD4 T cells at day 0, 1, and 3 after simulation with anti-CD3/CD28 mAbs. (B) ELISA for IL4, IL10, and IFNγ. The supernatants of the culture in A were assessed at day 1 and 3. The filled and open bars indicate control and Bach2−/− cells, respectively, and data are expressed as mean ± SD, n = 3. (C) Intracellular staining of IL4 and IFNγ. Naive CD4 T cells were stimulated for 3 d and restimulated with PMA plus ionomycin in the presence of brefeldin A for 5 h, followed by permeabilization and intracellular cytokine and analyzed by flow cytometry.
Restoration of the Bach2−/− T-Cell Phenotype by Bach2 Reexpression.
To examine whether Bach2-mediated regulation to maintain the naive status of T cells is cell-intrinsic, a Bach2 cDNA was transfected into Bach2−/− T cells. We found that forced Bach2 expression induced the suppression of ST-2, Blimp-1, IL-10, and S100a gene expression, suggesting that Bach2 regulates these effector memory-related genes in a T cell-intrinsic manner (Fig. S5A). Unlike ex vivo T cells (Fig. 5), IL-4 expression was not affected by Bach2 deficincy, probably because in vitro long culture (5 d) with exogenous IL-2 might increase IL-4 expression in WT T cells. Similarly, overexpression of Bach2 did not affect the expression of IL-4 (Fig. S5A). These results suggested that the up-regulation of the indicated genes in Bach2−/− cells was not due to autocrine effect of IL-4.
Because effector-memory T cells express a lower level of Bach2 than naive T cells, we examined whether forced expression of Bach2 suppresses those genes in normal effector-memory T cells. Indeed, Bach2 suppressed the expression of S100a, ST-2, Blimp-1, and IL10 genes in normal effector-memory T cells (Fig. S5C), suggesting that the down-regulation of Bach2 results in the up-regulation of these genes in normal effector-memory T cells and that, therefore, Bach2 contributes to the maintenance of the naive status of T cells.
Effects of Bach2 Deficiency in Immune Responses in Vivo.
To evaluate the role of Bach2 in T cells in vivo, we analyzed the effects of Bach2 deficiency in two disease models. First, we used the colitis model induced by the transfer of naive CD4 T cells into Rag−/− mice (Fig. S6 A and B). By comparing the loss of body weight, we found that Bach2 deficiency impaired the ability of naive CD4 T cells to induce colitis (Fig. S6A). Consistently, histological analysis demonstrated that, whereas mice transferred with control CD4 T cells showed severe infiltration of lymphocytes in the colon, the infiltration was reduced in Bach2−/− cell-transferred mice (Fig. S6B). The results suggest that Bach2−/− T cells are defective in generating colitogenic effector T cells.
Second, we exploited the Listeria monocytogenes infection model (Fig. S6 C–F). In this model, we used newly developed conditional Bach2-deficient mice [Bach2-flox/flox (fl/fl) crossed with lck-cre, hereafter referred to as Bach2-cKO] because Bach2-null mice have obvious defects in B-cell responses. We confirmed that CD4 T cells of the mice showed similar phenotypes to Bach2-null mice: the reduction of naive T cells (Fig. 6C) and up-regulation of IL-4 and IL-10 mRNA after TCR stimulation (Fig. 6D). By using these mice, we assessed the effect of Bach2 deficiency on immune response to L. monocytogenes infection. The number of viable bacteria after infection was increased in the spleen of Bach2-cKO mice (Fig. S6E). Analysis of cytokine production from splenocytes upon stimulation with L. monocytogenes antigen (LLO 189–201) showed significant reduction of IFNγ but not IL-4 by CD4 T cells from Bach2-cKO mice (Fig. 6F). These results indicate that Bach2−/− CD4 T cells were defective in immune responses in vivo, despite rapid responses to TCR stimulation in vitro.
