Abstract
The Bloom syndrome gene product, BLM, is a member of the highly conserved RecQ family. An emerging concept is the BLM helicase collaborates with the homologous recombination (HR) machinery to help avoid undesirable HR events and to achieve a high degree of fidelity during the HR reaction. However, exactly how such coordination occurs in vivo is poorly understood. Here, we identified a protein termed SPIDR (scaffolding protein involved in DNA repair) as the link between BLM and the HR machinery. SPIDR independently interacts with BLM and RAD51 and promotes the formation of a BLM/RAD51-containing complex of biological importance. Consistent with its role as a scaffolding protein for the assembly of BLM and RAD51 foci, cells depleted of SPIDR show increased rate of sister chromatid exchange and defects in HR. Moreover, SPIDR depletion leads to genome instability and causes hypersensitivity to DNA damaging agents. We propose that, through providing a scaffold for the cooperation of BLM and RAD51 in a multifunctional DNA-processing complex, SPIDR not only regulates the efficiency of HR, but also dictates the specific HR pathway.
Keywords: DNA double-strand breaks, noncrossover, double Holliday junction
Homologous recombination (HR) repair allows precise repair of DNA double-strand breaks and is critical for restarting stalled or collapsed DNA replication forks (1–4). The process of HR in eukaryotic cells requires several proteins, the central player of which is the RAD51 recombinase (1–4). HR repair is initiated by nuclease-mediated DNA end resection to generate 3′-single-stranded DNA (ssDNA) tails that are initially coated by the replication protein A (RPA) complex (1–4). In a subsequent step, recombination mediator proteins catalyze the replacement of RPA with RAD51, resulting in the formation of ssDNA-RAD51 filament (1–4). The ssDNA-RAD51 filament then catalyzes strand invasion into homologous duplex DNA, leading to the formation of a displacement loop (D-loop) (1–4). After DNA synthesis primed by the invading strand, the repair can bifurcate into two alternative subpathways referred to as synthesis-dependent strand annealing (SDSA) and double-strand break (DSB) repair (5–9). In SDSA, the extended D-loop can be dissolved by DNA helicases and the newly synthesized strand is annealed to the ssDNA tail on the other break end, which is followed by gap-filling DNA synthesis and ligation (5–9). The repair products from SDSA are always noncrossover (5–9). In DSB repair, the second DSB end is captured to form an intermediate with two Holliday junctions, called double Holliday junction (dHJ) (5–9). dHJ can be processed to yield crossover or noncrossover recombination products (5–9). There is mounting evidence that DNA repair mechanisms in mitotically proliferating cells avoid generating crossover recombination products, which can contribute significantly to genome instability (5–9).
In human cells, the Bloom syndrome gene product (BLM) helicase, inactivated in individuals with Bloom syndrome, associates with TopoIIIα and RMI1/2 (BLAP75/18) to form the BTR (BLM-TopoIIIα-RMI1/2) complex (10–15). This complex promotes the dissolution of dHJ to yield exclusively noncrossover recombination products (16–18). BLM can also drive HR toward the formation of noncrossover products through the utilization of the SDSA pathway (6, 7). The ability of BLM to yield noncrossover products is thought to play a critical role in the avoidance of chromosomal rearrangements during the homolog-directed repair of chromosomal lesions. As a result, cells defective for BLM exhibit elevated rates of sister chromatid exchange (SCE) (19–21).
Upon the occurrence of DNA damage, BLM is able to form discrete foci, where it colocalizes with other DNA repair proteins (22, 23). However, mechanistically how BLM is recruited to sites of DNA damage and how it collaborates with other proteins to mediate recombination repair remain largely unexplored. In this study, we used an affinity purification approach to isolate BLM-containing complex and identified a unique scaffolding protein KIAA0146, which we refer to as SPIDR (scaffolding protein involved in DNA repair). We demonstrate that SPIDR directly interacts with BLM and this interaction is required to target BLM to sites of DNA damage. Consistent with a role in BLM-mediated processes, cells depleted of SPIDR display an increased level of sister chromatid exchange. Moreover, we found that SPIDR promotes HR through a direct interaction with the recombinase RAD51. Finally, we show SPIDR promotes the formation of a BLM/RAD51-containing complex of biological importance. We therefore propose that, through licensing key cellular biochemical properties of BLM and RAD51, SPIDR not only regulates the efficiency of HR but also dictates the specific HR pathway.
Results
Identification of SPIDR as a Unique BLM-Binding Partner.