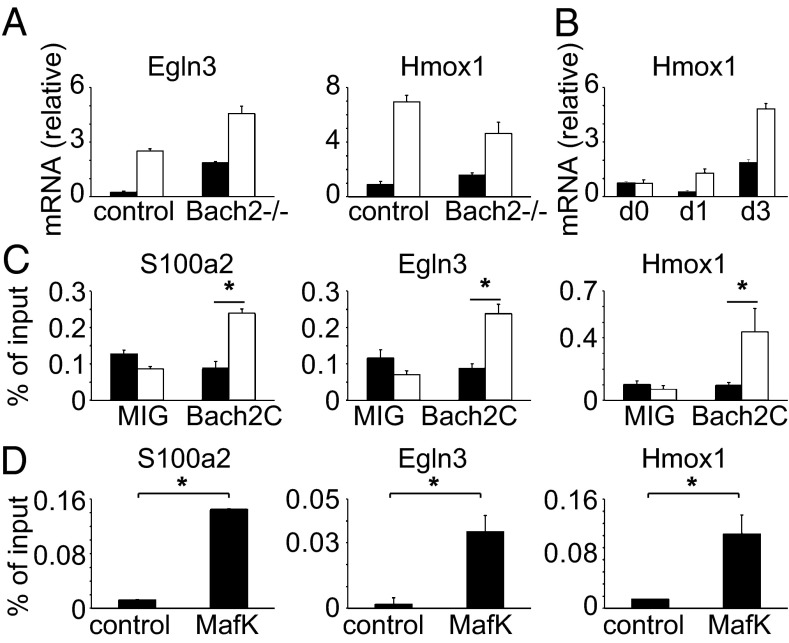
Fig. 6.
Identification of direct targets of Bach2. (A) PHD3 (Egln3) and Ho-1 (Hmox1) mRNA expression in WT and Bach2−/− CD4 T cells. The filled and open bars indicate naive and effector-memory CD4 T cells, respectively. (B) Ho-1 mRNA expression in WT and Bach2−/− naive CD4 T cells with or without stimulation with anti-CD3/CD28 Abs. The filled and open bars indicate control and Bach2−/− cells, respectively. (C) ChIP-qPCR for Bach2. 2B4 T hybridoma expressing FLAG-Bach2 or the control cells [pMXs-ires-EGFP (MIG)] were used for ChIP-qPCR assay with anti-FLAG Ab. The filled and open bars indicate control and anti-FLAG Abs, respectively. (D) Thy1+ splenocytes were used for ChIP-qPCR assay with anti-MafK Ab. Data are expressed as mean ± SD, n = 3, *P < 0.05 in A–D. MafK, musculoaponerotic fibrosarcoma oncogene family, protein K.
Direct Target Genes of Bach2 in T Cells.
To elucidate the mechanism of Bach2-mediated regulation of T-cell homeostasis, we attempted to identify direct targets of Bach2 in T cells. We conducted Chromatin Immunoprecipitation (ChIP)-seq analysis in the 2B4 T-cell hybridoma. Due to the lack of a good anti-Bach2 Ab for ChIP, we used the FLAG-tagged C-terminal half of Bach2 (Bach2C) containing the DNA binding domain. We specifically selected the peaks that included the ARE (the binding site of the CNC family and small maf proteins heterodimers), which is conserved between human and mouse and has no overlap with repetitive sequences. This process identified six peaks (Dataset S2) with the genes located adjacent to the elements. We further selected the genes whose expression was altered in Bach2−/− T cells, and three genes—egl nine homolog 3 [Egln3 encoding prolyl hydroxylase 3 (PHD3)], heme oxygenase 1 [Hmox1 encoding heme oxygenase 1 (HO-1)], and S100a—were finally identified as candidate Bach2 targets (Fig. 6 A and B and Fig. S7). Bach2 binding to these elements was confirmed by ChIP-qPCR (Fig. 6C). To examine whether these genes were endogenous targets in normal T cells, we analyzed binding of endogenous MafK (small Maf) as the partner of Bach2 by ChIP-qPCR using anti-MafK Ab (Fig. 6D). The result confirmed that endogenous MafK specifically bound to these three elements, suggesting that they are physiological targets of Bach2 in normal T cells. Although these genes are known to be involved in stress responses and innate immunity (23–25), it has not been known whether they are related to effector-memory T cells. Analysis of the expression of these genes in naive and effector-memory T cells indicates that they were all up-regulated in effector-memory T cells (Figs. 3 and 6A) and in naive Bach2−/− T cells, suggesting that Bach2 directly regulates effector memory-related genes in T cells.