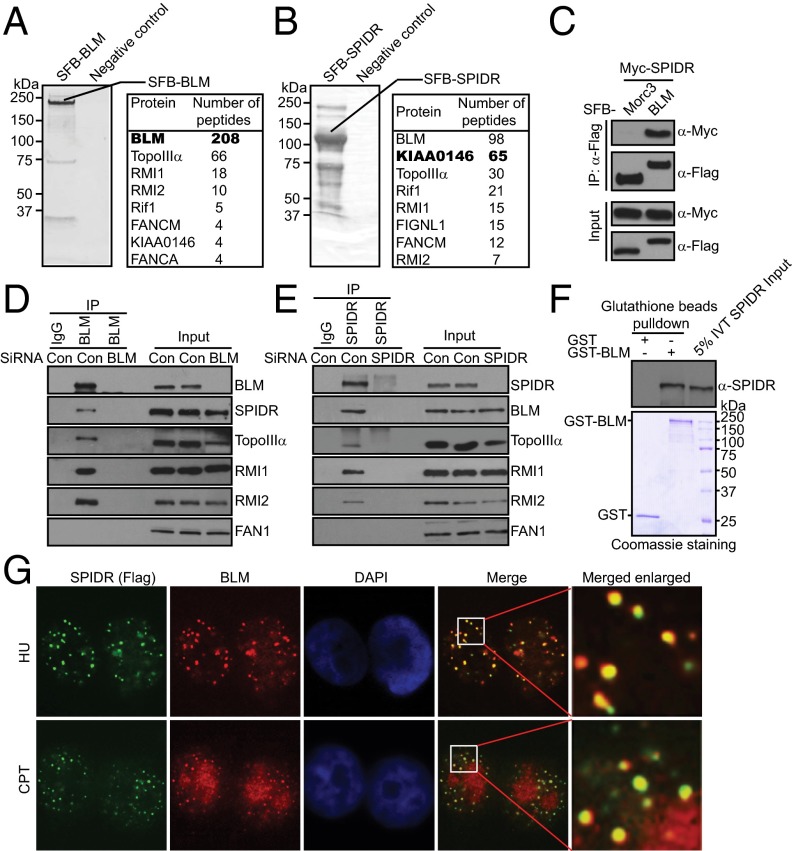
To search for previously undetected proteins present in BLM-containing complex, we performed tandem affinity purification (TAP) using 293T cells stably expressing streptavidin-flag-S protein (SFB)-tagged wild-type BLM for the identification of BLM-interacting proteins. Mass spectrometry analysis revealed several known BLM-associated proteins, including TopoIIIα, RMI1/2, Rif1, and FANCM (Fig. 1A). Interestingly, we also repeatedly identified a previously uncharacterized BLM-binding protein as KIAA0146 (Fig. 1A). To ensure that KIAA0146 indeed associates with BLM, we performed reverse TAP using a cell line stably expressing tagged KIAA0146, and identified BLM, TopoIIIα, Rif1, RMI1/2, and FANCM as major KIAA0146-associated proteins (Fig. 1B). These data strongly suggest that KIAA0146 is a bone fide BLM-binding protein. The KIAA0146 gene encodes a deduced polypeptide of 915 amino acids with a predicted molecular mass of 105 kDa. Database searches did not identify similarity to known proteins in any species or to any known functional motifs. On the basis of its functions described below, we designated this protein as SPIDR.
Fig. 1.
Identification of SPIDR as a BLM-binding partner. (A and B) 293T cells stably expressing SFB-tagged BLM or SPIDR were used for TAP of protein complexes. Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. (C) 293T cells were transiently transfected with plasmids encoding SFB-tagged BLM or Morc3 together with plasmids encoding Myc-tagged SPIDR. Coprecipitation and immunoblotting were carried out as indicated. (D and E) SiRNA-treated HeLa cells were lysed in the presence of benzonase, cell lysates were then incubated with protein A agarose beads conjugated with indicated antibodies and Western blot analysis was carried out as indicated. (F) Direct binding between recombinant GST-BLM purified from baculovirus-infected insect cells and in vitro translated SPIDR (IVT SPIDR). (Upper) SPIDR was detected by immunoblotting. (Lower) Input GST-proteins visualized by Coomassie staining. (G) SPIDR colocalizes with BLM. SFB-tagged SPIDR was expressed in HeLa cells. (Magnification: 100×.) Foci assembled by this fusion protein and by BLM following exposure to HU (2 mM) for 16 h or CPT (1 μM) for 3 h were detected by immunofluorescence using anti-Flag and anti-BLM antibodies, respectively. A merged image shows colocalization.
SPIDR Interacts with BLM in Vivo and in Vitro.
To validate our TAP results, we first performed coimmunoprecipitation experiments and found that Myc-tagged SPIDR interacts strongly with SFB-tagged BLM but not with the Morc3 control protein (Fig. 1C).
To examine the interaction between endogenous BLM and SPIDR, cell extracts from HeLa cells were immunoprecipitated with the anti-BLM antibody or with the control IgG. As expected, SPIDR was detected in the immunoprecipitations obtained with the anti-BLM antiserum but not with the control IgG (Fig. 1D). To prove the specificity of the BLM antibody, we performed coimmunoprecipitation in BLM-depleted HeLa cells treated with BLM-specific siRNA. Indeed, we failed to detect any SPIDR from the anti-BLM immunoprecipitations with these BLM-depleted cells (Fig. 1D). We also performed a reciprocal coimmunoprecipitation assay. As shown in Fig. 1E, the endogenous BLM complex was readily immunoprecipitated with the SPIDR-specific antibody, but not with the control IgG. In these experiments, benzonase was included in the lysis buffer to exclude the possibility that the interaction occurs indirectly via DNA/RNA bridging (Fig. 1 D and E).
To test whether there is a direct protein–protein interaction between SPIDR and BLM, we performed GST pull-down assays with recombinant GST-BLM fusion proteins purified from baculovirus-infected insect cells and in vitro-translated full-length SPIDR. As shown in Fig. 1F, BLM directly interacts with SPIDR in vitro.
Upon the occurrence of DNA damage, BLM could form large nuclear foci (22, 23). A physical interaction between SPIDR and BLM, as demonstrated above, raises the possibility that SPIDR may colocalize with BLM at sites of DNA damage. Indeed, discrete foci of SPIDR were readily detected in cells following hydroxyurea (HU) or camptothecin (CPT) treatment (Fig. 1G). Moreover, these foci colocalize with BLM foci, indicating that the localization of SPIDR, like that of BLM, is regulated in response to DNA damage (Fig. 1G).
To further define the binding between SPIDR and BLM, we sought to identify the regions within SPIDR responsible for its interaction with BLM. Coimmunoprecipitation experiments revealed that SPIDR associated with BLM through its entire C terminus (residues 451–915), because deletion mutants lacking any part of this region failed to coprecipitate with BLM (Fig. S1 A and B). Furthermore, pull-down experiments with recombinant proteins expressed in insect cells consolidated the above notion (Fig. S1C).
Conversely, using a series of overlapping BLM truncations and deletion mutants spanning its entire coding sequence, we mapped the SPIDR-binding region to residues 301–600 of BLM (Fig. S1 D–F).
SPIDR Is Required for BLM Foci Formation and Suppresses SCE.
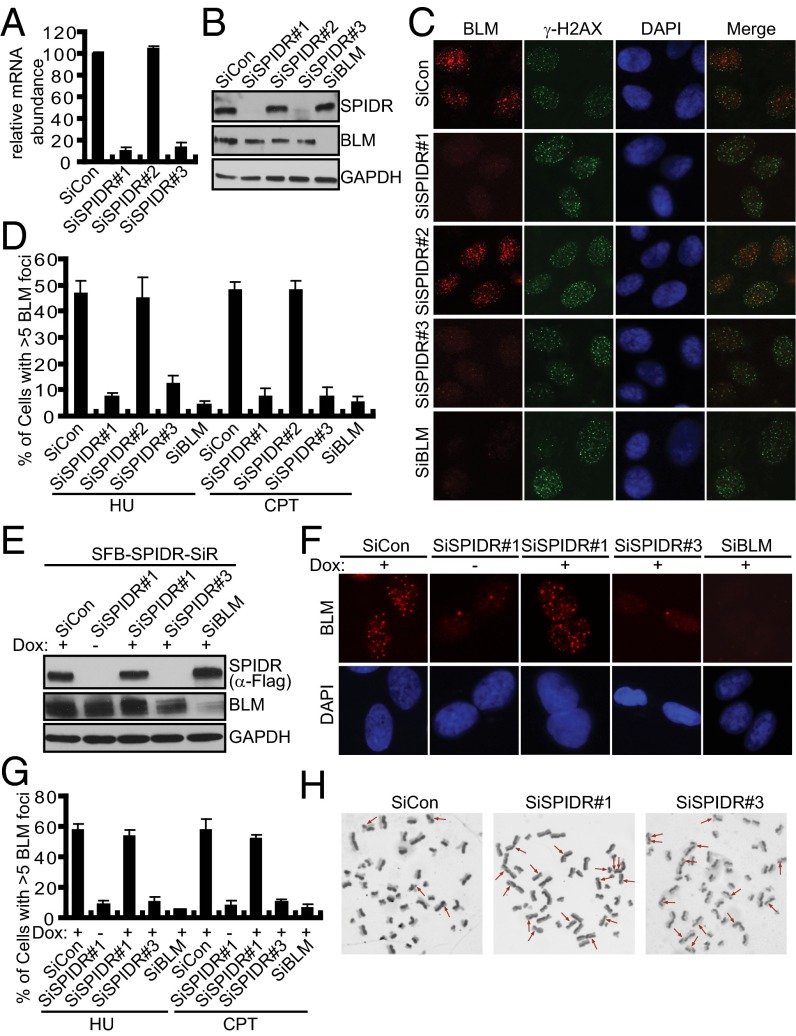
Because SPIDR exists in a complex with BLM, we sought to determine whether SPIDR has a role in the assembly of BLM foci. Thus, three different SPIDR-specific siRNA oligonucleotides were transfected into U2OS cells. Whereas siRNA#1 and siRNA#3 significantly reduced SPIDR expression, siRNA#2 resulted in being ineffective (Fig. 2 A and B). Moreover, the protein level of SPIDR remained the same in cells with BLM depletion and vice versa, suggesting that although SPIDR and BLM interact, they do not influence each other’s stability (Fig. 2B). Remarkably, HU- or CPT-induced BLM focus formation was severely impaired in SPIDR-depleted U2OS cells (Fig. 2 C and D). Similar results were obtained using HeLa cells (Fig. S2 A and B). In contrast, SPIDR depletion had no effect on γ-H2AX focus formation, indicating that SPIDR depletion does not generally interfere with DNA damage detection and signaling (Fig. 2C).
Fig. 2.