Discussion
Although Bach2 has been reported to have specific functions in B cells (5, 6), we show here that Bach2 is expressed in T cells, is up-regulated during differentiation, and functions to regulate homeostasis of peripheral T cells by suppressing expression of effector memory-related genes in naive T cells. Bach2 deficiency reduces naive T-cell numbers and enhances the expression of effector memory-related genes, particularly Th2-related genes and cytokines.
Although the expression of Bach2 mRNA in T and B cells was comparable, the protein expression in T cells was lower than that in B cells (Fig.1). It has not been clear what causes the difference. The difference may not be due to protein degradation because inhibition of degradation did not alter the difference. The difference in T and B cells might be caused by translational efficiency or differential sumoylation as Bach2 was reported to be sumoylated (26).
Bach2−/− mice had a reduction of naive T cells in secondary lymphoid tissues. A similar phenotype with decreased naive T cells was also reported in mice deficient in KLF2 and Foxo1 (2, 27). KLF2 positively regulates S1pr1 expression essential for thymocyte egress (2) and negatively regulates chemokine receptors for homing to nonlymphoid tissues (27). Consequently, KLF2−/− naive T cells were decreased in the periphery and accumulated in thymus and nonlymphoid tissues. On the other hand, Foxo1 positively regulates the expression of CD62L, CCR7, and IL7R genes required for homing/survival of T cells (1). The reduction of naive T cells in Foxo1−/− mice may be due to reduced homing to lymphoid tissues and impaired survival. Bach2 deficiency causes neither the accumulation in thymus nor the severe reduction of CD62L and CCR7 expression. In addition, the survival of naive Bach2−/− T cells was not altered regardless of the presence of IL-7 (Fig. S2D). Furthermore, the percentage and the suppressive activity of regulatory T cells were not affected by Bach2 deficiency (Fig. S2 E and F). By contrast, Bach2−/− T cells exhibited enhanced expression of CCR4 and CCR9, which may direct T cells toward nonlymphoid tissues, thus reducing their numbers in lymphoid tissues.
The gene ontology analysis of the microarray data revealed that Bach2 deficiency up-regulated genes related to stress and innate responses in T cells. Interestingly, some of these genes overlapped with those dysregulated in T cells of Itk−/− mice (Dataset S1) (19). Itk−/− T cells exhibit memory and innate lymphocyte-like phenotypes and rapid production of cytokines (28, 29). Notably, effector-memory lymphocytes and innate cells share common features of gene expression (20). Because Bach2 expression was lower in effector-memory T cells than in naive cells (Fig.1A) and the genes up-regulated in Bach2−/− naive cells were also highly expressed in normal effector-memory T cells (Fig.3 and Fig. S3A), we conclude that Bach2 functions to suppress effector memory-related genes in naive T cells.
Similar to Itk−/− mice, KLF2 and CBP−/− mice have a population of memory/innate-like lymphocytes (30) whose development is dependent on IL-4 secreted from PLZF+ T cells (3). By contrast, our results indicate that the effector memory-like phenotype of Bach2−/− T cells appears to be induced by different mechanism(s). The increased memory/innate CD8+ cells in mice deficient in Itk, KLF2, and CBP were found in the thymus, but this phenotype was not the case in Bach2−/− mice, and the expression of PLZF, as well as Itk, CBP, and KLF2, was not changed in Bach2−/− T cells. Instead, the effector-memory phenotype of Bach2−/− T cells is intrinsically induced, based on the observation that the overexpression of Bach2 suppressed effector-memory genes in Bach2−/− cells and that the direct targets of Bach2 were the genes up-regulated in effector-memory T cells.