SPIDR is required for BLM foci formation and suppresses SCE. (A) Quantitative RT-PCR showing SPIDR mRNA levels are down-regulated by siRNAs. Bars represent the average of three experiments, and error bars are SDs. (B) Representative immunoblotting of cells transfected with indicated siRNAs. (C and D) SPIDR is required for BLM foci formation. U2OS cells were transfected twice with control siRNA, or siRNAs specific for SPIDR or BLM. Forty-eight hours after transfection, cells were treated with HU (2 mM) for 16 h or CPT (1 μM) for 3 h before fixing and processed for BLM and γH2AX immunofluorescence. Representative BLM foci (HU treatment) are shown (C). (Magnification: 100×.) Quantification results were the average of three independent experiments and were presented as mean ± SEM (D). (E–G) Rescue of BLM foci formation by expression of a siRNA#1-resistant SPIDR cDNA. A U2OS cell line to express siRNA#1-resistant SPIDR (SFB-SPIDR-SiR) under the control of a tetracycline-inducible promoter was generated. The resulting cell line was transfected with indicated siRNAs and was either left uninduced (−) or was induced (+) by doxycycline (Dox) addition for 24 h before HU (2 mM) or CPT (1 μM) treatment. After 16 h of treatment with HU or 3 h of treatment with CPT, cells were fixed and processed for BLM immunofluorescence. The exogenous SPIDR expression was confirmed by immunoblotting using anti-Flag antibody (E). Representative BLM foci (HU treatment) were shown (F). (Magnification: 100×.) Quantification results were the average of three independent experiments and were presented as mean ± SEM (G). (H) SPIDR suppresses SCE. Representative metaphase spreads showing SCEs from HeLa cells transfected with control or SPIDR siRNAs (red arrows). (Magnification: 100×.)
To further ensure the specificity of the SPIDR-siRNA phenotype on BLM focus formation, we performed recovery experiments. We engineered a U2OS cell line to express siRNA#1-resistant form of SPIDR under the control of a tetracycline-inducible promoter and examined BLM focus formation as a readout of SPIDR function. Induced expression of siRNA#1-resistant SPIDR was no more decreased by siRNA#1, whereas it was reduced as expected upon transfection of SPIDR-siRNA#3 (Fig. 2E). Notably, the induced siRNA#1-resistant SPIDR was able to restore BLM focus formation conferred by siRNA#1, but not siRNA#3 (Fig. 2 F and G), showing that the effect on recovery is truly dependent on SPIDR.
The hallmark feature of BLM-deficient cells is their elevated frequency of SCEs. The observation that SPIDR is required for efficient recruitment of BLM to sites of DNA damage led us to propose that SPIDR may also be involved in SCE suppression. As expected, the SCE level in SPIDR-depleted HeLa cells was significantly elevated (Fig. 2H, and Fig. S2 C and D). The observed two- to threefold elevated frequency in SCE formation is equivalent to that seen following depletion of BLM or RMI1/BLAP75 in HeLa cells (11). Moreover, that depleting SPIDR and BLM together did not cause a further increase in SCE frequency compared with SPIDR or BLM depletion alone supports the idea that BLM and SPIDR may suppress SCE via a common pathway (Fig. S2 C and D). Flow cytometry analyses showed that cell-cycle distribution was not affected by SPIDR down-regulation (Fig. S2E), ruling out the possibility that the phenotypes observed in SPIDR-depleted cells may be a result of any change in cell-cycle distribution.
Direct Interaction Between SPIDR and BLM Is Required for BLM Foci Formation.
As shown above, SPIDR is involved in the recruitment of BLM to site of DNA damage. Therefore, it would be interesting to further test whether the binding to SPIDR might be required for BLM foci formation. Interestingly, whereas wild-type SPIDR successfully restored BLM focus formation to levels comparable to that of control cells, SPIDR deletion mutants defective in BLM binding failed to do so (Fig. S3 A–C). Consistently, although distinct nuclear foci of full-length BLM were readily detected in HU-treated cells, the Δ301–600 mutant, which does not bind to SPIDR, failed to form foci after HU treatment (Fig. S3D). Taken together, these results suggest that SPIDR and its interaction with BLM are required for the efficient accumulation of BLM at sites of DNA damage.
Strikingly, the Δ1–300 mutant of BLM also exhibited a defect in HU-induced foci formation (Fig. S3D). Early studies have shown the entire RMI (RecQ-mediated genome instability) complex is essential for BLM recruitment or retention at DNA damage sites primarily via a role in maintaining the protein stability of the BTR complex (11–13). Thus, it is possible that the RMI complex regulates the accumulation of BLM at sites of DNA damage through its interaction with the N-terminal region missing in the Δ1–300 mutant of BLM. We then mapped the RMI binding domain on BLM. Pull-down experiments revealed that BLM interacted with RMI1 via its N terminus, because deletion mutant lacking the 300 N-terminal amino acids (Δ1–300) failed to pull-down with RMI1 (Fig. S3E). Moreover, consistent with previous studies (24), we also found that BLM interacted with TopoIIIα via its N terminus (Fig. S3F). Taken together, these results suggest that both SPIDR and the RMI complex are required for optimal BLM foci formation.
Ability of BLM to Function in SCE Suppression Correlates with Its Association with SPIDR.
To further decipher the biological significance of the association between SPIDR and BLM, we used the SCE assay to investigate whether this interaction might be involved in suppressing SCE. Consistent with the above notion that the Δ301–600 mutant of BLM lost its focus forming ability, the Δ301–600 mutant failed to restore the elevated SCE phenotype in BLM-depleted HeLa cells (Fig. S3 G and H).
To rule out the possibility that the phenotypes observed in BLM-Δ301–600 reconstituted BLM-depleted cells may be caused by a failure in forming the BTR complex, we performed coimmunoprecipitation experiments. As shown in Fig. S3I, the interaction between TopoIIIα or RMI1 with Δ301–600 mutant is similar to that with wild-type BLM.