In this study, we identified the putative direct (S100a, PHD3, and HO-1) and T cell-intrinsic (ST-2, Blimp-1, and IL10) targets of Bach2 that are highly expressed in normal effector-memory T cells and enhanced in Bach2−/− naive T cells. These findings indicate that Bach2 directly and/or T-cell intrinsically suppresses effector memory-related genes in naive T cells. Interestingly, these are genes induced by cellular stresses; oxidative stress induces S100a and HO-1 expression (24, 31), and LPS and cellular stress induce Blimp-1 (32, 33), HO-1, and ST-2. Such stresses can also activate Nrf2 (7), which may also regulate these genes, although Nrf2 is a transcriptional activator whereas Bach2 is a repressor. In fact, HO-1 is an Nrf2 target and is regulated through the same ARE as Bach2 (Fig. S7) (34). Nrf2 also regulates Bach2-direct target gene, PHD3, and S100 (31, 34). Therefore, Nrf2 and Bach2 may regulate an overlapping set of genes with opposite outcomes such that the deficiency of Nrf2 and Bach2 results in the reduction (35) and augmentation of IL-10 (Fig.5), respectively.
Bach2−/−T cells cannot retain their naive status after expressing effector-memory genes and, consequently, exhibit preferential differentiation into Th2 effector cells. Recently, it was reported that Nrf2 also regulates the Th1/Th2 balance (36) by enhancing IFNγ and suppressing IL-4, IL-5, and IL-13, and that Nrf2−/− T cells had reduced IFNγ and increased Th2 cytokines (36). These results suggest that Bach2 and Nrf2 have opposite functions in Th1/Th2 differentiation by probably acting on the same cis-regulatory elements.
There are some similarities in the roles of Bach2 in T and B cells; in the Bach2−/− mice, both T and B cells showed a decrease in naive cells (Fig. 2) and rapid differentiation into effector cells upon stimulation (Th2 and plasma cells) (Fig. 5) (5, 6), suggesting a general role of Bach2 in maintaining homoeostasis of naive T and B cells. In addition, Bach2 has common target genes in T and B cells. Blimp-1 is a cell-intrinsic target of Bach2 and is up-regulated after T- (Figs. S4 and S5) and B-cell stimulation (5, 6). Blimp-1 negatively regulates proliferation and follicular helper T cells (Tfh) differentiation and augments the expression of effector molecules such as IL-10 and granzymes in T cells (37–39) whereas Blimp-1 suppresses proliferation and class switch recombination and enhances plasma cell differentiation in B cells (40). These findings indicate that Blimp-1 is essential for differentiation of both T and B effector cells and that Bach2 is critical in suppressing Blimp-1 function in naive cells. HO-1 is another common downstream Bach2 gene in T and B cells (Fig.6) (41). Its role in T- and B-cell function is not well known, but we reported that heme, a substrate of HO-1, induces Bach2 degradation and plasma-cell differentiation, suggesting that HO-1 may have a role in this process (41). Collectively, these results suggest that Bach2 regulates a common gene network to maintain the “naive-ness” of T and B cells.
Despite the effector memory-like phenotype, Bach2−/− T cells were found to be defective in immune responses in vivo (Fig. S6), suggesting that Bach2−/− T cells are not fully maturated as effector T cells. Similar phenotypes were reported on another type of innate-like CD4 T cell, which is induced by MHC class II-expressing thymocytes (42, 43). This type of innate-like CD4 T cells (T-CD4 T cells) showed rapid production of cytokines (especially Th2 cytokine) and exhibited reduced/suppressive activities of immune responses in vivo, resembling Bach2−/− CD4 T cells (42, 43). There might be a similar mechanism that determines the characteristics of Bach2-deficient and T-CD4 T cells.
In humans, genome-wide SNP analyses suggest that Bach2 is associated with type I diabetes, celiac disease, Crohn’s disease, and multiple sclerosis (44–47). Notably, we found that S100a protein was up-regulated in the serum of Bach2−/− mice (Fig. 4C), and it has been reported that S100a functions as a danger signal and that its expression level in serum and tissues is correlated with the severity of inflammatory diseases (48). These results suggest that Bach2 may regulate serum S100a levels and consequently play a role in the pathogenesis of these diseases.
Materials and Methods
Mice.
Mice were maintained under specific pathogen-free conditions. C57BL/6 mice were purchased from CLEA Japan. All experiments were carried out in accordance with the institutional guidelines of the animal facility of RIKEN Yokohama Institute. Bach2−/− mice have been previously described (6). Flox-Bach2 mice were generated by flanking ATG-containing exon 4 by two loxP sites (49). Neomycin resistant gene franked by FRT sequences was inserted into targeting vector for selecting ES cells. Homologous recombination with targeting vector was induced in Bruce4 ES cells, and the targeted ES cells were injected into blastocysts of BALB/c mice. Chimeric mice were crossed with C57BL/6 J mice to obtain germ line-transmitted mice. After removing neomycin resistant cassette by crossing with actin-FLPe mice, T cell-specific Bach2-deficient mice were obtained by crossing with Lck-Cre transgenic (Tg) mice.
Quantitative PCR.
Total RNA from sorted cells was extracted with an RNeasy Mini kit (Qiagen) and treated with DNase (Nippongene). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). qPCR was performed with the Fast Syber Green Master Mix (Applied Biosystems). Data were collected using the StepOnePlus real-time PCR system (Applied Biosystems).
Immunoblot Analysis.
Whole-cell extracts were resolved on 4–12% SDS/PAGE gels (ATTO) and transferred to a PVF membrane (Millipore). Abs to Bach2 (Cell Signaling) and Erk1/2 (Cell Signaling) were used as detection reagents.
Flow Cytometry.
Single-cell suspensions were prepared from spleen, lymph nodes, or thymus. For intracellular cytokine staining, cells were fixed, permeabilized with Cytofix/Cytoperm (BD Biosciences), stained with anti-cytokine Abs, and analyzed on FACSCalibur (BD Biosciences). Data were processed with CellQuest Pro software (BD Biosciences). Cells were sorted with a FACSAria (BD Biosciences). The following fluorochrome-labeled Abs purchased from BD Biosciences, BioLegend, or eBioscience were used: Abs against CD4 (GK1.5), CD8 (Ly2), CD62L (MEL-14), CD44 (IM7), HSA (30-F1), CD5 (53-7.3), CD69 (H1.2F3) and TCRβ (H57-597), B220 (RA3-6B2), CCR4 (2G12), IL4 (11B11), and IFNγ (XMG1.2).
Gene Expression Profiling.
Splenic CD44loCD62Lhi naive CD4+ T cells from WT and Bach2−/− mice at 6–8 wk old were sorted by FACS, and total RNA was purified. The RNA was labeled and hybridized to a Mouse Genome 430 2.0 array (Affymetrix). Expression values for each probe set were calculated using the GC-RMA method in the GeneSpring GX 7.3 software package (Agilent).
Chemotaxis Assays.
Migration of splenic T cells toward CCL22 and SDF1α was assessed with Transwell tissue culture inserts with a 3-μm pore size polycarbonate filter (Corning). The cells were suspended at 1 × 107 cells per mL in RPMI medium 1640 supplemented with 0.5% fatty acid-free BSA, and 50-μL aliquots were loaded into the upper inserts. Medium with or without CCL22 or SDF1α (R&D Systems) was placed in the lower wells. Chambers were incubated for 4 h, and cell migration was quantified by counting the cells by flow cytometry.
Transfection of a Bach2 cDNA into Bach2−/− T Cells.
Mouse Bach2 cDNA was cloned into the retroviral vector pMXs-IG (provided by T. Kitamura, University of Tokyo). Cells were infected with the retrovirus prepared in the Plat-E packaging cells by centrifugation at 1,200 × g in the presence of 10 μg/mL polybrene at day 1 and 2. The cells were cultured for an additional 3 d and analyzed by qPCR.
ChIP Assay.