SPIDR Is Required for RAD51 Foci Formation and Promotes Homologous Recombination.
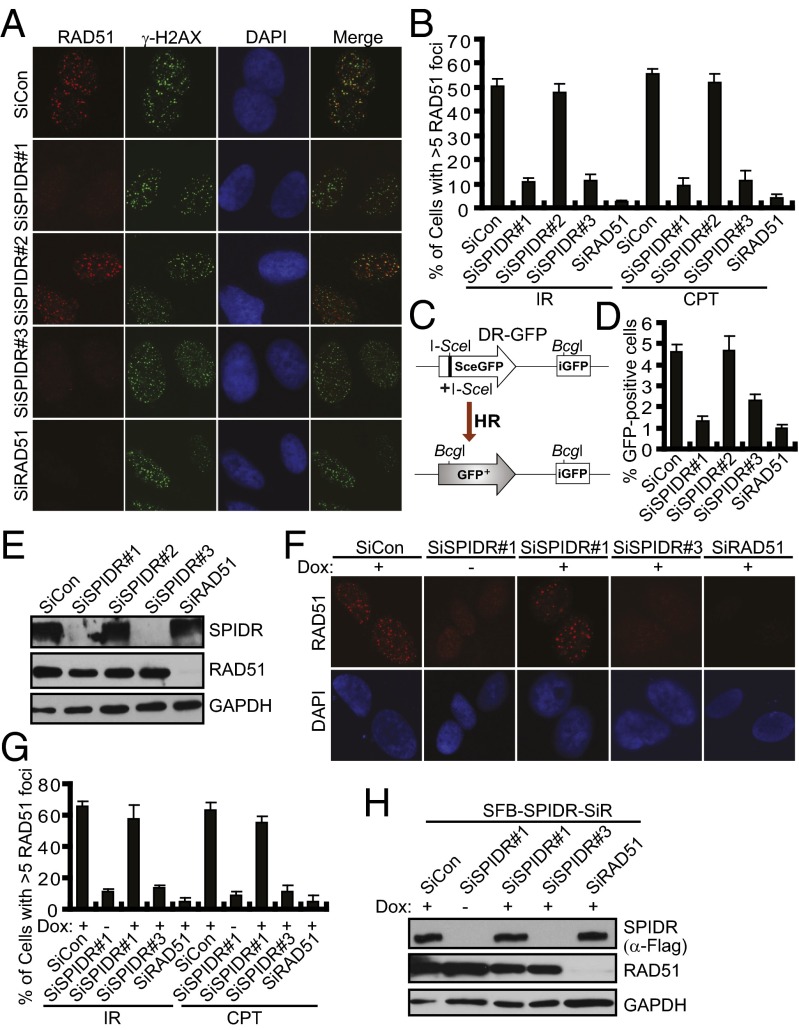
Given that multiple roles have been proposed for BLM in HR repair (25–27), it will be of interest to determine whether SPIDR plays a possible role in the early steps of HR. We then examined whether depletion of SPIDR in U2OS cells affected RAD51 focus formation in response to irradiation (IR) or CPT and found that in the absence of SPIDR, RAD51 focus formation was severely impaired (Fig. 3 A and B). Similar results were obtained using HeLa cells (Fig. S4 A and B). We did not observe an obvious decrease in end resection following SPIDR depletion, as measured by RPA2 focus formation, suggesting that SPIDR may promote RAD51 nucleofilament formation downstream of ssDNA generation (Fig. S4 C–E). To further confirm the role of SPIDR in HR, we performed a gene-conversion assay using the DR-GFP reporter system (28) (Fig. 3C). Consistent with a critical role in RAD51 recruitment, SPIDR depletion resulted in a significant reduction in HR (Fig. 3 D and E). Moreover, using the same method as described in Fig. 2F, we ruled out the possibility that the loss of RAD51 focus formation after SPIDR depletion was because of off-target effects of siRNAs (Fig. 3 F–H).
Fig. 3.
SPIDR promotes homologous recombination. (A and B) SPIDR is required for RAD51 foci formation. U2OS cells were treated with IR (10 Gy) or CPT (1 μM) for 3 h before fixing and processed for RAD51 and γH2AX immunofluorescence. Representative RAD51 foci (IR treatment) are shown (A). (Magnification: 100×.) Quantification results were the average of three independent experiments and were presented as mean ± SEM (B). (C) Schematic representation of HR assay. (D) U2OS DR-GFP cells were transfected with the indicated siRNA and 24 h later were electroporated with an I-SceI expression plasmid. Forty-eight hours after electroporation, cells were harvested and assayed for GFP expression by FACS analysis. Results were the average of three independent experiments and were presented as mean± SEM (E). Knockdown efficiency was confirmed by immunoblotting. (F and G) Rescue of RAD51 foci formation by expression of a siRNA#1-resistant SPIDR cDNA. Immunostaining experiments were performed as described in Fig. 2F. Representative RAD51 foci (IR treatment) are shown (F). (Magnification: 100×.) Quantification results were the average of three independent experiments and are presented as mean ± SEM (G). (H) The exogenous SPIDR expression was confirmed by immunoblotting.
Functional Interaction Between SPIDR and RAD51.