The C-terminal half of Bach2C (355-839 aa) from the full-length mouse Bach2 cDNA was subcloned into the pMXs-ires-EGFP retrovirus vector and tagged with 3× FLAG and streptavidin-binding peptide (Sigma). The 2B4 T-cell hybridoma was transfected by retrovirus transduction. ChIP was performed as previously described (50): the chromatin was precipitated with 5 μg of FLAG Ab (M2, Sigma) or control mouse IgG overnight. For deep sequencing, DNA samples were submitted to Takara Bio for sequencing with the Illumina GAIIx. Libraries were prepared according to Illumina's instructions accompanying the ChIP-seq sample preparation kit. Amplified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced on the Genome Analyzer following the manufacturer's protocols.
Statistical Analysis.
Standard two-tailed t tests assuming normal variance were used for all statistical calculations. All error bars and variances represent SEM, and asterisks on all graphs represent P < 0.05.
Supplementary Material
Acknowledgments
We thank H. Yamaguchi and S. Kato for secretarial assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306691110/-/DCSupplemental.
References
- 1.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10(2):176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 3.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11(8):709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33(6):890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muto A, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29(23):4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 7.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3(112):re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurokawa H, et al. Structural basis of alternative DNA recognition by Maf transcription factors. Mol Cell Biol. 2009;29(23):6232–6244. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8(1-2):107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 11.Oyake T, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16(11):6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki S, et al. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene. 2000;19(33):3739–3749. doi: 10.1038/sj.onc.1203716. [DOI] [PubMed] [Google Scholar]
- 13.Masopust D, Picker LJ. Hidden memories: Frontline memory T cells and early pathogen interception. J Immunol. 2012;188(12):5811–5817. doi: 10.4049/jimmunol.1102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nat Immunol. 2011;12(6):467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238(1):247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muto A, et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 1998;17(19):5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25(22):3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blomberg KEM, et al. Transcriptional signatures of Itk-deficient CD3+, CD4+ and CD8+ T-cells. BMC Genomics. 2009;10:233. doi: 10.1186/1471-2164-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 21.Cimmino L, et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol. 2008;181(4):2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 22.Kashiwada M, Cassel SL, Colgan JD, Rothman PB. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO J. 2011;30(10):2071–2082. doi: 10.1038/emboj.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 24.Gozzelino R, Jeney V, Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 25.Goyette J, Geczy CL. Inflammation-associated S100 proteins: New mechanisms that regulate function. Amino Acids. 2011;41(4):821–842. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- 26.Tashiro S, et al. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol Cell Biol. 2004;24(8):3473–3484. doi: 10.1128/MCB.24.8.3473-3484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 28.Atherly LO, et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25(1):79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25(1):93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32(2):50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leśniak W, Szczepańska A, Kuźnicki J. Calcyclin (S100A6) expression is stimulated by agents evoking oxidative stress via the antioxidant response element. Biochim Biophys Acta. 2005;1744(1):29–37. doi: 10.1016/j.bbamcr.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Doody GM, Stephenson S, Tooze RM. BLIMP-1 is a target of cellular stress and downstream of the unfolded protein response. Eur J Immunol. 2006;36(6):1572–1582. doi: 10.1002/eji.200535646. [DOI] [PubMed] [Google Scholar]
- 33.Kwon H, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31(6):941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malhotra D, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116(4):984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rockwell CE, Zhang M, Fields PE, Klaassen CD. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J Immunol. 2012;188(4):1630–1637. doi: 10.4049/jimmunol.1101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11(2):114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31(2):283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 39.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7(5):457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 40.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe-Matsui M, et al. Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood. 2011;117(20):5438–5448. doi: 10.1182/blood-2010-07-296483. [DOI] [PubMed] [Google Scholar]
- 42.Li W, et al. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007;204(9):2145–2157. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao Y, et al. Innate-like CD4 T cells selected by thymocytes suppress adaptive immune responses against bacterial infections. Open J Immunol. 2012;2(1):25–39. doi: 10.4236/oji.2012.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawcer S, et al. International Multiple Sclerosis Genetics Consortium Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86(3):557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 49.Kometani K, et al. Repression of Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity. 2013 doi: 10.1016/j.immuni.2013.06.011. in press. [DOI] [PubMed] [Google Scholar]
- 50.Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci USA. 2001;98(18):10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.