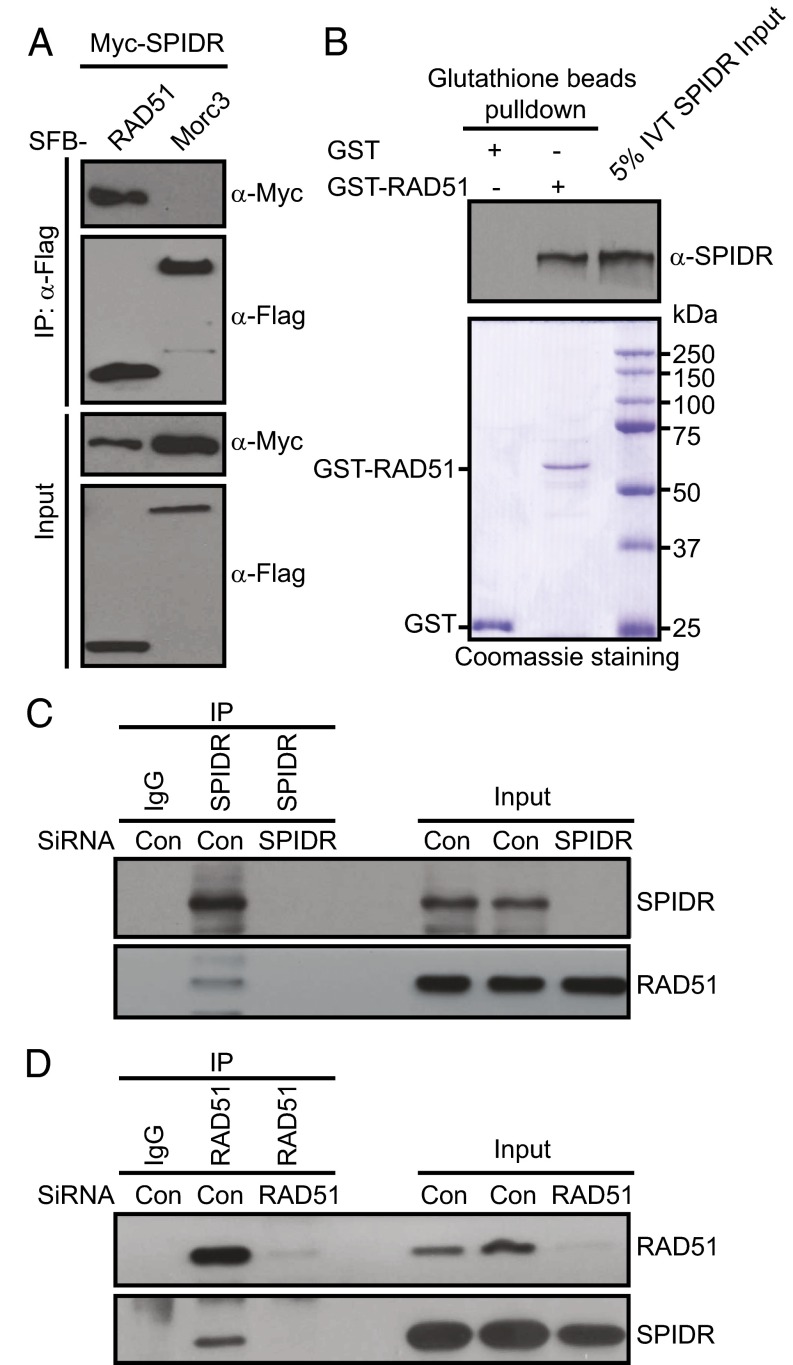
We next tested the possibility that the SPIDR might recruit RAD51 to sites of DNA damage through a direct SPIDR–RAD51 physical interaction. Coimmunoprecipitation experiments revealed that Myc-tagged SPIDR interacted strongly with SFB-tagged RAD51 (Fig. 4A). We also examined this interaction by using in vitro GST pull-down assays and found that GST-tagged human RAD51, obtained from bacteria, interacts with in vitro-translated SPIDR (Fig. 4B).
Fig. 4.
Functional interaction between SPIDR and RAD51. (A) 293T cells were transfected with plasmids encoding SFB-tagged RAD51 or Morc3 together with plasmids encoding Myc-tagged SPIDR. Coprecipitation and immunoblotting were carried out as indicated. (B) Direct binding between recombinant GST-RAD51 purified from bacteria and in vitro-translated SPIDR. (Upper) SPIDR was detected by immunoblotting. (Lower) Input GST-proteins visualized by Coomassie staining. (C and D) SiRNA-treated HeLa cells were lysed in the presence of benzonase, cell lysates were then incubated with protein A agarose beads conjugated with indicated antibodies, and Western blot analysis was carried out as indicated.
To further determine whether endogenous SPIDR and RAD51 interact, benzonase-treated HeLa cell lysates were prepared and subjected to immunoprecipitation with either a control or anti-SPIDR antibody. As expected, endogenous RAD51 was immunoprecipitated by anti-SPIDR antibody, but not by control antibody, and the reverse experiment confirmed this result (Fig. 4 C and D).
We next attempted to identify the regions within SPIDR responsible for its interaction with RAD51. Pull-down assays demonstrated that a domain spanning amino acids 151–450 of SPIDR is responsible for RAD51 binding (Fig. S5A).
To further investigate the biological significance of the SPIDR–RAD51 interaction, we performed rescue experiments similar to those described above to examine whether the RAD51-binding region on SPIDR is required for efficient RAD51 foci formation. Interestingly, whereas wild-type SPIDR successfully restored RAD51 focus formation, SPIDR deletion mutants defective in RAD51 binding failed to do so (Fig. S5 B–D), indicating that SPIDR may help to recruit RAD51 to DNA damage sites through a direct SPIDR–RAD51 interaction. In agreement with this hypothesis, the localization of BRCA2 or Palb2 was unaffected following SPIDR knockdown (Fig. S6).
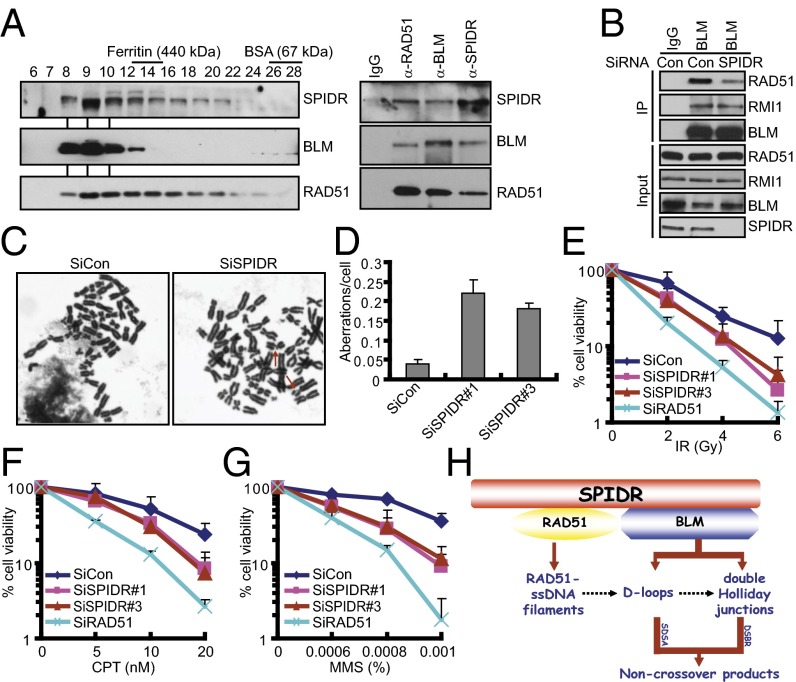
SPIDR Provides a Link Between the BLM Helicase and RAD51.
Previous studies have shown that BLM can form a complex with RAD51 (29). The ability of SPIDR to interact independently with BLM and RAD51 indicates that SPIDR, BLM, and RAD51 may form a ternary complex. To determine whether this is the case, HeLa extracts were resolved by gel filtration and the individual fractions were analyzed by immunoblotting (Fig. 5A). The fractions containing SPIDR, BLM, and RAD51 were then immunoprecipitated with anti-SPIDR, anti-BLM, or anti-RAD51 antibody. As shown in Fig. 5A, SPIDR, BLM, and RAD51 were present in all of the three immunoprecipitations, thereby demonstrating the formation of a trimeric complex. To further examine whether SPIDR may serve as a scaffolding protein to promote the BLM–RAD51 complex formation, we used siRNA to knock down SPIDR and examined the association between BLM and RAD51. As shown in Fig. 5B, when cells were treated with SPIDR siRNA, but not a control siRNA, the interaction between BLM and RAD51 was significantly reduced. These results suggested that SPIDR might mediate, at least in part, the interaction between the BLM helicase and RAD51.
Fig. 5.
SPIDR is required for maintaining genome integrity. (A) Endogenous SPIDR, BLM, and RAD51 are present in a trimeric complex. HeLa extracts were resolved by gel filtration and the fractions were analyzed by immunoblotting (Left). Anti-SPIDR, anti-BLM, and anti-RAD51 immunoprecipitaton of fractions containing SPIDR, BLM, and RAD51 were analyzed by immunoblotting with antibodies specific for SPIDR, BLM, and RAD51 (Right). (B) HeLa cells transfected with control or SPIDR siRNA were subjected to coimmunoprecipitation using anti-BLM antibody. Immunoblotting was performed using antibodies as indicated. (C) Representative images of metaphase spreads prepared from HeLa cells treated with the indicated siRNAs. Representative aberrations are marked by arrows. (Magnification: 100×.) (D) Quantification of chromosomal aberrations in control and SPIDR-depleted HeLa cells. The average of two experiments is shown; at least 50 cells were counted in each experiment. Error bars represent the SD. (E–G) Clonogenic survival assays in SPIDR-depleted HeLa cells following IR, CPT, or MMS treatment. Experiments were done in triplicates. Results shown are averages of three independent experiments. (H) A proposed model of SPIDR functions in DNA repair.
SPIDR Is Critical for Maintaining Genome Stability.
Given that SPIDR is required for proper DNA repair, we tested the effect of SPIDR on genome integrity. As shown in Fig. 5 C and D, SPIDR-depleted HeLa cells displayed a significant increase in chromosomal aberrations visualized in metaphase spreads. Furthermore, knockdown of SPIDR in HeLa cells conferred cellular hypersensitivity to IR, CPT, and methyl-methane sulfonate (MMS) (Fig. 5 E–G and Fig. S7A). More importantly, these cellular phenotypes can be rescued by reconstitution of SPIDR-depleted cells with wild-type SPIDR, but not with the deletion mutants defective in BLM or RAD51 binding, indicating that SPIDR plays a functionally important role in allowing cells to repair genotoxic damage and maintain chromosomal integrity, possibly through its direct interaction with BLM and RAD51 (Fig. S7 B–D).
Discussion
In this study, we report the identification of a unique protein designated SPIDR, which may serve as a scaffolding protein that helps to promote the formation of BLM/RAD51-containing complexes of biological importance.
It is well-established that accumulation of repair proteins on damaged chromosomes is required to restore genome stability. Like many DNA damage/repair proteins, the BLM helicase is able to accumulate into foci structures in response to DNA damage (22, 23). However, mechanistically how BLM is recruited to sites of DNA damage remains unclear. Previous studies have shown that FANCM can target the BLM complex to nuclear foci after treatment of cells with CPT or mitomycin-C (19). Targeting by FANCM is dependent on a direct interaction between the conserved MM2 domain of FANCM and the subunits RMI1 and TopoIIIα of the BLM complex (19). In addition, RMI1/2 has been shown to regulate BLM retention at sites of DNA damage primarily via a role in maintaining the protein stability of the BLM complex (11–13). In contrast, SPIDR recruits the BLM complex to sites of DNA damage through a direct interaction with BLM. It is also interesting to note that the MRN (MRE11, RAD50, and NBS1) complex can increase the affinity of BLM for DNA ends (30). The existence of multiple distinct regulatory mechanisms for the BLM complex recruitment in mammalian cells underscores the importance of this process in DNA repair.
RAD51 and BLM have been implicated in the regulation of HR at the early and late steps, respectively, according to the timing of respective involvement in the recombination process. To achieve maximal efficiency for HR repair, these steps must be tightly regulated and coordinated at the molecular level. However, the precise mechanisms for this coordination are poorly understood. Here, we have described one example of such a mechanism in which SPIDR coordinates both BLM and RAD51 functions following DNA damage. The SPIDR–RAD51 interaction might facilitate the loading of RAD51 on resected DNA to initiate HR, such that via its direct interaction with BLM, SPIDR may serve as a signaling platform to dictate the specific HR pathway (Fig. 5H).
It is noteworthy to point out that although cells lacking SPIDR or the breast ovarian cancer susceptibility (BRCA) proteins show similar defects in RAD51 focus formation, they have opposite effects on the frequency of SCE. Down-regulation of SPIDR exhibits an elevated SCE rate, whereas lack of BRCA1/2 leads to a reduction in the SCE frequency (31, 32). The simplest explanation for this difference is that SPIDR not only promotes assembly of subnuclear RAD51 foci, but also regulates the ratio of crossover-to-noncrossover recombinants via its ability to recruit the BLM helicase. As a result, although the overall HR efficiency is reduced in SPIDR-depleted cells, most of the residual HR intermediates are specifically resolved into crossover products, which in turn contribute, at least partially, to the elevated SCE frequency.
In summary, our study reveals a previously undescribed link between the BLM helicase and the HR machinery. Our results support the idea that SPIDR contributes to genomic integrity by licensing the assembly of the key DNA-processing enzymes BLM and RAD51 at DNA lesions. Defective HR repair is associated with genomic instability and cancer prone syndromes, and it will be particularly interesting to determine whether defects in SPIDR are also relevant to human diseases.
Supplementary Material
Acknowledgments
We thank all our colleagues in the J. Huang laboratory for insightful discussions and Dr. X. H. Feng for comments on the manuscript. This work was supported by National Basic Research Program of China Grants 2012CB944402 and 2013CB911003; National Natural Science Foundation of China Grant 31071243; Natural Science Foundation of Zhejiang Province Grant R2110569; and Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University Grant SKLCSB2013KF002. T.L. is a member of the X. H. Feng laboratory and supported by National Science Foundation of China Grants 31171347 and 31090360 and the Ministry of Science and Technology Grant 2012CB966600.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220921110/-/DCSupplemental.
References
- 1.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 3.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 4.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Colavito S, Prakash R, Sung P. Promotion and regulation of homologous recombination by DNA helicases. Methods. 2010;51(3):329–335. doi: 10.1016/j.ymeth.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299(5604):265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 7.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34(8):2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135(2):261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464(7290):937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272(49):30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 11.Yin J, et al. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24(7):1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22(20):2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seki M, et al. Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol. 2006;26(16):6299–6307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103(11):4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raynard S, et al. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem. 2008;283(23):15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282(43):31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 19.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36(6):943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Goss KH, et al. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297(5589):2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- 21.Gruber SB, et al. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297(5589):2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939. [PMC free article] [PubMed] [Google Scholar]
- 23.Bischof O, et al. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153(2):367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P, et al. Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum Mol Genet. 2001;10(12):1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 25.Chu WK, Hanada K, Kanaar R, Hickson ID. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010;29(33):4705–4714. doi: 10.1038/onc.2010.214. [DOI] [PubMed] [Google Scholar]
- 26.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21(23):3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276(22):19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 30.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20(17):4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Z, Zhang J. A dual role of BRCA1 in two distinct homologous recombination mediated repair in response to replication arrest. Nucleic Acids Res. 2012;40(2):726–738. doi: 10.1093/nar/gkr748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.